Downregulation of Enhancer of Zeste Homolog 2 (EZH2) is essential for the Induction of Autophagy and Apoptosis in Colorectal Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and DZNep
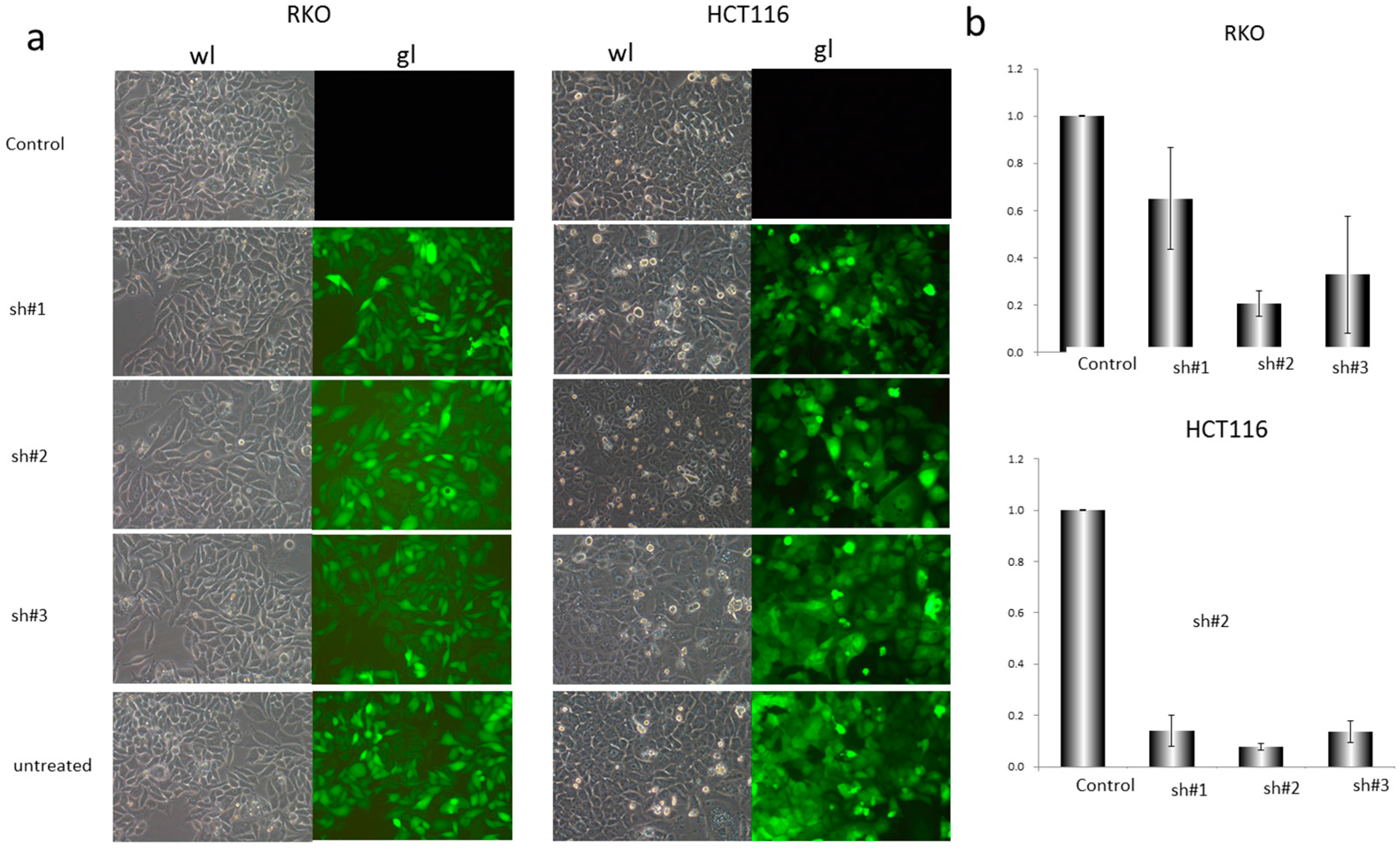
2.2. Construction of shRNA and Cell Transfection
2.3. Quantitative Real-Time Reverse Transcription PCR (QRT-PCR)
2.4. Western Blot
2.5. Cell Proliferation Assay
2.6. Cell Migration Assay
2.7. Cell Apoptosis Analysis
2.8. Cell Cycle Analysis
2.9. Statistical Analysis
3. Results
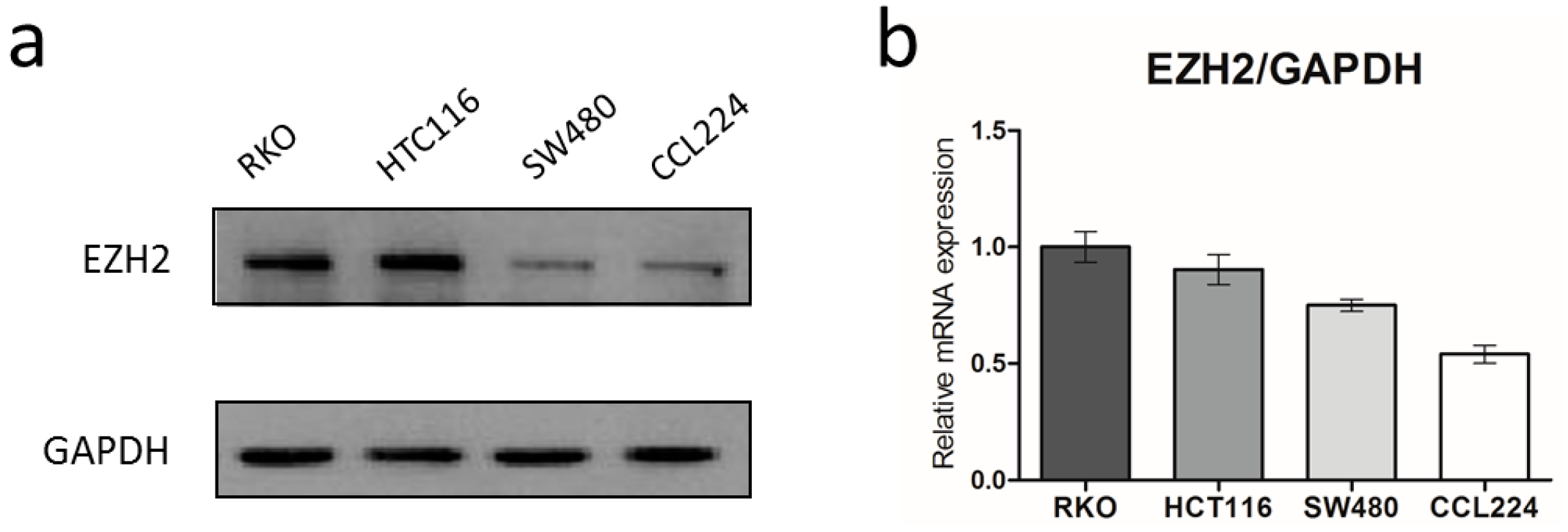
3.1. EZH2 Is Over-Expressed in CRC Cells
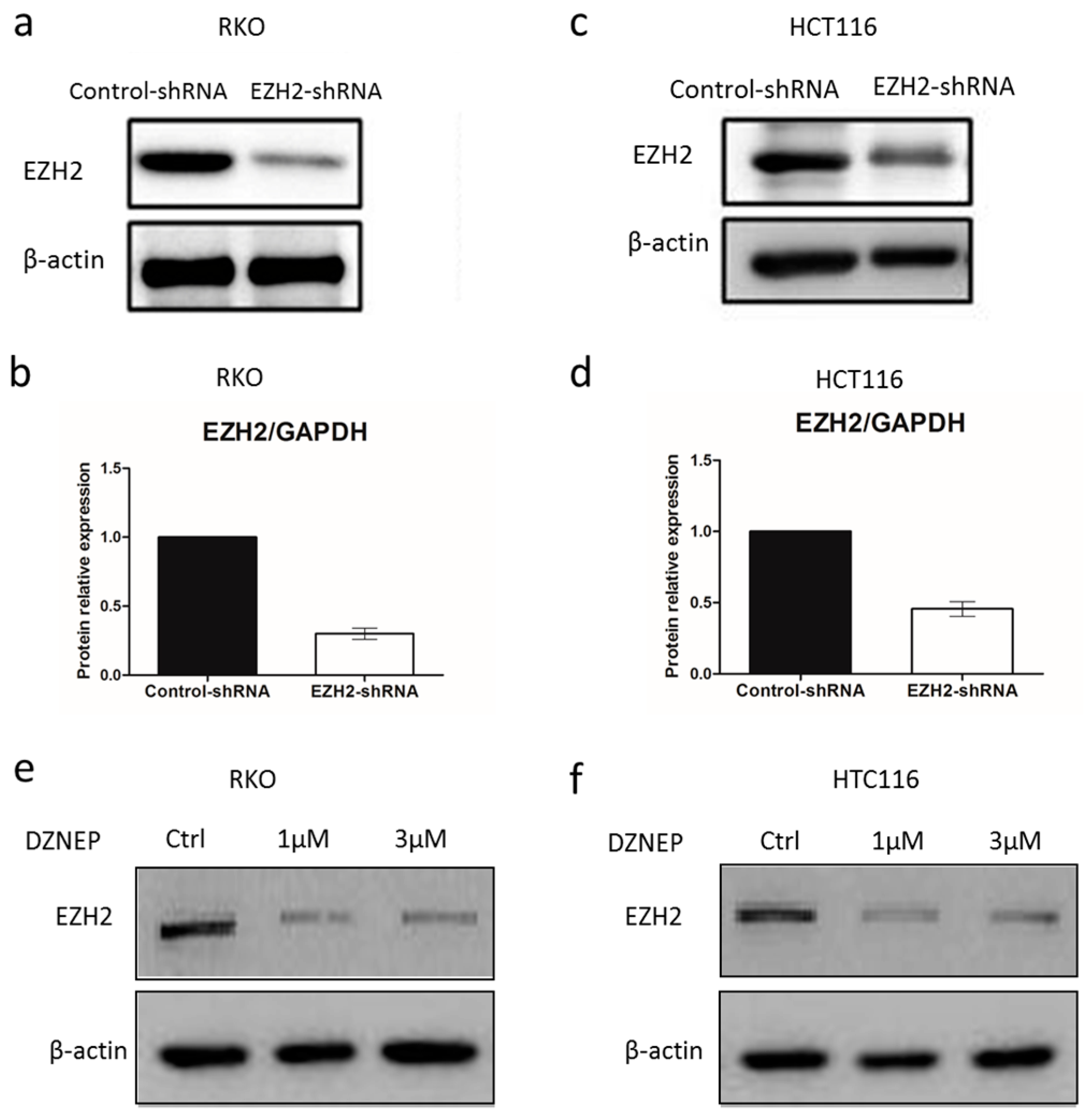
3.2. EZH2-shRNA and DZNep Inhibited the Expression of EZH2 mRNA and Protein in RKO and HCT116 Cells
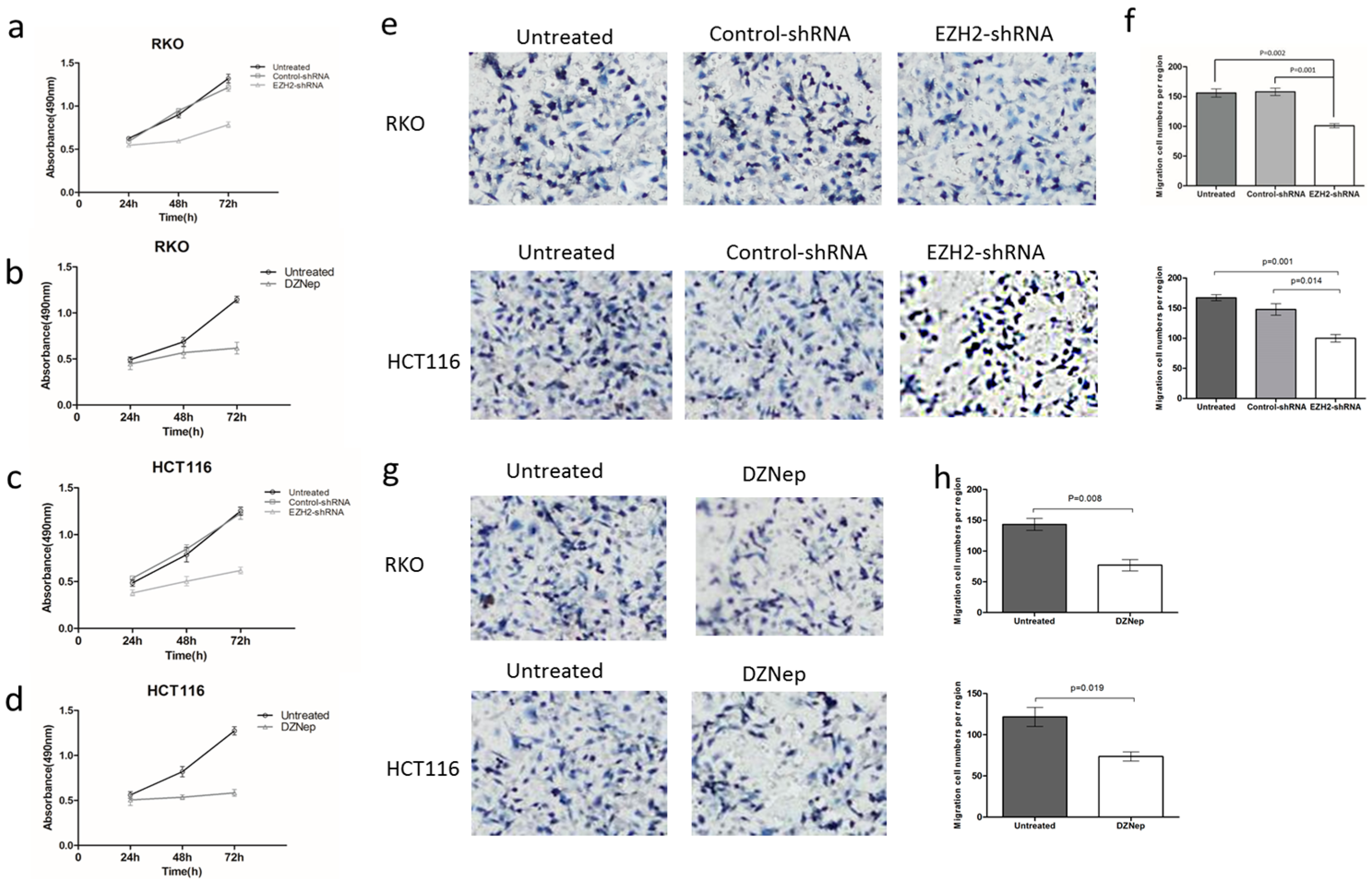
3.3. Effects of EZH2 Downregulation on the Growth and Migration of RKO and HCT116 Cells
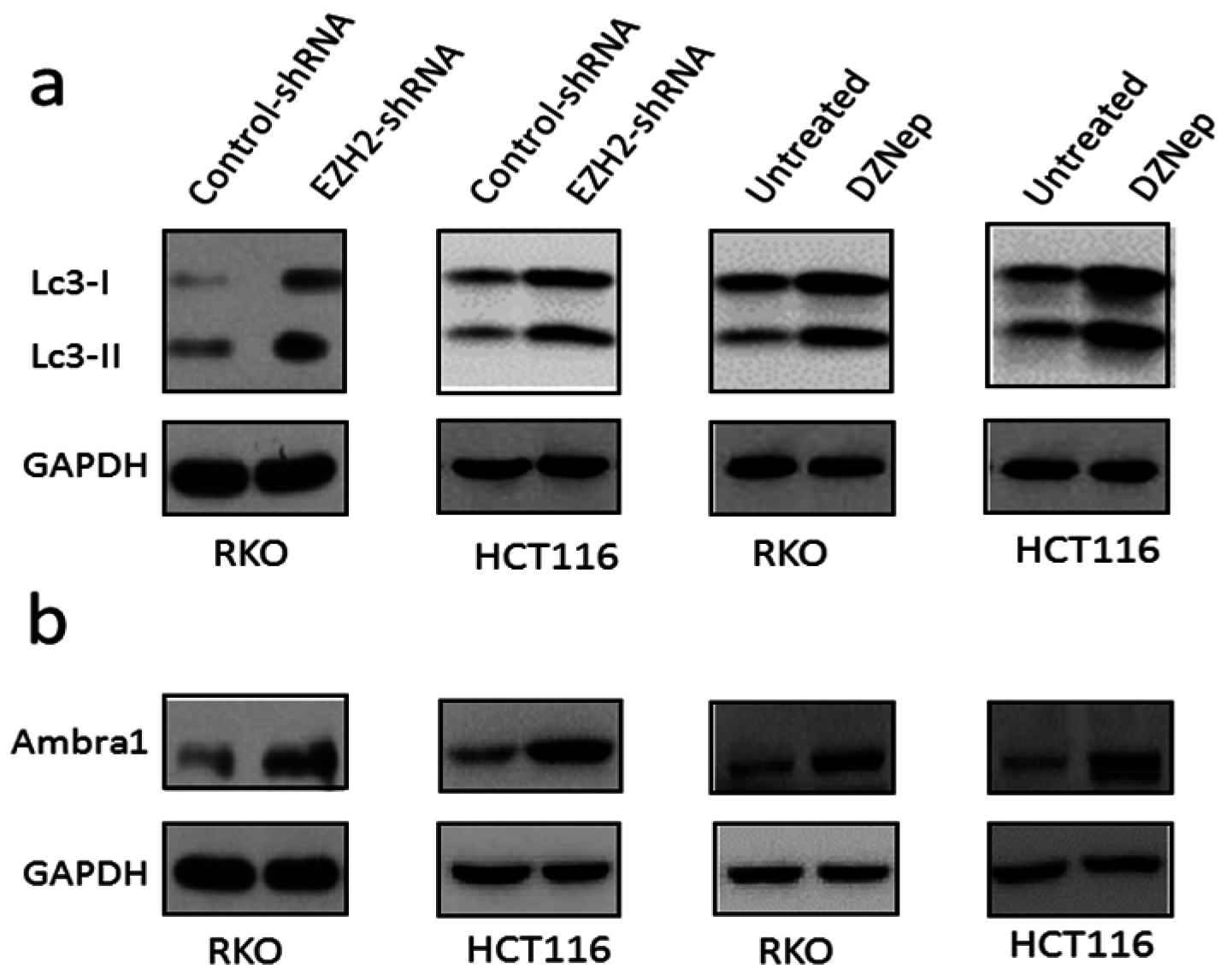
3.4. Downregulation of EZH2 Induced Autophagy in RKO and HCT116 Cells
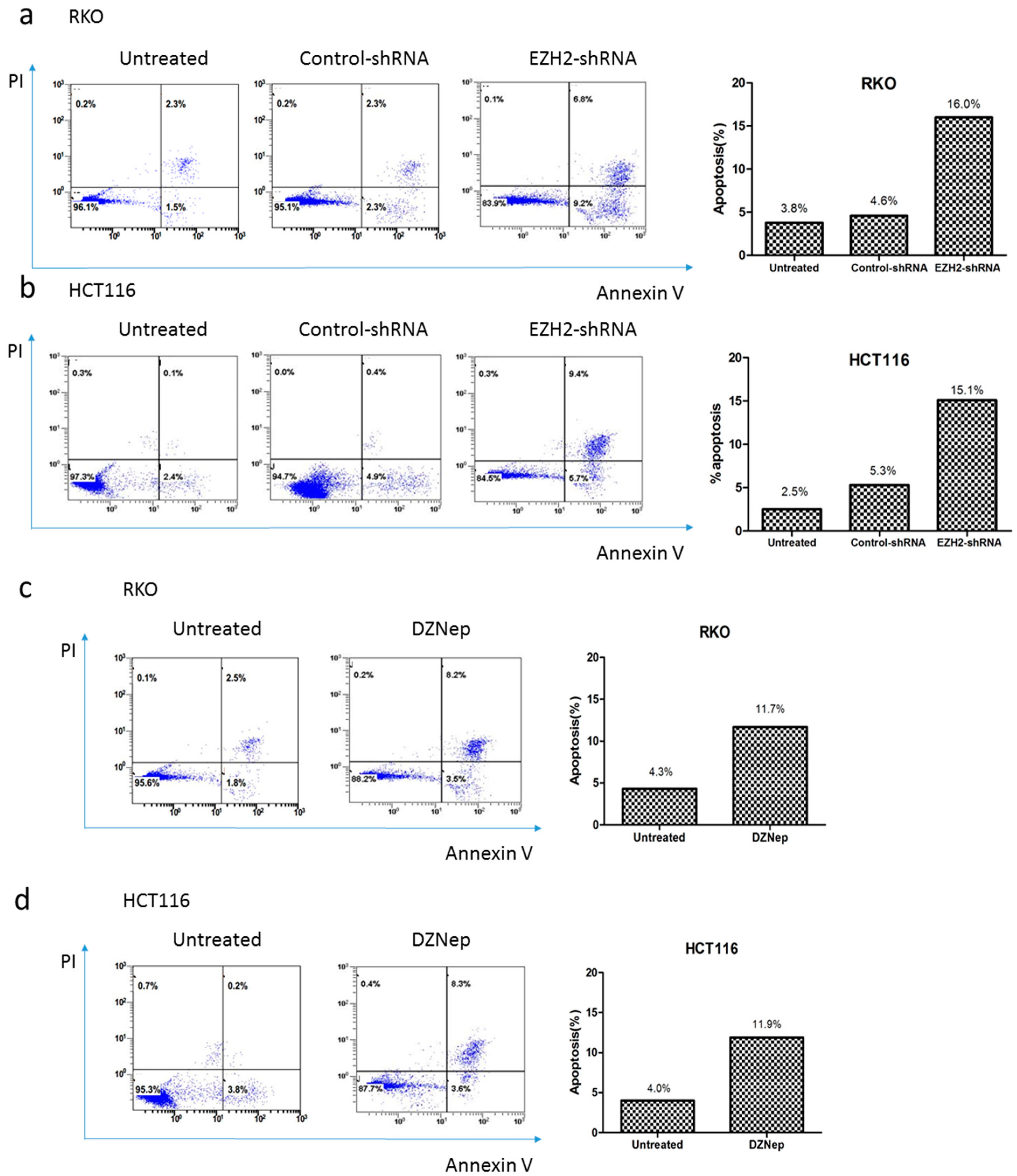
3.5. Downregulation of EZH2 Induced Apoptosis in RKO and HCT116 Cells
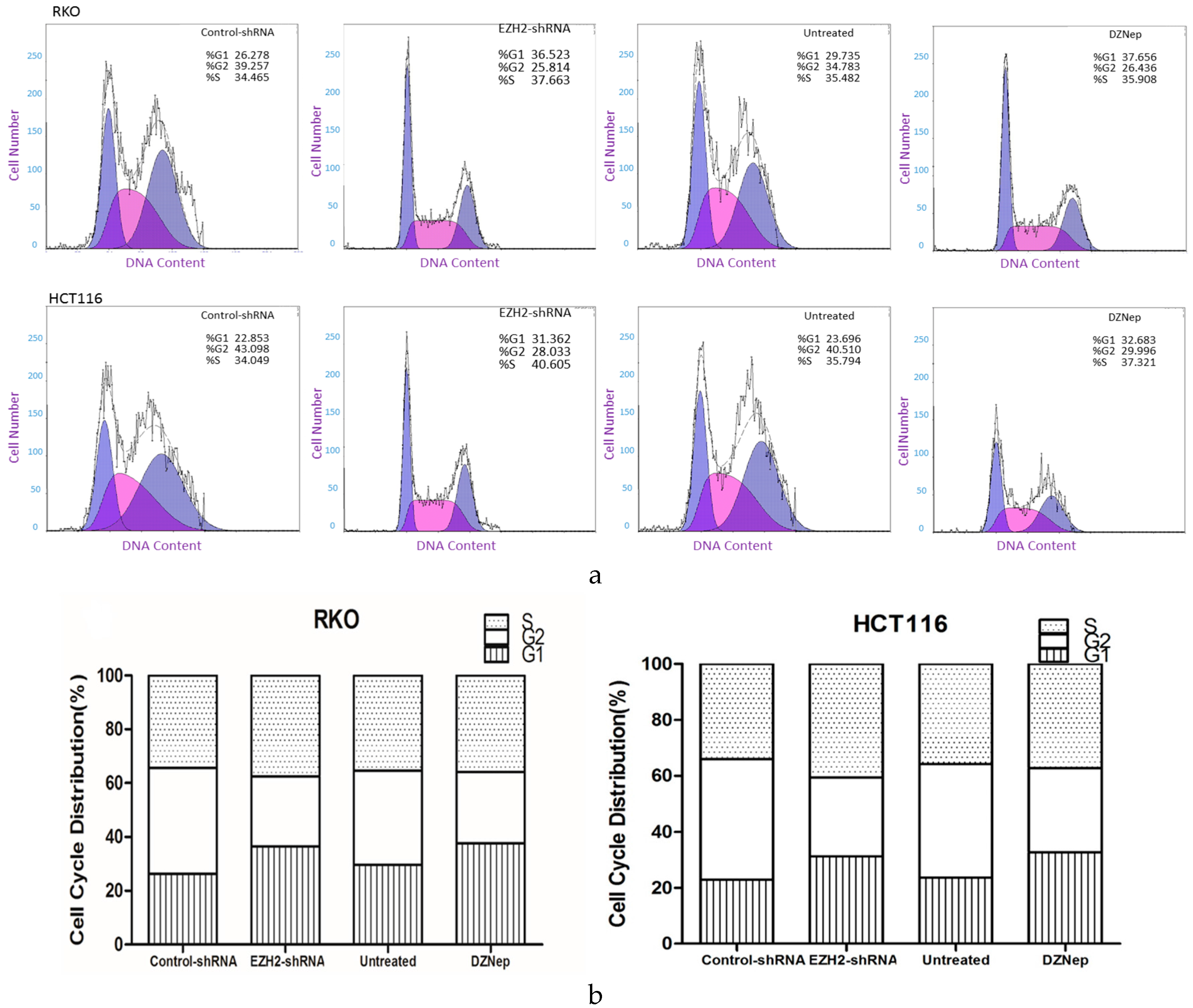
3.6. Downregulation of EZH2 Induced Cell Cycle Arrest in RKO and HCT116 Cells
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Viswanath, B.; Kim, S.; Lee, K. Recent insights into nanotechnology development for detection and treatment of colorectal cancer. Int. J. Nanomed. 2016, 11, 2491–2504. [Google Scholar]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, C.E.; Lin, C.C.; Mariotto, A.B.; Siegel, R.L.; Stein, K.D.; Kramer, J.L.; Alteri, R.; Robbins, A.S.; Jemal, A. Cancer treatment and survivorship statistics. CA Cancer J. Clin. 2014, 64, 252–271. [Google Scholar] [CrossRef] [PubMed]
- Weinstock, B.; Ward, S.C.; Harpaz, N.; Warner, R.R.; Itzkowitz, S.; Kim, M.K. Clinical and prognostic features of rectal neuroendocrine tumors. Neuroendocrinology 2013, 98, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.A.; Lange, C.A. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat. Res. 2008, 647, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Bracken, A.P.; Pasini, D.; Capra, M.; Prosperini, E.; Colli, E.; Helin, K. EZH2 is downstream of the pRB-E2f pathway, essential for proliferation and amplified in cancer. EMBO J. 2003, 22, 5323–5335. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.T.; Cai, M.Y.; Wang, X.G.; Kong, L.L.; Mai, S.J.; Liu, Y.H.; Zhang, H.B.; Liao, Y.J.; Zheng, F.; Zhu, W.; et al. EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and snail to inhibit e-cadherin. Oncogene 2012, 31, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Kleer, C.G.; Cao, Q.; Varambally, S.; Shen, R.; Ota, I.; Tomlins, S.A.; Ghosh, D.; Sewalt, R.G.; Otte, A.P.; Hayes, D.F.; et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. USA 2003, 100, 11606–11611. [Google Scholar] [CrossRef] [PubMed]
- Raman, J.D.; Mongan, N.P.; Tickoo, S.K.; Boorjian, S.A.; Scherr, D.S.; Gudas, L.J. Increased expression of the polycomb group gene, EZH2, in transitional cell carcinoma of the bladder. Clin. Cancer Res. 2005, 11, 8570–8576. [Google Scholar] [CrossRef] [PubMed]
- Collett, K.; Eide, G.E.; Arnes, J.; Stefansson, I.M.; Eide, J.; Braaten, A.; Aas, T.; Otte, A.P.; Akslen, L.A. Expression of enhancer of zeste homologue 2 is significantly associated with increased tumor cell proliferation and is a marker of aggressive breast cancer. Clin. Cancer Res. 2006, 12, 1168–1174. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, I.M.; Halvorsen, O.J.; Collett, K.; Stefansson, I.M.; Straume, O.; Haukaas, S.A.; Salvesen, H.B.; Otte, A.P.; Akslen, L.A. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol. 2006, 24, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Yamada, A.; Fujii, S.; Daiko, H.; Nishimura, M.; Chiba, T.; Ochiai, A. Aberrant expression of EZH2 is associated with a poor outcome and P53 alteration in squamous cell carcinoma of the esophagus. Int. J. Oncol. 2011, 38, 345–353. [Google Scholar] [PubMed]
- He, S.B.; Zhou, H.; Zhou, J.; Zhou, G.Q.; Han, T.; Wan, D.W.; Gu, W.; Gao, L.; Zhang, Y.; Xue, X.F.; et al. Inhibition of EZH2 expression is associated with the proliferation, apoptosis and migration of SW620 colorectal cancer cells in vitro. Exp. Biol. Med. 2015, 240, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Gao, T.; Wang, N.; Feng, Q.; You, X.; Ye, T.; Lei, Q.; Zhu, Y.; Xiong, M.; Xia, Y.; et al. Corrigendum: Selective inhibition of EZH2 by ZLD1039 blocks H3K27methylation and leads to potent anti-tumor activity in breast cancer. Sci. Rep. 2016. [Google Scholar] [CrossRef] [PubMed]
- Takashina, T.; Kinoshita, I.; Kikuchi, J.; Shimizu, Y.; Sakakibara-Konishi, J.; Oizumi, S.; Nishimura, M.; Dosaka-Akita, H. Combined inhibition of EZH2 and hdacs as a potential epigenetic therapy for non-small cell lung cancer cells. Cancer Sci. 2016. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, S.; Nagano, H.; Konno, M.; Eguchi, H.; Tomokuni, A.; Tomimaru, Y.; Asaoka, T.; Wada, H.; Hama, N.; Kawamoto, K.; et al. A crucial epithelial to mesenchymal transition regulator, Sox4/Ezh2 axis is closely related to the clinical outcome in pancreatic cancer patients. Int. J. Oncol. 2016, 48, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Cao, Q.; Mehra, R.; Laxman, B.; Yu, J.; Tomlins, S.A.; Creighton, C.J.; Dhanasekaran, S.M.; Shen, R.; Chen, G.; et al. Integrative genomics analysis reveals silencing of β-adrenergic signaling by polycomb in prostate cancer. Cancer Cell 2007, 12, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.; Chagraoui, J.; Krosl, J.; Gendron, P.; Wilhelm, B.; Lemieux, S.; Boucher, G.; Chagnon, P.; Drouin, S.; Lambert, R.; et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012, 26, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F.; Luo, X.; Xiang, L.S.; Li, H.T.; Zha, L.; Li, N.; He, J.M.; Xie, G.F.; Xie, X.; Liang, H.J. EZH2 promotes colorectal cancer stem-like cell expansion by activating p21cip1-Wnt/β-catenin signaling. Oncotarget 2016. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Sun, X.; Zhou, X.; Han, L.; Chen, L.; Shi, Z.; Zhang, A.; Ye, M.; Wang, Q.; Liu, C.; et al. Long non-coding RNA hotair promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget 2015, 6, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Okabe, H.; Sakamoto, Y.; Hayashi, H.; Hashimoto, D.; Yokoyama, N.; Sakamoto, K.; Kuroki, H.; Mima, K.; Nitta, H.; et al. Enhancer of zeste homolog 2 (EZH2) promotes progression of cholangiocarcinoma cells by regulating cell cycle and apoptosis. Ann. Surg. Oncol. 2013, 20, S667–S675. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Tang, J.; Wang, J.; Duan, G.; Zhou, L.; Zhou, X. Mir-138 acts as a tumor suppressor by targeting EZH2 and enhances cisplatin-induced apoptosis in osteosarcoma cells. PLoS ONE 2016, 11, e0150026. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Ren, Y.; Kong, L.; Cai, G.; Sun, S.; Song, W.; Wang, Y.; Jin, R.; Qi, L.; Mei, M.; et al. Targeting EZH2 regulates tumor growth and apoptosis through modulating mitochondria dependent cell-death pathway in hnscc. Oncotarget 2016, 6, 33720–33732. [Google Scholar]
- Zhang, H.; Gu, H.; Li, L.; Ren, Y.; Zhang, L. EZH2 mediates ato-induced apoptosis in acute myeloid leukemia cell lines through the wnt signaling pathway. Tumour Biol. 2016, 37, 5919–5923. [Google Scholar] [CrossRef] [PubMed]
- Smonskey, M.; Lasorsa, E.; Rosario, S.; Kirk, J.S.; Hernandez-Ilizaliturri, F.J.; Ellis, L. EZH2 inhibition re-sensitizes multidrug resistant B-cell lymphomas to etoposide mediated apoptosis. Oncoscience 2016, 3, 21–30. [Google Scholar] [PubMed]
- Wu, C.; Jin, X.; Yang, J.; Yang, Y.; He, Y.; Ding, L.; Pan, Y.; Chen, S.; Jiang, J.; Huang, H. Inhibition of EZH2 by chemo- and radiotherapy agents and small molecule inhibitors induces cell death in castration-resistant prostate cancer. Oncotarget. 2016, 7, 3440–3452. [Google Scholar] [PubMed]
- White, E. Deconvoluting the context-dependent role for autophagy in cancer. Nat. Rev. Cancer 2012, 12, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Maiuri, M.C.; Zalckvar, E.; Kimchi, A.; Kroemer, G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007, 8, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.Y.; Xia, B.; White, E. Autophagy-mediated tumor promotion. Cell 2013, 155, 1216–1219. [Google Scholar] [CrossRef] [PubMed]
- Kabeya, Y.; Mizushima, N.; Ueno, T.; Yamamoto, A.; Kirisako, T.; Noda, T.; Kominami, E.; Ohsumi, Y.; Yoshimori, T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000, 19, 5720–5728. [Google Scholar] [CrossRef] [PubMed]
- Fimia, G.M.; Stoykova, A.; Romagnoli, A.; Giunta, L.; Di Bartolomeo, S.; Nardacci, R.; Corazzari, M.; Fuoco, C.; Ucar, A.; Schwartz, P.; et al. Ambra1 regulates autophagy and development of the nervous system. Nature 2007, 447, 1121–1125. [Google Scholar] [PubMed]
- Gu, W.; Wan, D.; Qian, Q.; Yi, B.; He, Z.; Gu, Y.; Wang, L.; He, S. Ambra1 is an essential regulator of autophagy and apoptosis in SW620 cells: Pro-survival role of Ambra1. PLoS ONE 2014, 9, e90151. [Google Scholar] [CrossRef] [PubMed]
- Morselli, E.; Marino, G.; Bennetzen, M.V.; Eisenberg, T.; Megalou, E.; Schroeder, S.; Cabrera, S.; Benit, P.; Rustin, P.; Criollo, A.; et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J. Cell Biol. 2011, 192, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Fullgrabe, J.; Lynch-Day, M.A.; Heldring, N.; Li, W.; Struijk, R.B.; Ma, Q.; Hermanson, O.; Rosenfeld, M.G.; Klionsky, D.J.; Joseph, B. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature 2013, 500, 468–471. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.P.; Lo, H.L.; Wei, L.S.; Hsiao, H.H.; Yang, P.M. S-adenosyl-L-methionine-competitive inhibitors of the histone methyltransferase EZH2 induce autophagy and enhance drug sensitivity in cancer cells. Anticancer Drugs 2015, 26, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Qiu, S.; Ge, R.; He, L.; Li, M.; Li, Y.; Peng, Y. Mir-340 suppresses glioblastoma multiforme. Oncotarget 2015, 6, 9257–9270. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, L.; Shi, Z.; Zhao, S.; Wang, F.T.; Zhou, T.T.; Liu, B.; Bao, J.K. Programmed cell death pathways in cancer: A review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012, 45, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Rosengren, L.E.; Franlund, M.; Hamberger, A.; Haglid, K.G. BCL-2 expression regulates cell sensitivity to S100β-mediated apoptosis. Brain Res. Mol. Brain Res. 1999, 70, 167–176. [Google Scholar] [CrossRef]
- Wei, Y.; Pattingre, S.; Sinha, S.; Bassik, M.; Levine, B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell 2008, 30, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Amelio, I.; Melino, G.; Knight, R.A. Cell death pathology: Cross-talk with autophagy and its clinical implications. Biochem. Biophys. Res. Commun. 2011, 414, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Wirawan, E.; Vande Walle, L.; Kersse, K.; Cornelis, S.; Claerhout, S.; Vanoverberghe, I.; Roelandt, R.; De Rycke, R.; Verspurten, J.; Declercq, W.; et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, S.; Perozzo, R.; Schmid, I.; Ziemiecki, A.; Schaffner, T.; Scapozza, L.; Brunner, T.; Simon, H.U. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 2006, 8, 1124–1132. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M. Mutation spectra of histone methyltransferases with canonical set domains and EZH2-targeted therapy. Epigenomics 2016, 8, 285–305. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.T.; Zhu, F.; Lin, W.R.; Ying, R.B.; Yang, Y.P.; Zeng, L.H. The novel EZH2 inhibitor, GSK126, suppresses cell migration and angiogenesis via down-regulating VEGF-A. Cancer Chemother. Pharmacol. 2016, 77, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Girard, N.; Bazille, C.; Lhuissier, E.; Benateau, H.; Llombart-Bosch, A.; Boumediene, K.; Bauge, C. 3-Deazaneplanocin a (DZNep), an inhibitor of the histone methyltransferase EZH2, induces apoptosis and reduces cell migration in chondrosarcoma cells. PLoS ONE 2014, 9, e98176. [Google Scholar] [CrossRef] [PubMed]
- Koppens, M.A.; Bounova, G.; Cornelissen-Steijger, P.; de Vries, N.; Sansom, O.J.; Wessels, L.F.; van Lohuizen, M. Large variety in a panel of human colon cancer organoids in response to EZH2 inhibition. Oncotarget 2016. [Google Scholar] [CrossRef] [PubMed]
- Knutson, S.K.; Warholic, N.M.; Wigle, T.J.; Klaus, C.R.; Allain, C.J.; Raimondi, A.; Porter Scott, M.; Chesworth, R.; Moyer, M.P.; Copeland, R.A.; et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc. Natl. Acad. Sci. USA 2013, 110, 7922–7927. [Google Scholar] [CrossRef] [PubMed]
- Knutson, S.K.; Kawano, S.; Minoshima, Y.; Warholic, N.M.; Huang, K.C.; Xiao, Y.; Kadowaki, T.; Uesugi, M.; Kuznetsov, G.; Kumar, N.; et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-hodgkin lymphoma. Mol. Cancer Ther. 2014, 13, 842–854. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Hu, J.; Ding, H.; Chen, L.; Zhang, Y.; Liu, R.; Xu, P.; Du, D.; Lu, W.; Liu, J.; et al. Identification of novel EZH2 inhibitors through pharmacophore-based virtual screening and biological assays. Bioorg. Med. Chem. Lett. 2016, 26, 3813–3817. [Google Scholar] [CrossRef] [PubMed]
| Target sequence 1 | TAGGTTAATTGGGACCAAA |
| Sense strand 5’-3’ | CCGGGCTAGGTTAATTGGGACCAAACTCGAGTTTGGTCCCAATTAACC |
| Antisense strand 5’–3’ | AATTCAAAAAGCTAGGTTAATTGGGACCAAACTCGAGTTTGGTCCCAA |
| Target sequence 2 | CAACATAGATGGACCAAAT |
| Sense strand 5’-3’ | CCGGCCCAACATAGATGGACCAAATCTCGAGATTTGGTCCATCTATGTT |
| Antisense strand 5’–3’ | AATTCAAAAACCCAACATAGATGGACCAAATCTCGAGATTTGGTCCAT |
| Target sequence 3 | GAAATCTTAAACCAAGAAT |
| Sense strand 5’-3’ | CCGGCGGAAATCTTAAACCAAGAATCTCGAGATTCTTGGTTTAAG |
| Antisense strand 5’–3’ | AATTCAAAAACGGAAATCTTAAACCAAGAATCTCGAGATTCTTGGTTT |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, Y.; Hu, H.; Yang, Y.; Zhou, G.; Shang, Z.; Yang, X.; Sun, K.; Zhan, S.; Yu, Z.; Li, P.; et al. Downregulation of Enhancer of Zeste Homolog 2 (EZH2) is essential for the Induction of Autophagy and Apoptosis in Colorectal Cancer Cells. Genes 2016, 7, 83. https://doi.org/10.3390/genes7100083
Yao Y, Hu H, Yang Y, Zhou G, Shang Z, Yang X, Sun K, Zhan S, Yu Z, Li P, et al. Downregulation of Enhancer of Zeste Homolog 2 (EZH2) is essential for the Induction of Autophagy and Apoptosis in Colorectal Cancer Cells. Genes. 2016; 7(10):83. https://doi.org/10.3390/genes7100083
Chicago/Turabian StyleYao, Yizhou, Hao Hu, Yong Yang, Guoqiang Zhou, Zengfu Shang, Xiaodong Yang, Kang Sun, Shenghua Zhan, Zhengyuan Yu, Peiyao Li, and et al. 2016. "Downregulation of Enhancer of Zeste Homolog 2 (EZH2) is essential for the Induction of Autophagy and Apoptosis in Colorectal Cancer Cells" Genes 7, no. 10: 83. https://doi.org/10.3390/genes7100083
APA StyleYao, Y., Hu, H., Yang, Y., Zhou, G., Shang, Z., Yang, X., Sun, K., Zhan, S., Yu, Z., Li, P., Pan, G., Sun, L., Zhu, X., & He, S. (2016). Downregulation of Enhancer of Zeste Homolog 2 (EZH2) is essential for the Induction of Autophagy and Apoptosis in Colorectal Cancer Cells. Genes, 7(10), 83. https://doi.org/10.3390/genes7100083





