Allelic Expression Imbalance in the Human Retinal Transcriptome and Potential Impact on Inherited Retinal Diseases
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Eye Collection
2.2. DNA and RNA Isolation
2.3. RNA-sequencing
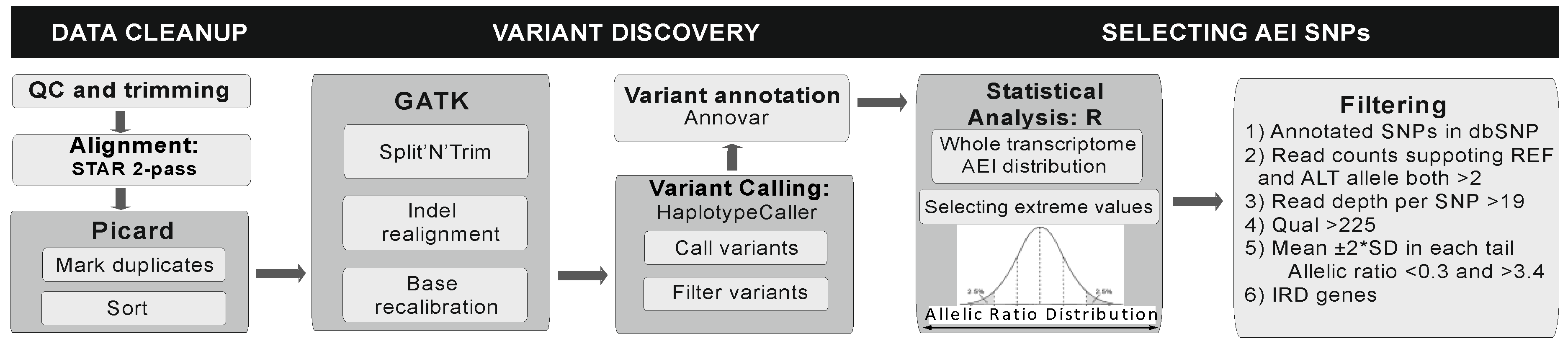
2.4. Allelic Expression Imbalance RNA-sequencing Analysis
2.5. Single Nucleotide PolymorphismHeterozygosity Confirmation by Sanger Sequencing
2.6. Pyrosequencing Assay Design
3. Results
3.1. RNA-sequencing
3.2. Allelic expression imbalanceAnalysis on RNA-sequencing Data
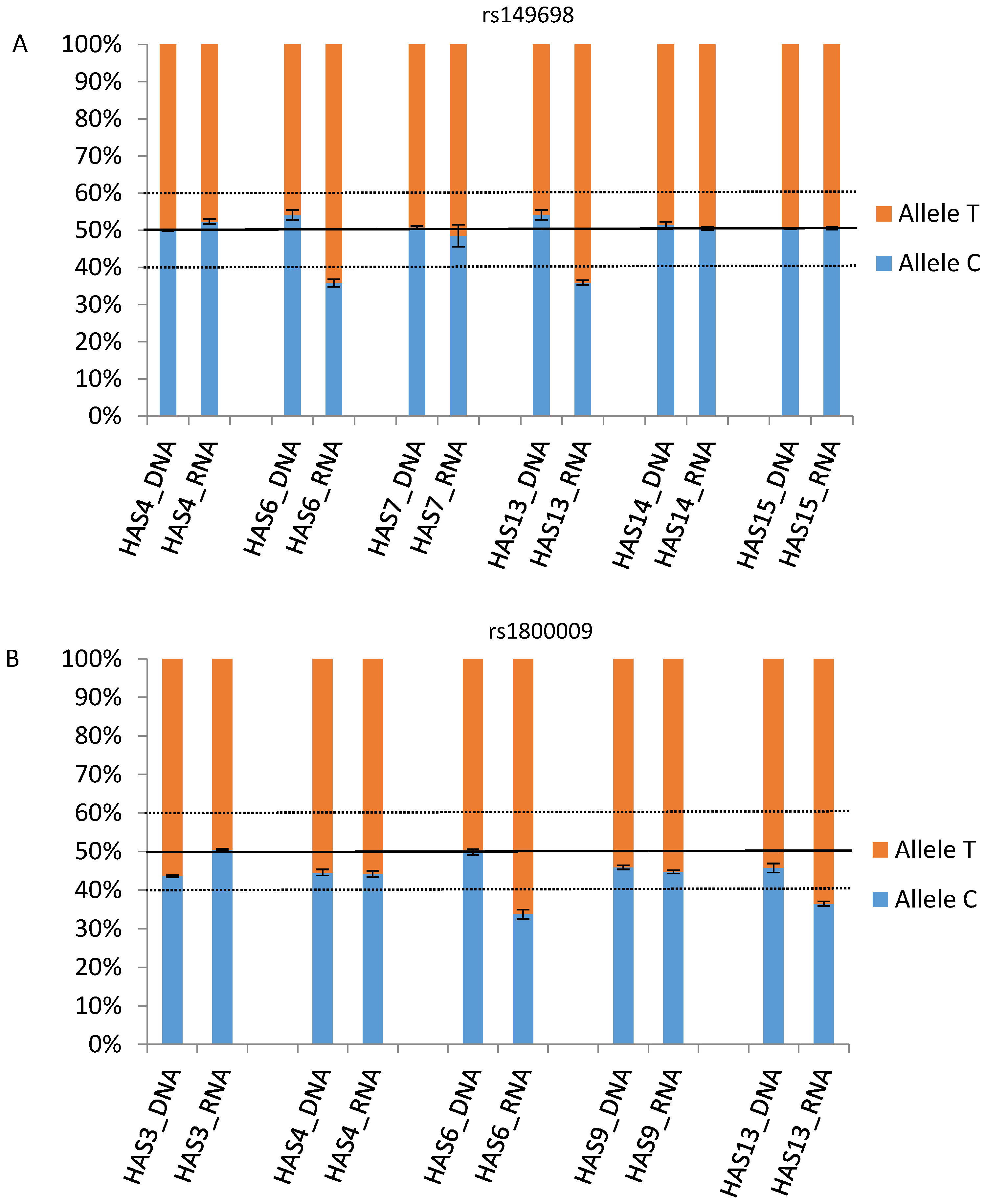
3.3. Pyrosequencing Confirmation
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- RetNet - Retinal Information Network. Available online: https://sph.uth.edu/retnet/ (accessed on 30 June 2017).
- Nash, B.M.; Wright, D.C.; Grigg, J.R.; Bennetts, B.; Jamieson, R.V. Retinal dystrophies, genomic applications in diagnosis and prospects for therapy. Transl. Pediatr. 2011, 4, 139–163. [Google Scholar] [CrossRef]
- Bravo-Gil, N.; Méndez-Vidal, C.; Romero-Pérez, L.; González-del Pozo, M.; Rodríguez-de la Rúa, E.; Dopazo, J.; Borrego, S.; Antiñolo, G. Improving the management of Inherited Retinal Dystrophies by targeted sequencing of a population-specific gene panel. Sci Rep. 2011, 6, 23910. [Google Scholar] [CrossRef] [PubMed]
- Louie, C.M.; Caridi, G.; Lopes, V.S.; Brancati, F.; Kispert, A.; Lancaster, M.A.; Schlossman, A.M.; Otto, E.A.; Leitges, M.; Gröne, H.-J.; et al. AHI1 is required for photoreceptor outer segment development and is a modifier for retinal degeneration in nephronophthisis. Nat. Genet. 2011, 42, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Cruz, N.M.; Yuan, Y.; Leehy, B.D.; Baid, R.; Kompella, U.; DeAngelis, M.M.; Escher, P.; Haider, N.B. Modifier genes as therapeutics: The nuclear hormone receptor Rev Erb alpha (Nr1d1) rescues Nr2e3 associated retinal disease. PLoS ONE 2011, 9. [Google Scholar] [CrossRef] [PubMed]
- Vollrath, D.; Yasumura, D.; Benchorin, G.; Matthes, M.T.; Feng, W.; Nguyen, N.M.; Sedano, C.D.; Calton, M.A.; LaVail, M.M. Tyro3 Modulates Mertk-Associated Retinal Degeneration. PLoS Genet. 2011, 11, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Gymrek, M.; Willems, T.; Guilmatre, A.; Zeng, H.; Markus, B.; Georgiev, S.; Daly, M.J.; Price, A.L.; Pritchard, J.; Sharp, A.; et al. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat. Genet. 2011, 48, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.; Powell, J.E.; Marigorta, U.M. Expression quantitative trait locus analysis for translational medicine. Genome Med. 2011, 7, 60. [Google Scholar] [CrossRef] [PubMed]
- Albert, F.W.; Kruglyak, L. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 2011, 16, 197–212. [Google Scholar] [CrossRef] [PubMed]
- Takata, A.; Matsumoto, N.; Kato, T. Genome-wide identification of splicing QTLs in the human brain and their enrichment among schizophrenia-associated loci. Nat. Commun. 2011, 8, 14519. [Google Scholar] [CrossRef] [PubMed]
- Castel, S.E.; Levy-Moonshine, A.; Mohammadi, P.; Banks, E.; Lappalainen, T. Tools and best practices for data processing in allelic expression analysis. Genome Biol. 2011, 16, 195. [Google Scholar] [CrossRef] [PubMed]
- Milani, L.; Gupta, M.; Andersen, M.; Dhar, S.; Fryknäs, M.; Isaksson, A.; Larsson, R.; Syvänen, A.C. Allelic imbalance in gene expression as a guide to cis-acting regulatory single nucleotide polymorphisms in cancer cells. Nucleic Acids Res. 2001, 35. [Google Scholar] [CrossRef] [PubMed]
- Rivolta, C.; McGee, T.L.; Frio, T.R.; Jensen, V.R.; Berson, E.L.; Dryja, T.P. Variation in retinitis pigmentosa-11 (PRPF31 or RP11) gene expression between symptomatic and asymptomatic patients with dominant RP11 mutations. Hum. Mutat. 2001, 27, 644–653. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.M.; Shah, A.Z.; Venturini, G.; Krishna, A.; Chakravarti, A.; Rivolta, C.; Bhattacharyaa, S.S. Transcriptional regulation of PRPF31 gene expression by MSR1 repeat elements causes incomplete penetrance in retinitis pigmentosa. Sci. Rep. 2011, 6, 19450. [Google Scholar] [CrossRef] [PubMed]
- Venturini, G.; Rose, A.M.; Shah, A.Z.; Bhattacharya, S.S.; Rivolta, C. CNOT3 Is a Modifier of PRPF31 Mutations in Retinitis Pigmentosa with Incomplete Penetrance. PLoS Genet. 2011, 8. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T.; Bartolomei, M.S. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 2011, 152, 1308–1323. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Smith, R.M.; Seweryn, M.; Wang, D.; Hartmann, K.; Webb, A.; Sadee, W.; Rempala, G.A. Analyzing allele specific RNA expression using mixture models. BMC Genom. 2011, 16, 566. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.M.; Webb, A.; Papp, A.C.; Newman, L.C.; Handelman, S.K.; Suhy, A.; Mascarenhas, R.; Oberdick, J.; Sadee, W. Whole transcriptome RNA-Seq allelic expression in human brain. BMC Genom. 2011, 14, 571. [Google Scholar] [CrossRef] [PubMed]
- Pinelli, M.; Carissimo, A.; Cutillo, L.; Lai, C.-H.; Mutarelli, M.; Moretti, M.N.; Singh, M.V.; Karali, M.; Carrella, D.; Russo, M.P.F.; et al. An atlas of gene expression and gene co-regulation in the human retina. Nucleic Acids Res. 2011, 44, 5773–5784. [Google Scholar] [CrossRef] [PubMed]
- UCSC Genome Browser Home. Available online: https://genome.ucsc.edu/ (accessed on 30 June 2017).
- Engström, P.G.; Steijger, T.; Sipos, B.; Grant, G.R.; Kahles, A.; Rätsch, G.; Goldman, N.; Hubbard, T.J.; Harrow, J.; Guigó, R.; et al. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat. Methods 2011, 10, 1185–1191. [Google Scholar] [CrossRef] [PubMed]
- dbSNP. Available online: https://www.ncbi.nlm.nih.gov/projects/SNP (accessed on 30 June 2017).
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Human BLAT Search. Available online: https://genome.ucsc.edu/cgi-bin/hgBlat (accessed on 30 June 2017).
- Ward, L.D.; Kellis, M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2011, 44, D877–D881. [Google Scholar] [CrossRef] [PubMed]
- Acke, F.R.; Malfait, F.; Vanakker, O.M.; Steyaertb, W.; de Leeneerb, K.; Mortierc, G.; Dhoogea, I.; de Paepeb, A.; de Leenheera, E.M.R.; Couckeb, P.J.; et al. Novel pathogenic COL11A1/COL11A2 variants in Stickler syndrome detected by targeted NGS and exome sequencing. Mol. Genet. Metab. 2011, 113, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Annunen, S.; Ko, J.; Czarny, M.; Warman, M.L.; Brunner, H.G.; Kääriäinen, H.; Mulliken, J.B.; Tranebjærg, L.; Brooks, D.G.; Cox, G.F.; et al. Splicing Mutations of 54-bp Exons in the COL11A1 Gene Cause Marshall Syndrome, but Other Mutations Cause Overlapping Marshall/Stickler Phenotypes. AJHG 1991, 1, 974–983. [Google Scholar] [CrossRef] [PubMed]
- Michaelides, M.; Holder, G.E.; Bradshaw, K.; Hunt, D.M.; Moore, A.T. Cone-rod dystrophy, intrafamilial variability, and incomplete penetrance associated with the R172W mutation in the peripherin/RDS gene. Ophthalmology 2001, 112, 1592–1598. [Google Scholar] [CrossRef] [PubMed]
- Shankar, S.P.; Hughbanks-Wheaton, D.K.; Birch, D.G.; Sullivan, L.S.; Conneely, K.N.; Bowne, S.J.; Stone, E.M.; Daiger, S.P. Autosomal dominant retinal dystrophies caused by a founder splice site mutation, c.828+3A>T, in PRPH2 and protein haplotypes in trans as modifiers. Investig. Ophthalmol. Vis. Sci. 2011, 57, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Arrigoni, F.I.; Matarin, M.; Thompson, P.J.; Michaelides, M.; McClements, M.E.; Redmond, E.; Clarke, L.; Ellins, E.; Mohamed, S.; Pavord, I.; et al. Extended extraocular phenotype of PROM1 mutation in kindreds with known autosomal dominant macular dystrophy. Eur. J. Hum. Genet. 2011, 19, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Audo, I.; Mohand-Said, S.; Dhaenens, C.-M.; Germain, A.; Orhan, E.; Antonio, A.; Hamel, C.; Sahel, J.-A.; Bhattacharya, S.S.; Zeitz, C. RP1 and autosomal dominant rod-cone dystrophy: Novel mutations, a review of published variants, and genotype-phenotype correlation. Hum. Mutat. 2011, 33, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Sodi, A.; Passerini, I.; Murro, V.; Caputo, R.; Bacci, G.M.; bodoj, M.; Torricelli, F.; Menchini, U. BEST1 sequence variants in Italian patients with vitelliform macular dystrophy. Mol. Vis. 2011, 18, 2736–2748. [Google Scholar]
- Cohn, A.C.; Turnbull, C.; Ruddle, J.B.; Guymer, R.H.; Kearns, S.; Staffieri, S.; Daggett, H.T.; Hewitt, A.W.; Mackey, D.A. Best’s macular dystrophy in Australia: Phenotypic profile and identification of novel BEST1 mutations. Eye (Lond.) 2011, 25, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Khan, K.; Kasilian, M.L.; Strauss, R.W.; Hplder, G.E.; Ronson, A.G.; Thompson, D.A.; Morre, A.T.; Michaelides, M. Unilateral BEST1-Associated Retinopathy. Am. J. Ophthalmol. 2011, 169, 24–32. [Google Scholar] [CrossRef] [PubMed]
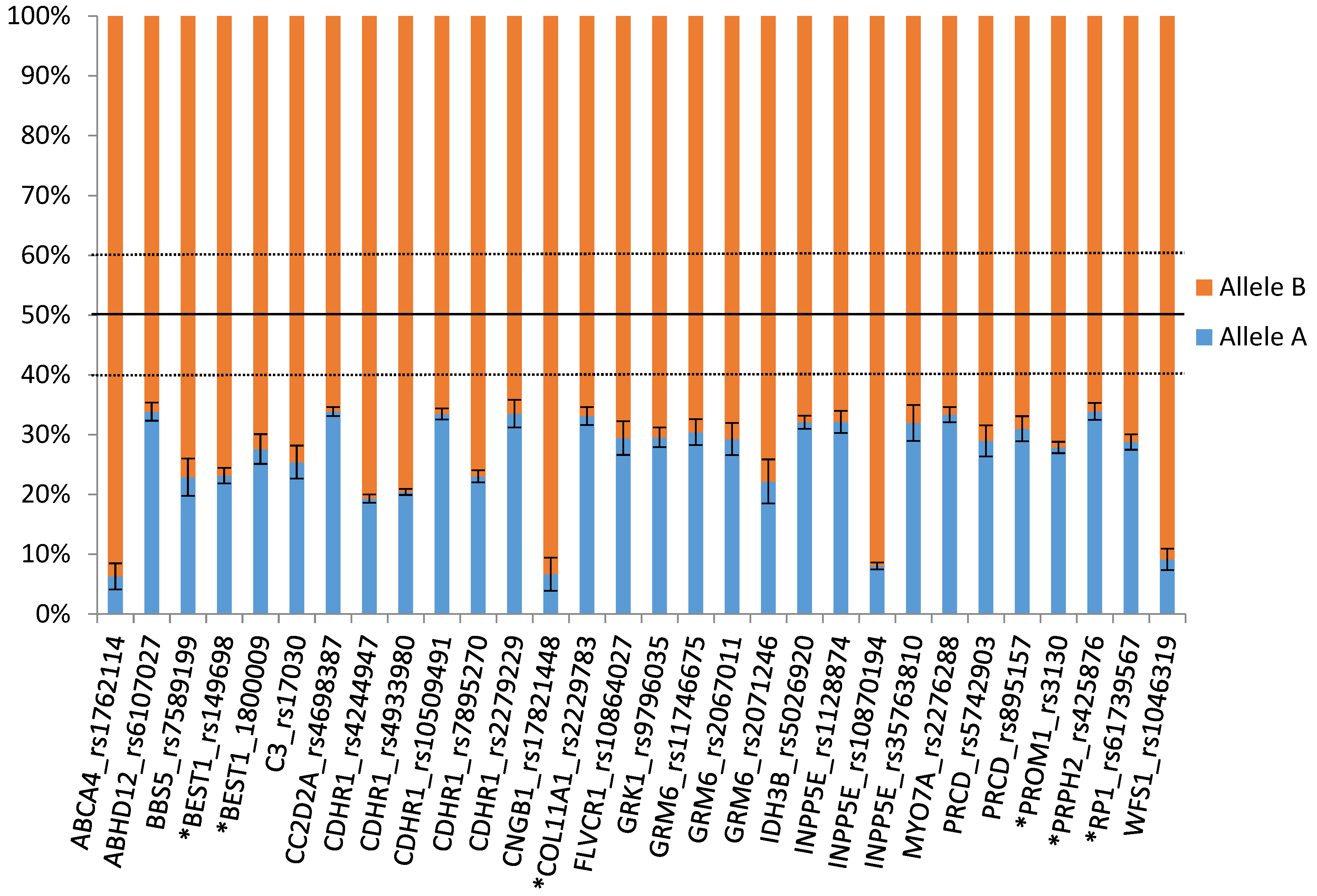
| Gene Symbol_ SNP | Samples in Top Mean ±2*SD Ratios | Heterozygous Samples | Allelic Ratio <0.66 | Allelic Ratio >1.5 | Sum Imbalanced Samples | AEI Frequency in Heterozygous Samples | AEI Frequency in 52 Samples | Absolute Mean Allelic Ratio | SEM Allelic Ratio | Kurtosis | SEK |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ABCA4_rs1762114 | 7 | 16 | 0 | 8 | 8 | 50.0% | 15.4% | 0.090 | 0.069 | 7.636 | 1.481 |
| ABHD12_rs6107027 | 1 | 20 | 11 | 1 | 12 | 60.0% | 23.1% | 0.536 | 0.050 | 8.709 | 1.232 |
| BBS5_rs7589199 | 3 | 11 | 5 | 3 | 8 | 72.7% | 15.4% | 0.354 | 0.101 | −2.19 | 1.481 |
| *BEST1_rs149698 | 4 | 22 | 1 | 6 | 7 | 31.8% | 13.5% | 0.310 | 0.044 | 0.4463 | 1.587 |
| *BEST1_1800009 | 2 | 20 | 4 | 2 | 6 | 30.0% | 11.5% | 0.412 | 0.091 | −1.865 | 1.741 |
| C3_rs17030 | 2 | 29 | 2 | 6 | 8 | 27.6% | 15.4% | 0.385 | 0.087 | −0.5529 | 1.481 |
| CC2D2A_rs4698387 | 1 | 17 | 11 | 0 | 11 | 64.7% | 21.2% | 0.519 | 0.032 | 0.3474 | 1.279 |
| CDHR1_rs4244947 | 16 | 23 | 21 | 2 | 23 | 100.0% | 44.2% | 0.247 | 0.020 | 0.9735 | 0.9348 |
| CDHR1_rs4933980 | 17 | 21 | 21 | 0 | 21 | 100.0% | 40.4% | 0.260 | 0.016 | 0.3758 | 0.9719 |
| CDHR1_rs10509491 | 1 | 20 | 11 | 0 | 11 | 55.0% | 21.2% | 0.513 | 0.039 | −0.1936 | 1.279 |
| CDHR1_rs7895270 | 6 | 16 | 10 | 0 | 10 | 62.5% | 19.2% | 0.308 | 0.038 | 1.261 | 1.334 |
| CDHR1_rs2279229 | 1 | 15 | 7 | 1 | 8 | 53.3% | 15.4% | 0.541 | 0.076 | 7.562 | 1.481 |
| CNGB1_rs17821448 | 10 | 30 | 1 | 10 | 11 | 36.7% | 21.2% | 0.158 | 0.147 | 11 | 1.279 |
| *COL11A1_rs2229783 | 2 | 19 | 2 | 7 | 9 | 47.4% | 17.3% | 0.516 | 0.058 | 1.01 | 1.4 |
| FLVCR1_rs10864027 | 1 | 24 | 5 | 1 | 6 | 25.0% | 11.5% | 0.454 | 0.095 | 1.819 | 1.741 |
| GRK1_rs9796035 | 3 | 21 | 10 | 2 | 12 | 57.1% | 23.1% | 0.449 | 0.058 | 0.5881 | 1.232 |
| GRM6_rs11746675 | 2 | 32 | 5 | 4 | 9 | 28.1% | 17.3% | 0.474 | 0.074 | 1.466 | 1.4 |
| GRM6_rs2067011 | 1 | 29 | 4 | 3 | 7 | 24.1% | 13.5% | 0.455 | 0.091 | 0.6681 | 1.587 |
| GRM6_rs2071246 | 3 | 22 | 2 | 4 | 6 | 27.3% | 11.5% | 0.344 | 0.125 | −3.026 | 1.741 |
| IDH3B_rs5026920 | 1 | 12 | 8 | 0 | 8 | 66.7% | 15.4% | 0.482 | 0.045 | 0.2172 | 1.481 |
| INPP5E_rs1128874 | 2 | 27 | 8 | 1 | 9 | 33.3% | 17.3% | 0.500 | 0.062 | 5.798 | 1.154 |
| INPP5E_rs10870194 | 2 | 24 | 0 | 24 | 24 | 100.0% | 46.2% | 0.092 | 0.016 | 0.3604 | 1.4 |
| INPP5E_rs35763810 | 3 | 20 | 0 | 6 | 6 | 30.0% | 11.5% | 0.513 | 0.099 | 6.436 | 1.587 |
| MYO7A_rs2276288 | 3 | 17 | 13 | 1 | 14 | 82.4% | 26.9% | 0.522 | 0.044 | 2.193 | 1.014 |
| PRCD_rs5742903 | 1 | 25 | 7 | 2 | 9 | 36.0% | 17.3% | 0.457 | 0.085 | 0.9821 | 1.481 |
| PRCD_rs895157 | 1 | 17 | 6 | 1 | 7 | 41.2% | 13.5% | 0.474 | 0.069 | 1.585 | 1.481 |
| *PROM1_rs3130 | 8 | 20 | 0 | 19 | 19 | 95.0% | 36.5% | 0.401 | 0.032 | −1.368 | 1.587 |
| *PRPH2_rs425876 | 6 | 14 | 1 | 7 | 8 | 57.1% | 15.4% | 0.529 | 0.057 | 5.791 | 1.4 |
| *RP1_rs61739567 | 2 | 11 | 0 | 8 | 8 | 72.7% | 15.4% | 0.438 | 0.049 | 10.86 | 0.9178 |
| WFS1_rs1046319 | 1 | 12 | 0 | 7 | 7 | 58.3% | 13.5% | 0.111 | 0.046 | 5.809 | 1.741 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Llavona, P.; Pinelli, M.; Mutarelli, M.; Marwah, V.S.; Schimpf-Linzenbold, S.; Thaler, S.; Yoeruek, E.; Vetter, J.; Kohl, S.; Wissinger, B. Allelic Expression Imbalance in the Human Retinal Transcriptome and Potential Impact on Inherited Retinal Diseases. Genes 2017, 8, 283. https://doi.org/10.3390/genes8100283
Llavona P, Pinelli M, Mutarelli M, Marwah VS, Schimpf-Linzenbold S, Thaler S, Yoeruek E, Vetter J, Kohl S, Wissinger B. Allelic Expression Imbalance in the Human Retinal Transcriptome and Potential Impact on Inherited Retinal Diseases. Genes. 2017; 8(10):283. https://doi.org/10.3390/genes8100283
Chicago/Turabian StyleLlavona, Pablo, Michele Pinelli, Margherita Mutarelli, Veer Singh Marwah, Simone Schimpf-Linzenbold, Sebastian Thaler, Efdal Yoeruek, Jan Vetter, Susanne Kohl, and Bernd Wissinger. 2017. "Allelic Expression Imbalance in the Human Retinal Transcriptome and Potential Impact on Inherited Retinal Diseases" Genes 8, no. 10: 283. https://doi.org/10.3390/genes8100283
APA StyleLlavona, P., Pinelli, M., Mutarelli, M., Marwah, V. S., Schimpf-Linzenbold, S., Thaler, S., Yoeruek, E., Vetter, J., Kohl, S., & Wissinger, B. (2017). Allelic Expression Imbalance in the Human Retinal Transcriptome and Potential Impact on Inherited Retinal Diseases. Genes, 8(10), 283. https://doi.org/10.3390/genes8100283





