Identification and Characterization of TALE Homeobox Genes in the Endangered Fern Vandenboschia speciosa
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Next Generation Sequencing, Transcriptome Assembly, Annotation, and Comparative In Silico Analysis of Gene Expression Levels
2.3. Sequence Analysis
2.4. Expression Analysis
2.4.1. RNA Extraction and Complementary DNA Synthesis
2.4.2. Target and Housekeeping Genes
2.4.3. Relative Gene Expression Quantification
2.4.4. Statistical Analyses
3. Results
3.1. Identification and Sequence Analysis of TALE Homeobox Genes Contigs in V. Speciosa
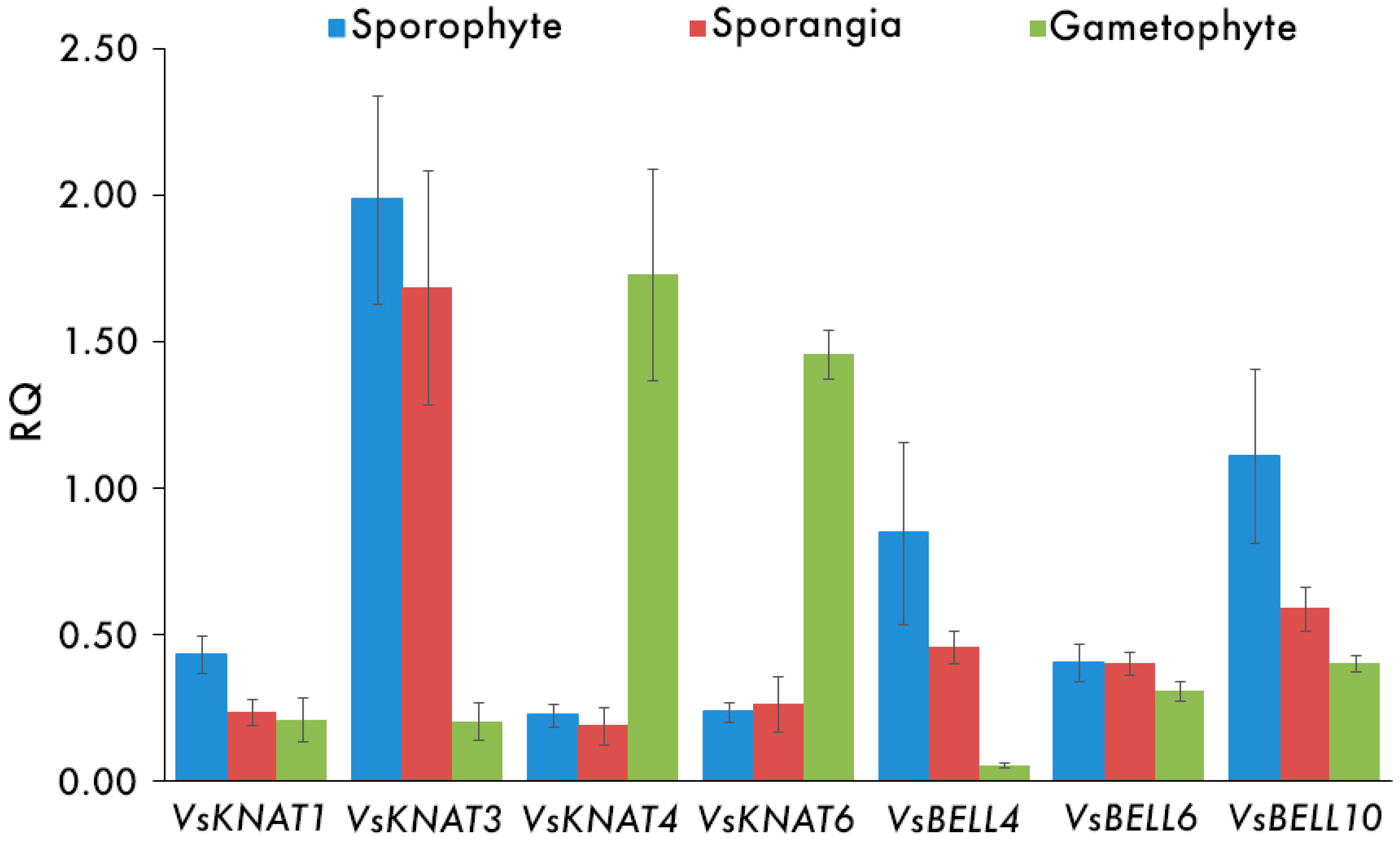
3.2. Expression Analysis of TALE Homeobox Genes in V. speciosa
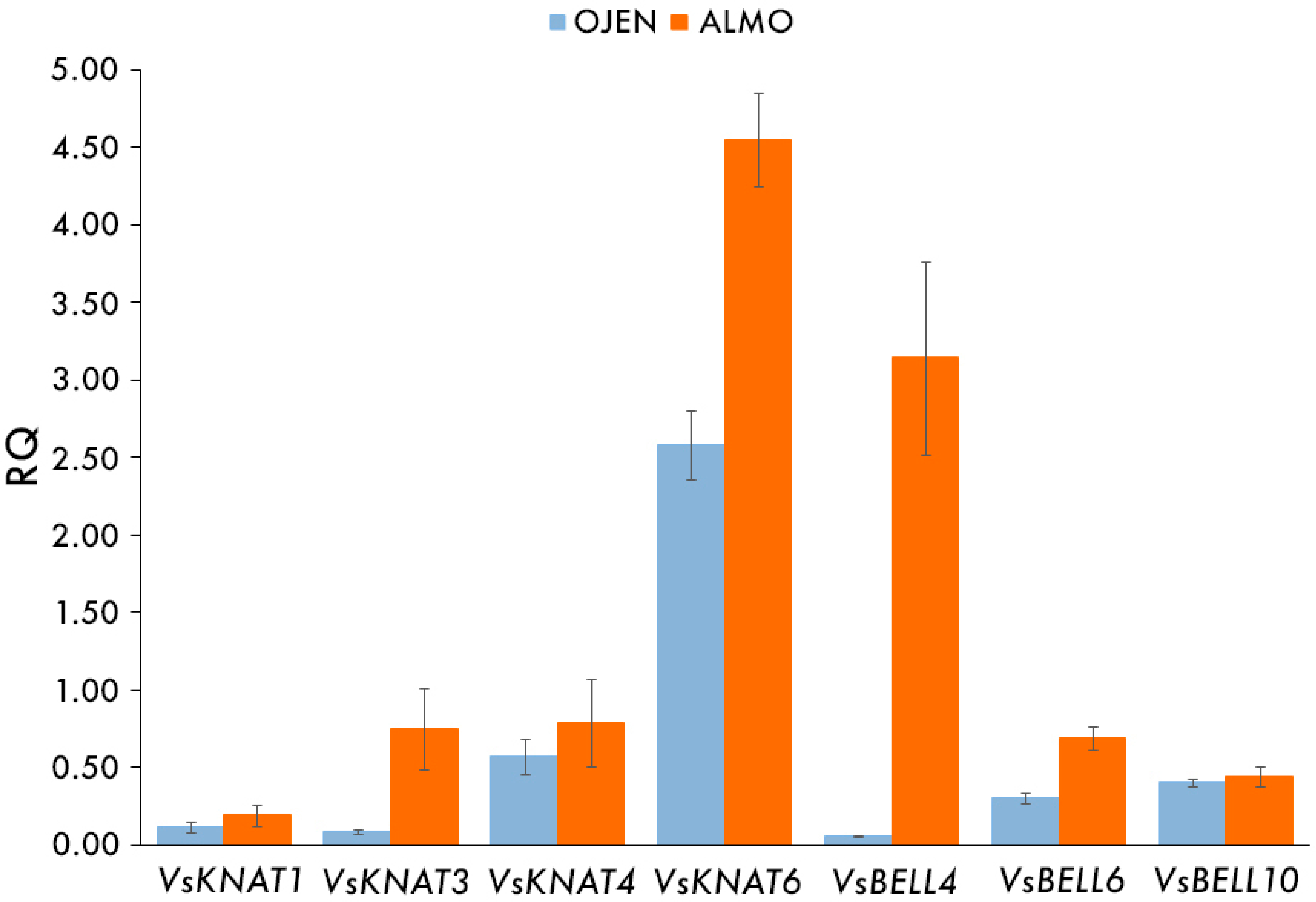
3.3. Expression Analysis of TALE Homeobox Genes in La Almoraima, a Population Composed Only by Independent Gametophytes
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pryer, K.M.; Schneider, H.; Smith, A.R.; Cranfill, R.; Wolf, P.G.; Hunt, J.S.; Sipes, S.D. Horsetails and ferns are a monophyletic group and the closest living relatives to seed plants. Nature 2001, 409, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Pryer, K.M.; Schneider, H.; Zimmer, E.A.; Banks, J.A. Deciding among green plants for whole genome studies. Trends Plant Sci. 2002, 7, 550–554. [Google Scholar] [CrossRef]
- Schneider, H.; Pryer, K.M.; Cranfill, R.; Smith, A.R.; Wolf, P.G. Evolution of vascular plant body plans: A phylogenetic perspective. In Developmental Genetics and Plant Evolution; Cronk, Q.C.B., Bateman, R.M., Harris, J.A., Eds.; Taylor and Francis: Philadelphia, PA, USA, 2002; pp. 330–364. [Google Scholar]
- Banks, J.A.; Nishiyama, T.; Hasebe, M.; Bowman, J.L.; Gribskov, M.; Albert, V.A.; Aono, N.; Aoyama, T.; Ambrose, B.A.; Ashton, N.W.; et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 2011, 332, 960–963. [Google Scholar] [CrossRef] [PubMed]
- Schneider, H. Evolutionary morphology of ferns (monilophytes). Annu. Plant Rev. Evol. Plant Form 2013, 45, 115–140. [Google Scholar]
- Huang, Q.; Li, W.; Fan, R.; Chang, Y. New MADS-box gene in fern: Cloning and expression analysis of DfMADS1 from Dryopteris fragrans. PLoS ONE 2014, 9, e86349. [Google Scholar] [CrossRef] [PubMed]
- Plackett, A.R.G.; Di Stilio, V.S.; Langdale, J.A. Ferns: The missing link in shoot evolution and development. Front. Plant Sci. 2015, 6, 972. [Google Scholar] [CrossRef] [PubMed]
- Rumsey, F.J.; Vogel, J.C.; Russell, S.J.; Barrett, J.A.; Gibby, M. Population genetics and conservation biology of the endangered fern Trichomanes speciosum (Hymenophyllaceae) in Scotland. Biol. J. Linn. Soc. 1999, 66, 333–344. [Google Scholar]
- Johnson, G.N.; Rumsey, F.J.; Headley, A.D.; Sheffield, E. Adaptations to extreme low light in the fern Trichomanes speciosum. New Phytol. 2000, 148, 423–431. [Google Scholar] [CrossRef]
- Makgomol, K.; Sheffield, E. Gametophyte morphology and ultrastructure of the extremely deep shade fern, Trichomanes speciosum. New Phytol. 2001, 151, 243–255. [Google Scholar] [CrossRef]
- Mukherjee, K.; Brocchieri, L.; Bürglin, T.R. A comprehensive classification and evolutionary analysis of plant Homeobox genes. Mol. Biol. Evol. 2009, 26, 2775–2794. [Google Scholar] [CrossRef] [PubMed]
- Bürglin, T.R. Homeodomain subtypes and functional diversity. In A Handbook of Transcription Factors; Springer: New York, NY, USA, 2011. [Google Scholar]
- Bürglin, T.R.; Affolter, M. Homeodomain proteins: An update. Chromosoma 2016, 125, 497–521. [Google Scholar] [CrossRef] [PubMed]
- Hamant, O.; Pautot, V. Plant development: A TALE story. C. R. Biol. 2010, 333, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Di Giacomo, E.; Iannelli, M.A.; Frugis, G. TALE and shape: How to make a leaf different. Plants 2013, 2, 317–342. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Lin, T.; Grandellis, C.; Yu, M.; Hannapel, D.J. The BEL1-like family of transcription factors in potato. J. Exp. Bot. 2014, 65, 709–723. [Google Scholar] [CrossRef] [PubMed]
- Ghate, T.H.; Sharma, P.; Kondhare, K.R.; Hannapel, D.J.; Banerjee, A.K. The mobile RNAs, StBEL11 and StBEL29, suppress growth of tubers in potato. Plant Mol. Biol. 2017, 93, 563–578. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.; Tsiantis, M. A KNOX family TALE. Curr. Opin. Plant Biol. 2009, 12, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Hannapel, D.J. A model system of development regulated by the long-distance transport of mRNA. J. Integr. Plant Biol. 2010, 52, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Sharma, P.; Gonzalez, D.H.; Viola, I.L.; Hannapel, D.J. The impact of the long-distance transport of a BEL1-like messenger RNA on development. Plant Physiol. 2013, 161, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; You, S.; Taylor-Teeples, M.; Li, W.L.; Schuetz, M.; Brady, S.M.; Douglas, C.J. BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 interact and regulate secondary cell wall formation via repression of REVOLUTA. Plant Cell 2014, 26, 4843–4861. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Yang, X.; Zhao, W.; Lang, T.; Samuelsson, T. Evolution, diversification, and expression of KNOX proteins in plants. Front. Plant Sci. 2015, 6, 882. [Google Scholar] [CrossRef] [PubMed]
- Horst, N.A.; Katz, A.; Pereman, I.; Decker, E.L.; Ohad, N.; Reski, R. A single homeobox gene triggers phase transition, embryogenesis and asexual reproduction. Nat. Plants 2016, 2. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, K.; Ando, S.; Yi, H.K.; Tamada, Y.; Hiwatashi, Y.; Murata, T.; Deguchi, H.; Hasebe, M.; Bowman, J.L. KNOX2 genes regulate the haploid-to-diploid morphological transition in land plants. Science 2013, 339, 1067–1070. [Google Scholar] [CrossRef] [PubMed]
- Furumizu, C.; Alvarez, J.P.; Sakakibara, K.; Bowman, J.L. Antagonistic roles for KNOX1 and KNOX2 genes in patterning the land plant body plan following an ancient gene duplication. PLoS Genet. 2015, 11, e1004980. [Google Scholar] [CrossRef] [PubMed]
- Kerstetter, R.; Vollbrecht, E.; Lowe, B.; Veit, B.; Yamaguchi, J.; Hake, S. Sequence-Analysis and Expression Patterns Divide the Maize Knotted1-Like Homeobox Genes into 2 Classes. Plant Cell 1994, 6, 1877–1887. [Google Scholar] [CrossRef] [PubMed]
- Bharathan, G.; Janssen, B.J.; Kellogg, E.A.; Sinha, N. Did homeodomain proteins duplicate before the origin of angiosperms, fungi, and metazoa? Proc. Natl. Acad. Sci. USA 1997, 94, 13749–13753. [Google Scholar] [CrossRef] [PubMed]
- Raven, J.A. Comparative physiology of plant and arthropod land adaptation. Philos. Trans. R. Soc. Lond. B 1985, 309, 273–288. [Google Scholar] [CrossRef]
- Raven, J.A. Plant responses to high O2 concentrations: Relevance to previous high O2 episode. Palaeogeogr. Palaeoclimatol. Palaeoecol. 1991, 97, 19–38. [Google Scholar] [CrossRef]
- Edwards, D.; Fanning, U.; Richardson, J.B. Stomata and sterome in early land plants. Nature 1986, 323, 438–440. [Google Scholar] [CrossRef]
- Sztein, A.E.; Cohen, J.D.; Slovin, J.P.; Cooke, T.J. Auxin metabolism in representative land plants. Am. J. Bot. 1995, 82, 1514–1521. [Google Scholar] [CrossRef]
- Tsuda, K.; Hake, S. Diverse functions of KNOX transcription factors in the diploid body plan of plants. Curr. Opin. Plant Biol. 2015, 27, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, A.D.; Lang, D.; Buchta, K.; Rombauts, S.; Nishiyama, T.; Hasebe, M.; Van de Peer, Y.; Rensing, S.A.; Reski, R. Reannotation and extended community resources for the genome of the non-seed plant Physcomitrella patens provide insights into the evolution of plant gene structures and functions. BMC Genom. 2013, 14, 498. [Google Scholar] [CrossRef] [PubMed]
- Salmi, M.L.; Bushart, T.J.; Stout, S.C.; Roux, S.J. Profile and analysis of gene expression changes during early development in germinating spores of Ceratopteris richardii. Plant Physiol. 2005, 138, 1734–1745. [Google Scholar] [CrossRef] [PubMed]
- Matasci, N.; Hung, L.H.; Yan, Z.; Carpenter, E.J.; Wickett, N.J.; Mirarab, S.; Nguyen, N.; Warnow, T.; Ayyampalayam, S.; Barker, M.; et al. Data access for the 1000 Plants (1KP) project. GigaScience 2014, 3, 17. [Google Scholar] [CrossRef] [PubMed]
- Wolf, P.G.; Sessa, E.B.; Marchant, D.B.; Li, F.W.; Rothfels, C.J.; Sigel, E.M.; Gitzendanner, M.A.; Visger, C.J.; Banks, J.A.; Soltis, D.E.; et al. An exploration into fern genome space. Genome Biol. Evol. 2015, 7, 2533–2544. [Google Scholar] [CrossRef] [PubMed]
- Frank, M.H.; Edwards, M.B.; Schultz, E.R.; McKain, M.R.; Fei, Z.; Sørensen, I.; Rose, J.K.C.; Scanlon, M.J. Dissecting the molecular signatures of apical cell-type shoot meristems from two ancient land plant lineages. New Phytol. 2015, 207, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Harrison, C.J.; Corley, S.B.; Moylan, E.C.; Alexander, D.L.; Scotland, R.W.; Langdale, J.A. Independent recruitment of a conserved developmental mechanism during leaf evolution. Nature 2005, 434, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Sano, R.; Juárez, C.M.; Hass, B.; Sakakibara, K.; Ito, M.; Banks, J.A.; Hasebe, M. KNOX homeobox genes potentially have similar function in both diploid unicellular and multicellular meristems, but not in haploid meristems. Evol. Dev. 2005, 7, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Ambrose, B.A.; Vasco, A. Bringing the multicellular fern meristem into focus. New Phytol. 2016, 210, 790–793. [Google Scholar] [CrossRef] [PubMed]
- FastQC. A quality control tool for high throughput sequence data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 16 October 2017).
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.; Birol, I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Robertson, G.; Schein, J.; Chiu, R.; Corbett, R.; Field, M.; Jackman, S.D.; Mungall, K.; Lee, S.; Okada, H.M.; Qian, J.Q.; et al. De novo assembly and analysis of RNA-seq data. Nat. Methods 2010, 7, 909–912. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Madan, A. CAP3: A DNA Sequence Assembly Program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2014, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B. 1995, 57, 289–300. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Higgins, D.G.; Sharp, P.M. CLUSTAL: A package for performing multiple sequence alignment on a microcomputer. Gene 1988, 73, 237–244. [Google Scholar] [CrossRef]
- Le Bail, A.; Scholz, S.; Kost, B. Evaluation of Reference Genes for RT qPCR analyses of structure-specific and hormone regulated gene expression in Physcomitrella patens Gametophytes. PLoS ONE 2013, 8, e70998. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef]
- TIBCO Statistica. Available online: http://statistica.io/products/ (accessed on 17 October 2017).
- GeneBank. Available online: https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 17 October 2017).
- The European Nucleotide Archive (ENA). Available online: https://www.ebi.ac.uk/ena (accessed on 17 October 2017).
- Sakakibara, K.; Nishiyama, T.; Deguchi, H.; Hasebe, M. Class 1 KNOX genes are not involved in shoot development in the moss Physcomitrella patens but do function in sporophyte development. Evol. Dev. 2008, 10, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Byrne, M.E.; Simorowski, J.; Martienssen, R.A. ASYMMETRIC LEAVES1 reveals KNOX gene redundancy in Arabidopsis. Development 2002, 129, 1957–1965. [Google Scholar] [PubMed]
- Douglas, S.J.; Chuck, G.; Dengler, R.E.; Pelecanda, L.; Riggs, C.D. KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell 2002, 14, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Venglat, S.P.; Dumonceaux, T.; Rozwadowski, K.; Parnell, L.; Babic, V.; Keller, W.; Martienssen, R.; Selvaraj, G.; Datla, R. The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 4730–4735. [Google Scholar] [CrossRef] [PubMed]
- Ragni, L.; Belles-Boix, E.; Günl, M.; Pautot, V. Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. Plant Cell 2008, 20, 888–900. [Google Scholar] [CrossRef] [PubMed]
- Harrison, C.J. Development and genetics in the evolution of land plant body plans. Philos. Trans. R. Soc. B 2017, 372. [Google Scholar] [CrossRef] [PubMed]
- Roeder, A.H.; Ferrandiz, C.; Yanofsky, M.F. The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 2003, 13, 1630–1635. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Cho, Y.H.; Ryu, H.; Kim, Y.; Kim, T.H.; Hwang, I. BLH1 and KNAT3 modulate ABA responses during germination and early seedling development in Arabidopsis. Plant J. 2013, 75, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Rosin, F.M.; Prat, S.; Hannapel, D.J. Interacting transcription factors from the three-amino acid loop extension superclass regulate tuber formation. Plant Physiol. 2003, 132, 1391–1404. [Google Scholar] [CrossRef] [PubMed]


| Ferns (1) | Lycophytes (2) | Moss (3) | Spermatophytes | |
|---|---|---|---|---|
| VsKNAT1 | 63–69% | 54% | -- | 44–60% |
| VsKNAT3 | 49% | 43–45% | 39–40% | 48–88% |
| VsKNAT4 | 43% | 41–46% | 39–45% | 46–49% |
| VsKNAT6 | 70–75% | -- | -- | 51–65% |
| VsBELL4 | -- | 40–59% | 46–48% | 37–43% |
| VsBELL6 | -- | 45–52% | 45–46% | 45–52% |
| VsBELL10 | -- | 47% | 48–50% | 47–54% |
| Developmental Phase | VsKNAT1 | VsKNAT3 | VsKNAT4 | VsKNAT6 | VsBELL4 | VsBELL6 | VsBELL10 |
|---|---|---|---|---|---|---|---|
| Sporophyte | 0.43 (0.06) | 1.98 (0.35) | 0.23 (0.04) | 0.24 (0.03) | 0.85 (0.31) | 0.40 (0.06) | 1.11 (0.30) |
| Sporangia | 0.23 (0.04) | 1.68 (0.40) | 0.19 (0.06) | 0.26 (0.10) | 0.46 (0.06) | 0.40 (0.04) | 0.59 (0.08) |
| Gametophyte | 0.21 (0.07) | 0.21 (0.07) | 1.73 (0.36) | 1.46 (0.08) | 0.05 (0.01) | 0.31 (0.03) | 0.40 (0.03) |
| Developmental Phase | VsKNAT1 | VsKNAT3 | VsKNAT4 | VsKNAT6 | VsBELL4 | VsBELL6 | VsBELL10 |
|---|---|---|---|---|---|---|---|
| Sporophyte–Gametophyte | 0.076 | 0.009 * | 0.009 * | 0.009 * | 0.009 * | 0.175 | 0.028 * |
| Sporangia–Gametophyte | 0.465 | 0.009 * | 0.009 * | 0.009 * | 0.009 * | 0.117 | 0.047 * |
| Sporophyte–Sporangia | 0.047 * | 0.465 | 0.601 | 0.917 | 0.602 | 0.917 | 0.251 |
| Population | Phase | VsKNAT1 | VsKNAT3 | VsKNAT4 | VsKNAT6 | VsBELL4 | VsBELL6 | VsBELL10 |
|---|---|---|---|---|---|---|---|---|
| OJEN | Gametophyte | 0.11 (0.04) | 0.09 (0.01) | 0.57 (0.11) | 2.58 (0.22) | 0.05 (0.01) | 0.31 (0.03) | 0.40 (0.03) |
| ALMO | Gametophyte | 0.19 (0.07) | 0.75 (0.26) | 0.79 (0.28) | 4.55 (0.30) | 3.14 (0.63) | 0.69 (0.07) | 0.44 (0.07) |
| OJEN–ALMO | Gametophyte | 0.347 | 0.009 * | 0.917 | 0.009 * | 0.014 * | 0.014 * | 0.806 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-Estévez, M.; Bakkali, M.; Martín-Blázquez, R.; Garrido-Ramos, M.A. Identification and Characterization of TALE Homeobox Genes in the Endangered Fern Vandenboschia speciosa. Genes 2017, 8, 275. https://doi.org/10.3390/genes8100275
Ruiz-Estévez M, Bakkali M, Martín-Blázquez R, Garrido-Ramos MA. Identification and Characterization of TALE Homeobox Genes in the Endangered Fern Vandenboschia speciosa. Genes. 2017; 8(10):275. https://doi.org/10.3390/genes8100275
Chicago/Turabian StyleRuiz-Estévez, Mercedes, Mohammed Bakkali, Rubén Martín-Blázquez, and Manuel A. Garrido-Ramos. 2017. "Identification and Characterization of TALE Homeobox Genes in the Endangered Fern Vandenboschia speciosa" Genes 8, no. 10: 275. https://doi.org/10.3390/genes8100275
APA StyleRuiz-Estévez, M., Bakkali, M., Martín-Blázquez, R., & Garrido-Ramos, M. A. (2017). Identification and Characterization of TALE Homeobox Genes in the Endangered Fern Vandenboschia speciosa. Genes, 8(10), 275. https://doi.org/10.3390/genes8100275





