Comparative Genomics of Non-TNL Disease Resistance Genes from Six Plant Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Hidden Markov Model (HMM) Profiling and Sequence Identification
2.2. Sequence Alignment, Phylogenetic Analysis and Conserved Motifs Assessment
2.3. Gene Clustering, Structural and Functional Variation
3. Results
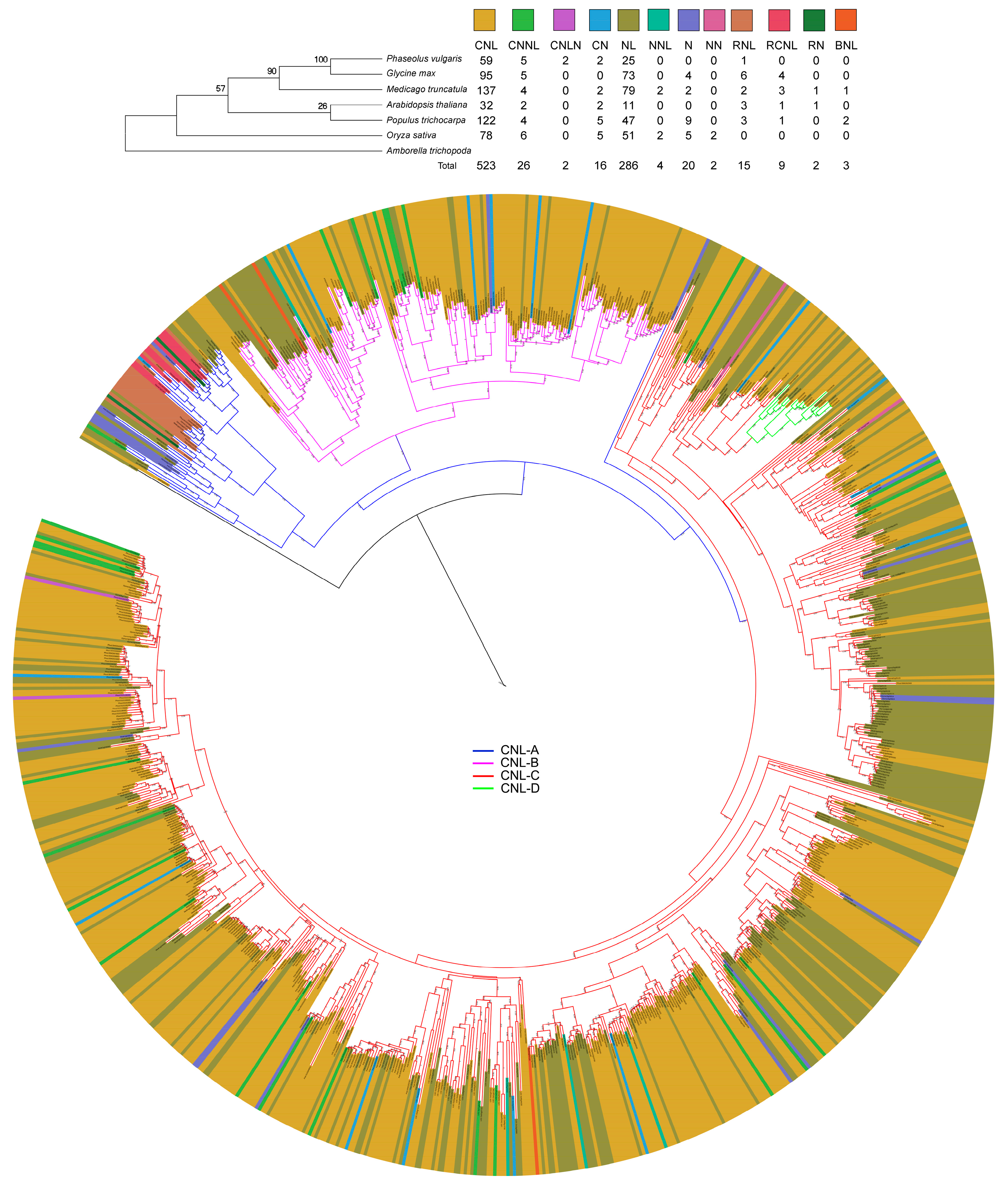
3.1. Diversity and Phylogenetic Relationship of the Identified nTNL Genes
3.2. Chromosomal Distribution of nTNL Genes, Gene Clustering and Selection Pressure
3.3. Gene Expression Analysis
4. Discussion
4.1. Phylogenetic History and Motif Structure of the nTNL Genes
4.2. Chromosomal Distribution of nTNL Gene Clustering
4.3. Assessing Gene Expression Data
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Marone, D.; Russo, M.A.; Laido, G.; De Leonardis, A.M.; Mastrangelo, A.M. Plant Nucleotide Binding Site-Leucine-Rich Repeat (NBS-LRR) Genes: Active Guardians in Host Defense Responses. Int. J. Mol. Sci. 2013, 14, 7302–7326. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Chen, X.; Lin, W.; Chen, S.; Lu, D.; Niu, Y.; Li, L.; Cheng, C.; McCormack, M.; Sheen, J. Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Flor, H.H. Inheritance of pathogenicity in Melampsora lini. Phytopathology 1942, 32, 653–669. [Google Scholar]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Dorrance, A.; McClure, S.; DeSilva, A. Pathogenic diversity of Phytophthora sojae in Ohio soybean fields. Plant Dis. 2003, 87, 139–146. [Google Scholar] [CrossRef]
- Costamilan, L.M.; Clebsch, C.C.; Soares, R.M.; Seixas, C.D.S.; Godoy, C.V.; Dorrance, A.E. Pathogenic diversity of Phytophthora sojae pathotypes from Brazil. Eur. J. Plant Pathol. 2013, 135, 845–853. [Google Scholar] [CrossRef]
- Mackey, D.; Belkhadir, Y.; Alonso, J.M.; Ecker, J.R.; Dangl, J.L. Arabidopsis RIN4 Is a Target of the Type III Virulence Effector AvrRpt2 and Modulates RPS2-Mediated Resistance. Cell 2003, 112, 379–389. [Google Scholar] [CrossRef]
- Liu, J.; Elmore, J.M.; Lin, Z.-J.D.; Coaker, G. A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 2011, 9, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.-Q.; Zhang, Y.-M.; Hang, Y.-Y.; Xue, J.-Y.; Zhou, G.-C.; Wu, P.; Wu, X.-Y.; Wu, X.-Z.; Wang, Q.; Wang, B. Long-Term Evolution of Nucleotide-Binding Site-Leucine-Rich Repeat (NBS-LRR) Genes: Understandings Gained From and Beyond the Legume Family. Plant Physiol. 2014, 166, 217–234. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cheng, Y.; Ma, W.; Zhao, Y.; Jiang, H.; Zhang, M. Identification and characterization of NBS-encoding disease resistance genes in Lotus japonicus. Plant Syst. Evol. 2010, 289, 101–110. [Google Scholar] [CrossRef]
- Kang, Y.; Kim, K.; Shim, S.; Yoon, M.; Sun, S.; Kim, M.; Van, K.; Lee, S.-H. Genome-wide mapping of NBS-LRR genes and their association with disease resistance in soybean. BMC Plant Biol. 2012, 12. [Google Scholar] [CrossRef] [PubMed]
- Ashfield, T.; Egan, A.N.; Pfeil, B.E.; Chen, N.W.; Podicheti, R.; Ratnaparkhe, M.B.; Ameline-Torregrosa, C.; Denny, R.; Cannon, S.; Doyle, J.J. Evolution of a complex disease resistance gene cluster in diploid Phaseolus and tetraploid Glycine. Plant Physiol. 2012, 159, 336–354. [Google Scholar] [CrossRef] [PubMed]
- Dangl, J.L.; Jones, J.D. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Sanseverino, W.; Hermoso, A.; D’Alessandro, R.; Vlasova, A.; Andolfo, G.; Frusciante, L.; Lowy, E.; Roma, G.; Ercolano, M.R. PRGdb 2.0: Towards a community-based database model for the analysis of R-genes in plants. Nucleic Acids Res. 2013, 41, D1167–D1171. [Google Scholar] [CrossRef] [PubMed]
- Meyers, B.C.; Dickerman, A.W.; Michelmore, R.W.; Sivaramakrishnan, S.; Sobral, B.W.; Young, N.D. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 1999, 20, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.J.; Sornaraj, P.; deCourcy-Ireland, E.; Menz, R.I.; Kobe, B.; Ellis, J.G.; Dodds, P.N.; Anderson, P.A. An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol. Plant. Microbe Interact. 2011, 24, 897–906. [Google Scholar] [CrossRef] [PubMed]
- Andersen, E.J.; Nepal, M.P. Genetic diversity of disease resistance genes in foxtail millet (Setaria italica L.). Plant Gene 2017, 10, 8–16. [Google Scholar] [CrossRef]
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.; Michelmore, R.W. Genome-wide analysis of NBS-LRR–encoding genes in Arabidopsis. Plant Cell Online 2003, 15, 809–834. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, R.; Kuang, H.; Meyers, B.C. The diversification of plant NBS-LRR defense genes directs the evolution of microRNAs that target them. Mol. Biol. Evol. 2016, 33, 2692–2705. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.Q.; Xue, J.Y.; Wu, P.; Zhang, Y.M.; Wu, Y.; Hang, Y.Y.; Wang, B.; Chen, J.Q. Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol. 2016, 170, 2095–2109. [Google Scholar] [CrossRef] [PubMed]
- Nepal, M.P.; Benson, B.V. CNL disease resistance genes in soybean and their evolutionary divergence. Evol. Bioinf. 2015, 11, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Zhang, Y.; Hu, Q.; Chen, J.; Li, K.; Lu, C.; Liu, H.; Wang, W.; Kuang, H. Dynamic nucleotide-binding site and leucine-rich repeat-encoding genes in the grass family. Plant Physiol. 2012, 159, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Michelmore, R.W.; Christopoulou, M.; Caldwell, K.S. Impacts of resistance gene genetics, function, and evolution on a durable future. Annu. Rev. Phytopathol. 2013, 51, 291–319. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Tian, Z.; Sui, Y.; Zhao, M.; Song, Q.; Cannon, S.B.; Cregan, P.; Ma, J. Pericentromeric effects shape the patterns of divergence, retention, and expression of duplicated genes in the paleopolyploid soybean. Plant Cell Online 2012, 24, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2013, D222–D230. [Google Scholar] [CrossRef]
- Benson, B.V. Disease resistance genes and their evolutionary history in six plant species. Master’s Thesis, South Dakota State University, Brookings, SD, USA, 2014. [Google Scholar]
- O’Kane, S.L., Jr.; Schaal, B.A.; Al-Shehbaz, I.A. The origins of Arabidopsis suecica (Brassicaceae) as indicated by nuclear rDNA sequences. Syst. Bot. 1996, 21, 559–566. [Google Scholar] [CrossRef]
- Carter, T.E.; Nelson, R.L.; Sneller, C.H.; Cui, Z. Genetic Diversity in Soybean. In Soybeans: Improvement, Production, and Uses; Boerma, H.R., Specht, J.E., Eds.; American Society of Agronomy, Inc.: Madison, WI, USA; Crop Science Society of America, Inc.: Madison, WI, USA; Soil Science Society of America, Inc.: Madison, WI, USA, 2004; pp. 303–416. [Google Scholar]
- Vitte, C.; Ishii, T.; Lamy, F.; Brar, D.; Panaud, O. Genomic paleontology provides evidence for two distinct origins of Asian rice (Oryza sativa L.). Mol. Genet. Genom. 2004, 272, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.R. Medicago truncatula—A model in the making!: Commentary. Curr. Opin. Plant Biol. 1999, 2, 301–304. [Google Scholar] [CrossRef]
- Pickersgill, B.; Debouck, D. Domestication patterns in common bean (Phaseolus vulgaris L.) and the origin of the Mesoamerican and Andean cultivated races. Theor. Appl. Genet. 2005, 110, 432–444. [Google Scholar]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Ashton, B.; Buxton, S.; Cheung, M.; Cooper, A.; Duran, C.; Field, M.; Heled, J.; Kearse, M.; Markowitz, S.; et al. Geneious 5.4. 2011. Available online: http://www.geneious.com/ (accessed on 16 June 2013).
- Rambaldi, D.; Ciccarelli, F.D. FancyGene: Dynamic visualization of gene structures and protein domain architectures on genomic loci. Bioinformatics 2009, 25, 2281–2282. [Google Scholar] [CrossRef] [PubMed]
- Soderlund, C.; Bomhoff, M.; Nelson, W.M. SyMAP v3. 4: A turnkey synteny system with application to plant genomes. Nucleic Acids Res. 2011. [Google Scholar] [CrossRef] [PubMed]
- Dash, S.; Van Hemert, J.; Hong, L.; Wise, R.P.; Dickerson, J.A. PLEXdb: Gene expression resources for plants and plant pathogens. Nucleic Acids Res. 2011, 40, D1194–D1201. [Google Scholar] [CrossRef] [PubMed]
- De Vos, M.; Van Oosten, V.R.; Van Poecke, R.M.; Van Pelt, J.A.; Pozo, M.J.; Mueller, M.J.; Buchala, A.J.; Métraux, J.-P.; Van Loon, L.; Dicke, M. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant-Microbe Interact. 2005, 18, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.T.; Van DE Mortel, M.; Bancroft, T.J.; Braun, E.; Nettleton, D.; Nelson, R.T.; Frederick, R.D.; Baum, T.J.; Graham, M.A.; Whitham, S.A. Biphasic gene expression changes elicited by Phakopsora pachyrhizi in soybean correlate with fungal penetration and haustoria formation. Plant Physiol. 2011, 157, 355–371. [Google Scholar] [CrossRef] [PubMed]
- Moreau, S.; Fromentin, J.; Vailleau, F.; Vernié, T.; Huguet, S.; Balzergue, S.; Frugier, F.; Gamas, P.; Jardinaud, M.F. The symbiotic transcription factor MtEFD and cytokinins are positively acting in the Medicago truncatula and Ralstonia solanacearum pathogenic interaction. New Phytol. 2014, 201, 1343–1357. [Google Scholar] [CrossRef] [PubMed]
- Hochreiter, S.; Clevert, D.-A.; Obermayer, K. A new summarization method for Affymetrix probe level data. Bioinformatics 2006, 22, 943–949. [Google Scholar] [CrossRef] [PubMed]
- Battke, F.; Symons, S.; Nieselt, K. Mayday—Integrative analytics for expression data. BMC Bioinf. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Friedman, A.R.; Baker, B.J. The evolution of resistance genes in multi-protein plant resistance systems. Curr. Opin. Genet. Dev. 2007, 17, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, M.J.; Thorne, J.L.; Wikström, N.; Bremer, K. Molecular evidence on plant divergence times. Am. J. Bot. 2004, 91, 1656–1665. [Google Scholar] [CrossRef] [PubMed]
- Krom, N.; Ramakrishna, W. Comparative analysis of divergent and convergent gene pairs and their expression patterns in rice, Arabidopsis, and Populus. Plant Physiol. 2008, 147, 1763–1773. [Google Scholar] [CrossRef] [PubMed]
- Stefanovic, S.; Pfeil, B.E.; Palmer, J.D.; Doyle, J.J. Relationships among phaseoloid legumes based on sequences from eight chloroplast regions. Syst. Bot. 2009, 34, 115–128. [Google Scholar] [CrossRef]
- Qi, D.; DeYoung, B.J.; Innes, R.W. Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol. 2012, 158, 1819–1832. [Google Scholar] [CrossRef] [PubMed]
- Peart, J.R.; Mestre, P.; Lu, R.; Malcuit, I.; Baulcombe, D.C. NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr. Biol. 2005, 15, 968–973. [Google Scholar] [CrossRef] [PubMed]
- Collier, S.M.; Hamel, L.-P.; Moffett, P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol. Plant-Microbe Interact. 2011, 24, 918–931. [Google Scholar] [CrossRef] [PubMed]
- Michelmore, R.W.; Meyers, B.C. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998, 8, 1113–1130. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.-M.; Hu, J.; Yin, W.-B.; Chen, Y.-H.; Wang, R.R.-C.; Hu, Z.-M. Cloning of resistance gene analogs located on the alien chromosome in an addition line of wheat-Thinopyrum intermedium. Theor. Appl. Genet. 2005, 111, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Meyers, B.C.; Morgante, M.; Michelmore, R.W. TIR-X and TIR-NBS proteins: Two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J. 2002, 32, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Tarr, D.E.K.; Alexander, H.M. TIR-NBS-LRR genes are rare in monocots: Evidence from diverse monocot orders. BMC Res. Notes 2009, 2. [Google Scholar] [CrossRef] [PubMed]
- Mohr, T.J.; Mammarella, N.D.; Hoff, T.; Woffenden, B.J.; Jelesko, J.G.; McDowell, J.M. The Arabidopsis downy mildew resistance gene RPP8 is induced by pathogens and salicylic acid and is regulated by W box cis elements. Mol. Plant-Microbe Interact. 2010, 23, 1303–1315. [Google Scholar] [CrossRef] [PubMed]
- McDowell, J.M.; Cuzick, A.; Can, C.; Beynon, J.; Dangl, J.L.; Holub, E.B. Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J. 2000, 22, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Bittner-Eddy, P.D.; Beynon, J.L. The Arabidopsis downy mildew resistance gene, RPP13-Nd, functions independently of NDR1 and EDS1 and does not require the accumulation of salicylic acid. Mol. Plant-Microbe Interact. 2001, 14, 416–421. [Google Scholar] [CrossRef] [PubMed]
- Kuang, H.; Woo, S.S.; Meyers, B.C.; Nevo, E.; Michelmore, R.W. Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell 2004, 16, 2870–2894. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.A.; Donoghue, M.J. Rates of molecular evolution are linked to life history in flowering plants. Science 2008, 322, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Chen, J.; Kuang, H. Dramatic Number Variation of R Genes in Solanaceae Species Accounted for by a Few R Gene Subfamilies. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Tehrim, S.; Zhang, F.; Tong, C.; Huang, J.; Cheng, X.; Dong, C.; Zhou, Y.; Qin, R.; Hua, W. Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genom. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.; Fazio, G.; Altman, N.; Praul, C.; McNellis, T. Mapping in an apple (Malus x domestica) F1 segregating population based on physical clustering of differentially expressed genes. BMC Genom. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, B. Plant domestication and resistance to herbivory. Int. J. Plant Genom. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Wong, G.K.-S.; Wang, J.; Tao, L.; Tan, J.; Zhang, J.; Passey, D.A.; Yu, J. Compositional gradients in Gramineae genes. Genome Res. 2002, 12, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Sémon, M.; Mouchiroud, D.; Duret, L. Relationship between gene expression and GC-content in mammals: Statistical significance and biological relevance. Hum. Mol. Genet. 2005, 14, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.B. Intron-Mediated Regulation of Gene Expression. In Nuclear pre-mRNA Processing in Plants; Reddy, A.N., Golovkin, M., Eds.; Springer: Berlin, Germany, 2008; Volume 326, pp. 277–290. [Google Scholar]
- Adams, K.L.; Wendel, J.F. Exploring the genomic mysteries of polyploidy in cotton. Biol. J. Linn. Soc. Lond. 2004, 82, 573–581. [Google Scholar] [CrossRef]
- Blanc, G.; Wolfe, K.H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 2004, 16, 1679–1691. [Google Scholar] [CrossRef] [PubMed]

| Plant Species | Domestication (Years Ago) | Native Range | Chromosome Number (N) | Genome Size |
|---|---|---|---|---|
| Arabidopsis thaliana | None | Eurasia (adaptive elsewhere) | 5 | 135 MB |
| Glycine max | 6000–9000 | Central or Northern China | 20 | 1100 MB |
| Oryza sativa | 11,000–12,000 | Southern China | 12 | 383 MB |
| Medicago truncatula | None | Mediterranean basin | 8 | 465 MB |
| Phaseolus vulgaris | 4400 | Americas (Argentina, Guatemala) | 11 | 650 MB |
| Populus trichocarpa | Literature unclear before 1977 | North Western United States ranging from Alaska to California and as far east as Western North Dakota | 19 | 423 MB |
| Clades | At | Gm | Mt | Os | Pv | Pt | Total |
|---|---|---|---|---|---|---|---|
| CNL-A | 8 | 14 | 19 | 0 | 1 | 7 | 49 |
| CNL-B | 25 | 37 | 22 | 3 | 17 | 61 | 165 |
| CNL-C | 7 | 134 | 191 | 145 | 75 | 124 | 676 |
| CNL-D | 12 | 2 | 1 | 0 | 1 | 1 | 17 |
| Unnested | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| Total | 52 a | 187 | 233 | 149 b | 94 | 193 | 908 |
| Species | CNL-A | CNL-B | CNL-C | CNL-D | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Max | Min | Avg. | Max | Min | Avg. | Max | Min | Avg. | Max | Min | Avg. | |
| At | 10 | 3 | 5.6 | 4 | 1 | 1.5 | 2 | 1 | 1.1 | 6 | 3 | 4.3 |
| Gm | 7 | 2 | 5.2 | 15 | 1 | 7.1 | 9 | 1 | 2.7 | 4 | 2 | 3.0 |
| Mt | 7 | 4 | 5.4 | 14 | 1 | 6.3 | 17 | 1 | 2.7 | 2 | 2 | 2.0 |
| Os | 0 | 0 | 0.0 | 2 | 1 | 1.7 | 8 | 1 | 2.6 | 0 | 0 | 0.0 |
| Pt | 8 | 2 | 4.7 | 11 | 1 | 3.7 | 10 | 1 | 2.4 | 2 | 2 | 2.0 |
| Pv | 5 | 5 | 5.0 | 16 | 2 | 8 | 7 | 1 | 2.3 | 2 | 2 | 2.0 |
| Species | Count of Clusters Observed | Genes in Clusters | Total nTNL Genes in Clusters (%) | Genes in Largest Cluster | Average Genes Per Cluster |
|---|---|---|---|---|---|
| At | 10 | 29 | 58% | 5 | 2.90 |
| Gm | 33 | 134 | 71% | 11 | 4.06 |
| Mt | 35 | 190 | 81% | 15 | 5.43 |
| Os | 29 | 74 | 50% | 7 | 2.64 |
| Pv | 15 | 82 | 87% | 26 | 5.47 |
| Pt | 34 | 156 | 80% | 18 | 4.59 |
| Sub Clade (Number of Gene Members) | Minimum Ka/Ks | Maximum Ka/Ks | Average Ka/Ks |
|---|---|---|---|
| CNL-A2 (14) | 0.09 | 0.43 | 0.20 |
| CNL-B3 (32) | 0.10 | 0.81 | 0.27 |
| CNL-C13 (23) | 0.12 | 0.55 | 0.30 |
| CNL-D (17) | 0.76 | 2.29 | 1.55 |
| Highly expressed Genes | AT3G46710, AT3G14470, Glyma20g33740, Glyma09g02401, Glyma12g14700, Potri.001G025400, LOC_Os01g23380, LOC_Os01g25720, LOC_Os01g33684, LOC_Os01g57270, LOC_Os01g57280, LOC_Os02g16270, LOC_Os02g17304, LOC_Os02g25900, LOC_Os03g40194, LOC_Os03g63150, LOC_Os04g02110, LOC_Os05g30220, LOC_Os05g31570, LOC_Os05g34220, LOC_Os05g34230, LOC_Os07g17250, LOC_Os07g29820, LOC_Os08g07330, LOC_Os08g10260, LOC_Os08g31800, LOC_Os08g43010, LOC_Os10g10360, LOC_Os11g12000, LOC_Os11g12340, LOC_Os11g37740, LOC_Os11g37759, LOC_Os11g42070, LOC_Os11g44580, LOC_Os11g44960, LOC_Os11g45790, LOC_Os12g32660, LOC_Os12g33160. LOC_Os12g37760. Medtr1g016210, Medtr2g014820, Medtr3g014080, Medtr3g015260, Medtr3g027420, Medtr3g032150, Medtr3g055720, Medtr3g056190, Medtr3g056300, Medtr3g070590, Medtr5g021080, Medtr6g046930, Medtr6g047210, Medtr6g052390, Medtr7g089080, Medtr7g091110, Medtr8g011280, Medtr8g038590Potri.001G134700, Potri.001G261300, Potri.003G149800, Potri.003G201800, Potri.005G119800, Potri.006G014400, Potri.006G271800, Potri.007G137100, Potri.008G212200, Potri.011G040800, Potri.012G121900, Potri.017G121500, and Potri.017G136400 |
| Genes expressed at the basal level | Glyma01g35120, Glyma02g03010, Glyma02g03520, Glyma03g05772, Glyma05g08621, Glyma13g26000, Glyma13g26141, Glyma13g26380, LOC_Os01g25630, LOC_Os03g36920, LOC_Os03g50150, LOC_Os05g12770, LOC_Os05g31550, LOC_Os05g40150, LOC_Os09g13820, LOC_Os10g03570, LOC_Os10g07400, LOC_Os11g45160, LOC_Os11g45970, LOC_Os12g29280 and LOC_Os12g31620, Medtr1g023600, Medtr2g039010, Medtr3g035480, Medtr3g086070, Medtr5g035240, Medtr5g070470, Medtr6g046440, Medtr8g011590, Medtr8g011600, Potri.003G099000, Potri.004G195200, Potri.012G122200, Potri.017G127000, Potri.017G143400, Potri.017G143500, and Potri.018G017900 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nepal, M.P.; Andersen, E.J.; Neupane, S.; Benson, B.V. Comparative Genomics of Non-TNL Disease Resistance Genes from Six Plant Species. Genes 2017, 8, 249. https://doi.org/10.3390/genes8100249
Nepal MP, Andersen EJ, Neupane S, Benson BV. Comparative Genomics of Non-TNL Disease Resistance Genes from Six Plant Species. Genes. 2017; 8(10):249. https://doi.org/10.3390/genes8100249
Chicago/Turabian StyleNepal, Madhav P., Ethan J. Andersen, Surendra Neupane, and Benjamin V. Benson. 2017. "Comparative Genomics of Non-TNL Disease Resistance Genes from Six Plant Species" Genes 8, no. 10: 249. https://doi.org/10.3390/genes8100249
APA StyleNepal, M. P., Andersen, E. J., Neupane, S., & Benson, B. V. (2017). Comparative Genomics of Non-TNL Disease Resistance Genes from Six Plant Species. Genes, 8(10), 249. https://doi.org/10.3390/genes8100249






