Down-Regulation of miR-129-5p and the let-7 Family in Neuroendocrine Tumors and Metastases Leads to Up-Regulation of Their Targets Egr1, G3bp1, Hmga2 and Bach1
Abstract
:1. Introduction
2. Experimental
2.1. Clinical Samples
2.2. Cell Culture
2.3. RNA Extraction
2.4. miRNA Microarray Analyses
2.5. qPCR
2.6. In Situ Hybridization
2.7. Immunohistochemistry
2.8. Transfection Studies and Cell Growth Analyses
2.9. mRNA Microarray Analysis
2.10. Evaluating Global Down-Regulation of microRNA Target Genes
2.12. Gene Set Enrichment Analysis
2.13. 3'UTR Luciferase Assay
2.14. Western Blotting
2.15. Statistical Analyses
3. Results
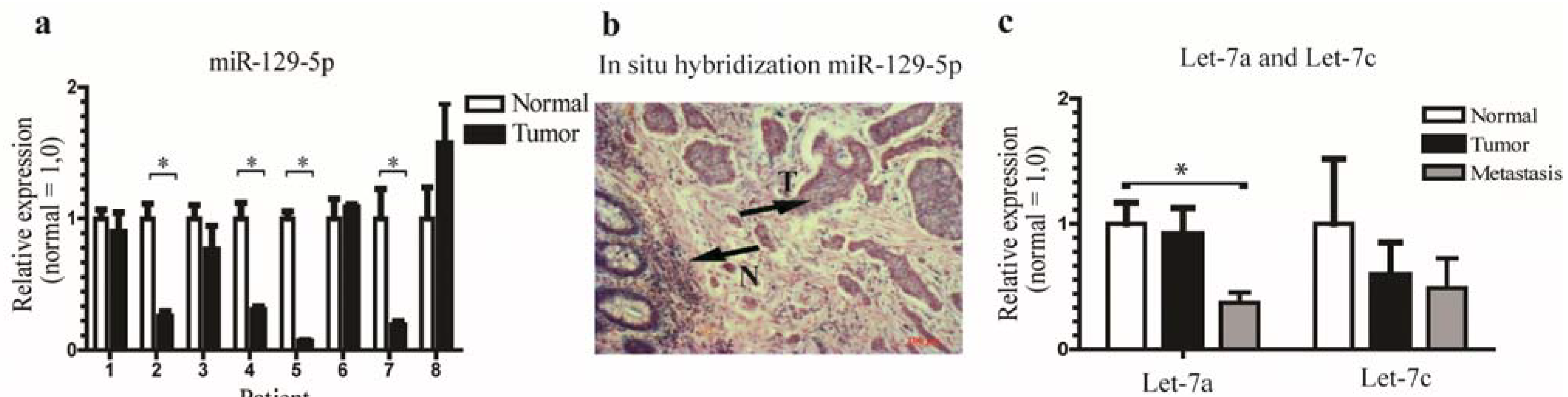
3.1. miR-129-5p is Down-Regulated in Neuroendocrine Tumors and the let-7 Family is Down-Regulated in Neuroendocrine Tumor Metastases
| T + M vs. N | Log FC | FC | p-value |
|---|---|---|---|
| hsa-miR-129-5p | −1.3 | 0.4 | 0.01 |
| hsa-let-7a | −1.7 | 0.3 | 0.002 |
| hsa-let-7c | −1.7 | 0.3 | 0.001 |
| hsa-let-7b | −1.5 | 0.3 | 0.0003 |
| hsa-let-7d | −1.5 | 0.3 | 0,003 |
| hsa-let-7e | −1.4 | 0.4 | 0.002 |
| hsa-let-7f | −1.4 | 0.4 | 0.003 |
| hsa-let-7g | −1.1 | 0.5 | 0.01 |
| hsa-let-7i | −1.3 | 0.4 | 0.001 |
| M vs. N | Log FC | FC | p-value |
| hsa-let-7a | −1.7 | 0.3 | 0.002 |
| hsa-let-7c | −1.7 | 0.3 | 0.001 |
| hsa-let-7b | −1.5 | 0.3 | 0.0003 |
| hsa-let-7d | −1.5 | 0.3 | 0.003 |
| hsa-let-7e | −1.4 | 0.4 | 0.002 |
| hsa-let-7f | −1.4 | 0.4 | 0.003 |
| hsa-let-7g | −1.1 | 0.5 | 0.01 |
| hsa-let-7i | −1.3 | 0.4 | 0.001 |
| M vs. T | Log FC | FC | p-value |
| hsa-let-7a | −0.03 | 0.8 | 0.3 |
| hsa-let-7c | −0.02 | 1.0 | 0.9 |
| hsa-let-7b | −0.3 | 0.8 | 0.3 |
| hsa-let-7d | −0.2 | 0.9 | 0.6 |
| hsa-let-7e | −0.1 | 0.9 | 0.6 |
| hsa-let-7f | −0.1 | 0.9 | 0.8 |
| hsa-let-7g | −0.1 | 0.9 | 0.8 |
| hsa-let-7i | −0.02 | 1.0 | 0.9 |
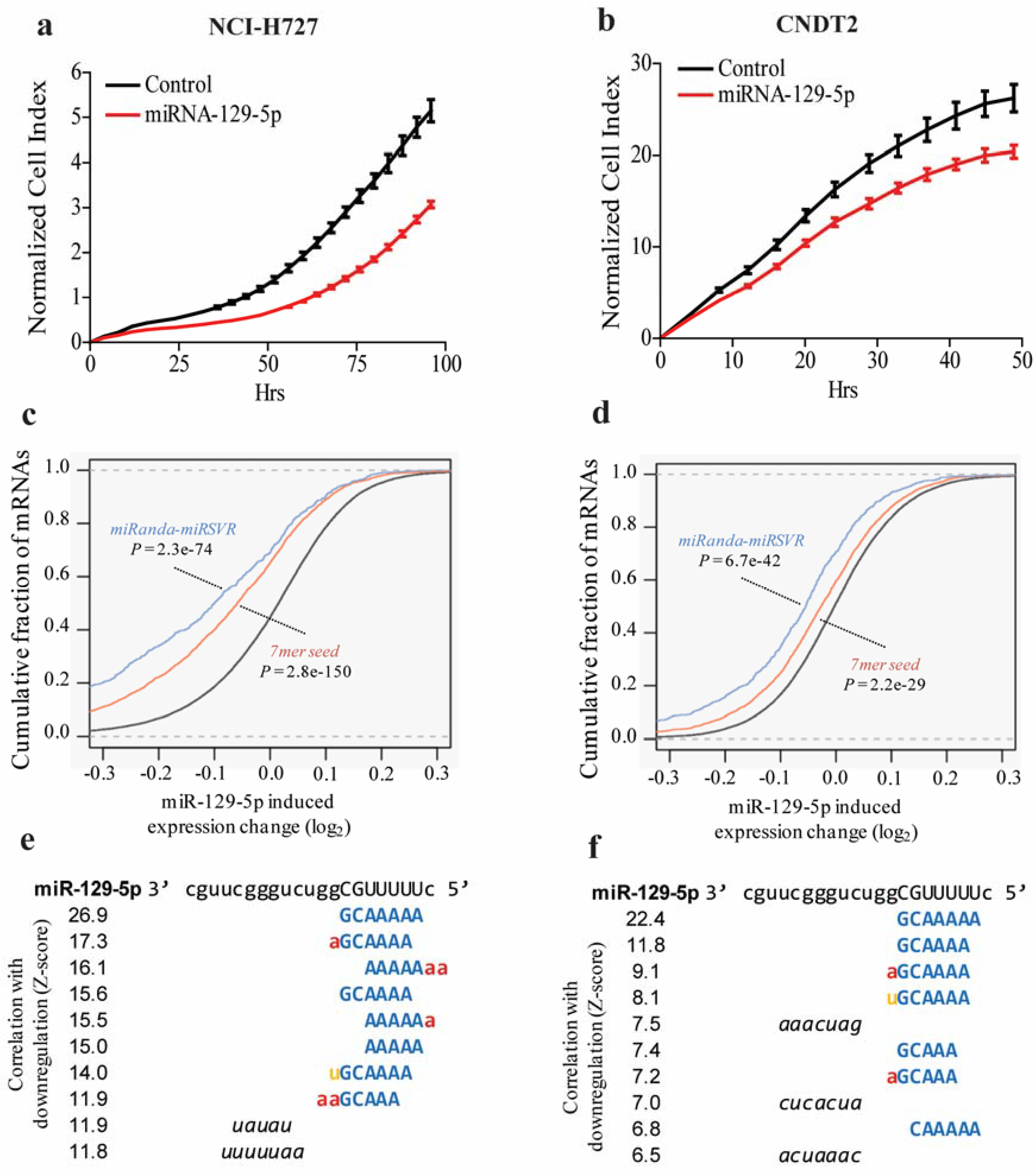
3.2. miR-129-5p Inhibits Growth of the Carcinoid Cell Lines NCI-H727 and CNDT2
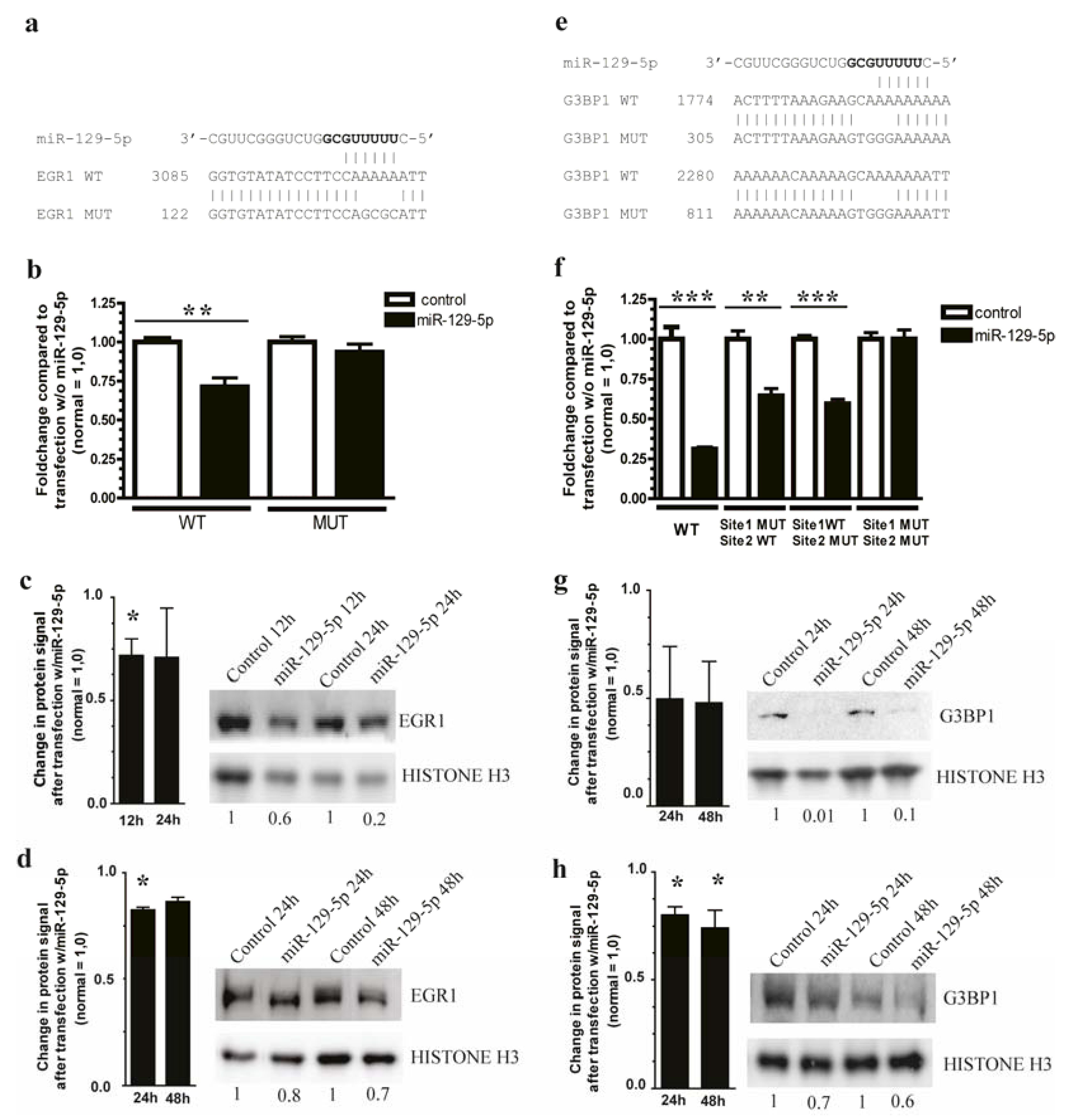
3.3. miR-129-5p Targets EGR1 and G3BP1 in Carcinoid Cell Lines NCI-H727 and CNDT2
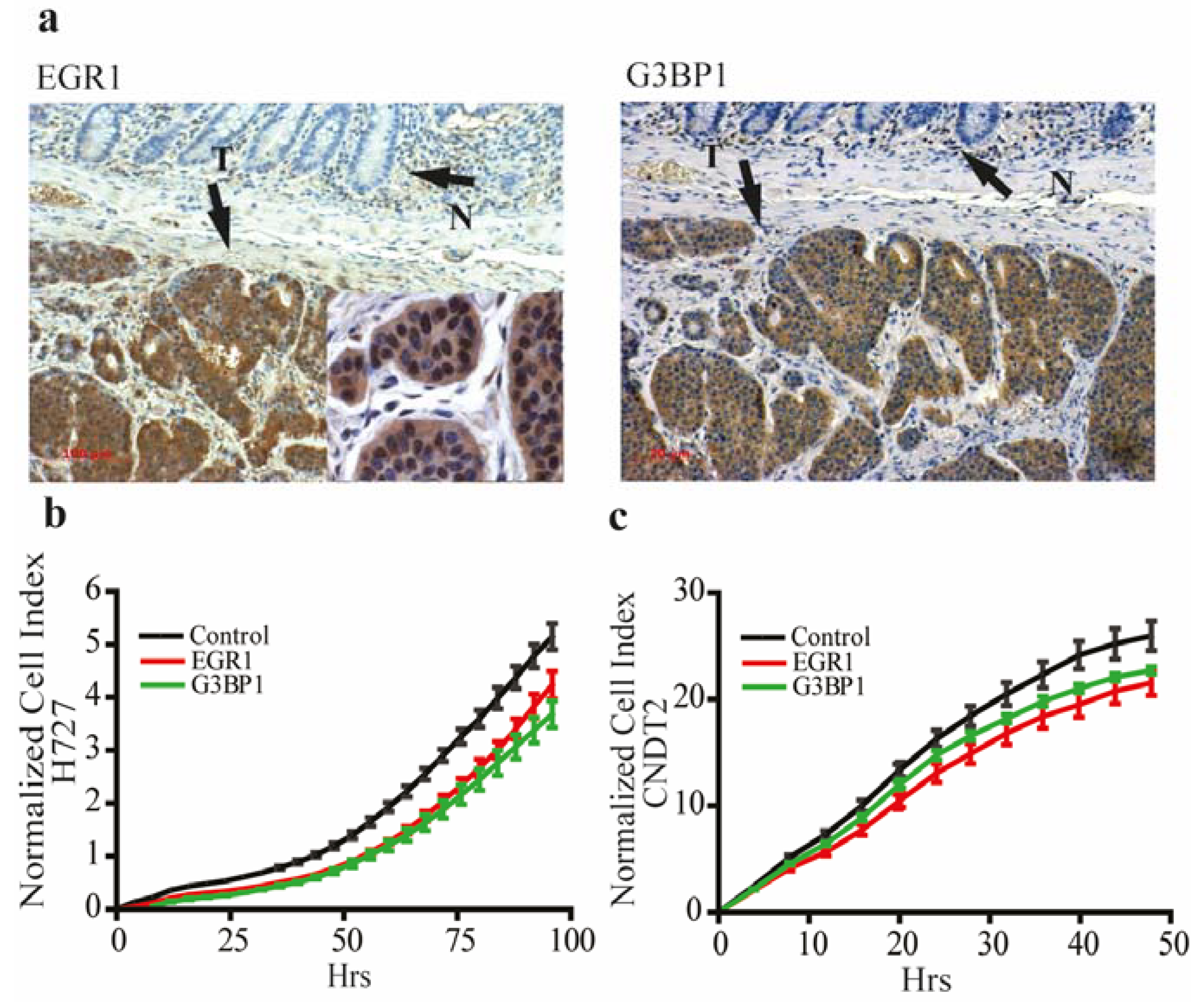
3.4. EGR1 and G3BP1 are Up-Regulated in Neuroendocrine Tumors and Promote Growth in Carcinoid Cell Lines
3.5. The let-7 Family is Involved in the Neuroendocrine Tumor Metastatic Process by Targeting HMGA2 and BACH1
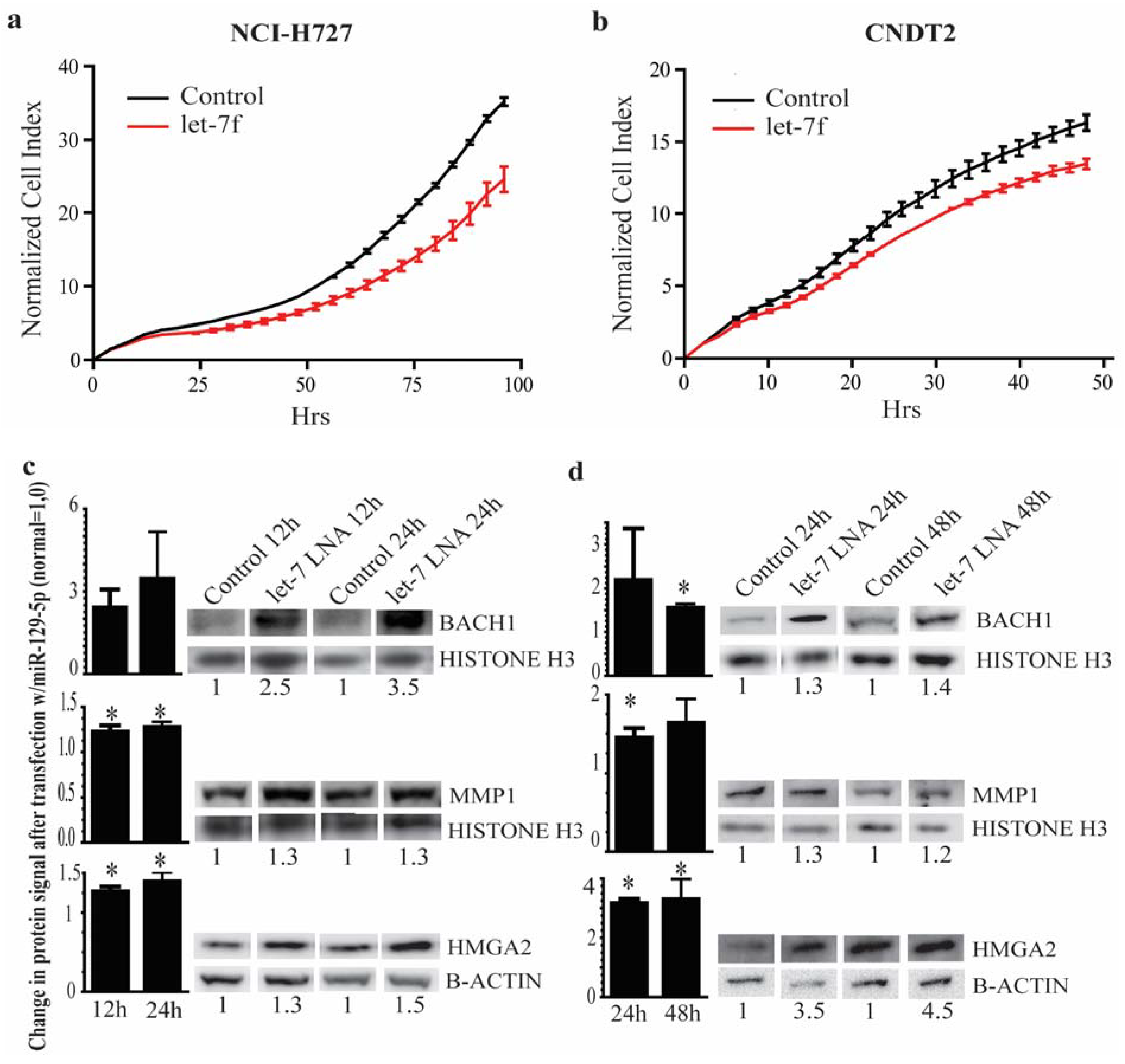
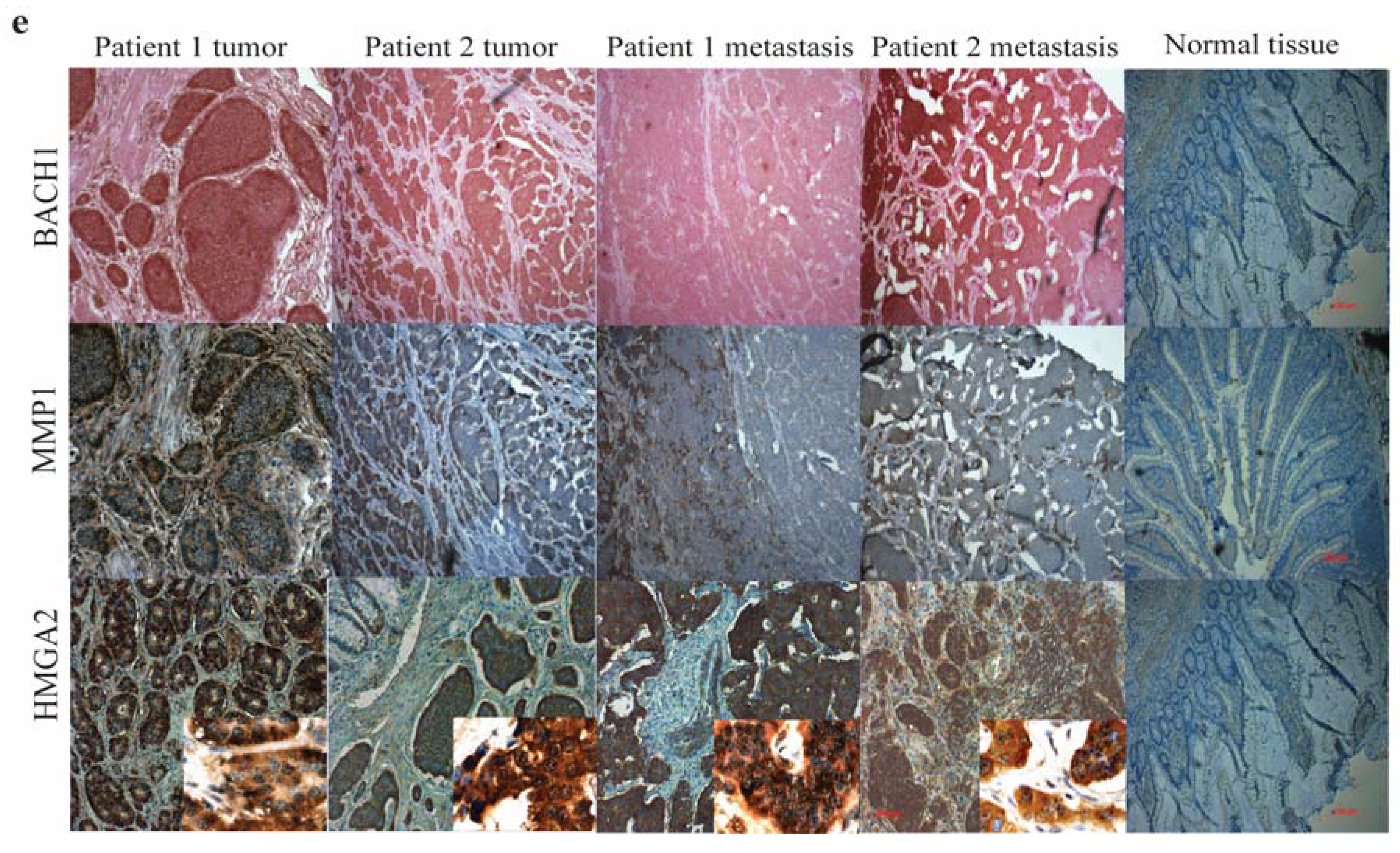
4. Discussion
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Modlin, I.M.; Oberg, K.; Chung, D.C.; Jensen, R.T.; de Herder, W.W.; Thakker, R.V.; Caplin, M.; Delle Fave, G.; Kaltsas, G.A.; Krenning, E.P.; et al. Gastroenteropancreatic neuroendocrine tumours. Lancet Oncol. 2008, 9, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Kloppel, G. Classification and pathology of gastroenteropancreatic neuroendocrine neoplasms. Endocr.-Relat. Cancer 2011, 18 (Suppl 1), S1–S16. [Google Scholar] [CrossRef]
- Lee, D.J.; Modlin, R.L. DNA transportation authority. Nat. Med. 2008, 14, 1319–1320. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Sebolt-Leopold, J.S. Advances in the development of cancer therapeutics directed against the ras-mitogen-activated protein kinase pathway. Clin. Cancer Res. 2008, 14, 3651–3656. [Google Scholar] [CrossRef] [PubMed]
- Gysin, S.; Salt, M.; Young, A.; McCormick, F. Therapeutic strategies for targeting ras proteins. Genes Cancer 2011, 2, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Grewal, T.; Koese, M.; Tebar, F.; Enrich, C. Differential regulation of rasgaps in cancer. Genes Cancer 2011, 2, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Pazman, C.; Mayes, C.A.; Fanto, M.; Haynes, S.R.; Mlodzik, M. Rasputin, the drosophila homologue of the rasgap sh3 binding protein, functions in ras- and rho-mediated signaling. Development 2000, 127, 1715–1725. [Google Scholar] [PubMed]
- Irvine, K.; Stirling, R.; Hume, D.; Kennedy, D. Rasputin, more promiscuous than ever: A review of g3bp. Int. J. Dev. Biol. 2004, 48, 1065–1077. [Google Scholar] [CrossRef] [PubMed]
- Li, W.M.; Barnes, T.; Lee, C.H. Endoribonucleases—Enzymes gaining spotlight in mRNA metabolism. FEBS J. 2010, 277, 627–641. [Google Scholar] [CrossRef] [PubMed]
- Pagel, J.I.; Deindl, E. Early growth response 1—a transcription factor in the crossfire of signal tranduction cascades. Indian J. Biochem. Biophys. 2011, 48, 226–235. [Google Scholar] [PubMed]
- Arvidsson, Y.; Bergstrom, A.; Arvidsson, L.; Kristiansson, E.; Ahlman, H.; Nilsson, O. Hypoxia stimulates cxcr4 signalling in ileal carcinoids. Endoc.-Relat. Cancer 2010, 17, 303–316. [Google Scholar] [CrossRef]
- Dangi-Garimella, S.; Yun, J.; Eves, E.M.; Newman, M.; Erkeland, S.J.; Hammond, S.M.; Minn, A.J.; Rosner, M.R. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving lin28 and let-7. EMBO J. 2009, 28, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.; Frankenberger, C.A.; Kuo, W.L.; Boelens, M.C.; Eves, E.M.; Cheng, N.; Liang, H.; Li, W.H.; Ishwaran, H.; Minn, A.J.; et al. Signalling pathway for rkip and let-7 regulates and predicts metastatic breast cancer. EMBO J. 2011, 30, 4500–4514. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Slack, F.J. MicroRNAs: Small molecules with big roles—C. Elegans to human cancer. Biol. Cell 2008, 100, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, W.C. Oncomirs: The discovery and progress of microRNAs in cancers. Mol. Cancer 2007, 6, e60. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—MicroRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Van Buren, G., 2nd; Rashid, A.; Yang, A.D.; Abdalla, E.K.; Gray, M.J.; Liu, W.; Somcio, R.; Fan, F.; Camp, E.R.; Yao, J.C.; et al. The development and characterization of a human midgut carcinoid cell line. Clin. Cancer Res. 2007, 13, 4704–4712. [Google Scholar]
- Peltier, H.J.; Latham, G.J. Normalization of microRNA expression levels in quantitative rt-pcr assays: Identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA 2008, 14, 844–852. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, S.; Baker, A.; Moller, S.; Nielsen, B.S. Robust one-day in situ hybridization protocol for detection of microRNAs in paraffin samples using lna probes. Methods 2010, 52, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Kustermann, S.; Boess, F.; Buness, A.; Schmitz, M.; Watzele, M.; Weiser, T.; Singer, T.; Suter, L.; Roth, A.B. A label-free, impedance-based real time assay to identify drug-induced toxicities and differentiate cytostatic from cytotoxic effects. Toxicol. In Vitro 2012, 27, 1589–1595. [Google Scholar] [CrossRef] [PubMed]
- Ke, N.; Wang, X.; Xu, X.; Abassi, Y.A. The xcelligence system for real-time and label-free monitoring of cell viability. Methods Mol. Biol. 2011, 740, 33–43. [Google Scholar] [PubMed]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, eR80. [Google Scholar] [CrossRef] [Green Version]
- Gregersen, L.H.; Jacobsen, A.B.; Frankel, L.B.; Wen, J.; Krogh, A.; Lund, A.H. MicroRNA-145 targets yes and stat1 in colon cancer cells. PLOS ONE 2010, 5, e8836. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, A.; Wen, J.; Marks, D.S.; Krogh, A. Signatures of RNA binding proteins globally coupled to effective microRNA target sites. Genome Res. 2010, 20, 1010–1019. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.W.; Wentzel, E.A.; Mendell, J.T. Cell-cell contact globally activates microRNA biogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 7016–7021. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Yasuda, T.; Suzuki, K.; Kanamori, M.; Kimura, T. Skeletal metastasis of carcinoid tumors: Two case reports and review of the literature. Oncol. Lett. 2012, 3, 1105–1108. [Google Scholar] [PubMed]
- Baron, V.; Adamson, E.D.; Calogero, A.; Ragona, G.; Mercola, D. The transcription factor egr1 is a direct regulator of multiple tumor suppressors including tgfbeta1, pten, p53, and fibronectin. Cancer Gene Ther. 2006, 13, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Grimson, A.; Farh, K.K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.C.; Hassan, M.; Phan, A.; Dagohoy, C.; Leary, C.; Mares, J.E.; Abdalla, E.K.; Fleming, J.B.; Vauthey, J.N.; Rashid, A.; et al. One hundred years after "carcinoid": Epidemiology of and prognostic factors for neuroendocrine tumors in 35,825 cases in the united states. J. Clin. Oncol. 2008, 26, 3063–3072. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, E.; Frankenberger, C.A.; Rosner, M.R. Rkip suppresses breast cancer metastasis to the bone by regulating stroma-associated genes. Int. J. Breast Cancer 2012, 2012, e124704. [Google Scholar] [CrossRef]
- Ruebel, K.; Leontovich, A.A.; Stilling, G.A.; Zhang, S.; Righi, A.; Jin, L.; Lloyd, R.V. MicroRNA expression in ileal carcinoid tumors: Downregulation of microRNA-133a with tumor progression. Modern Pathol. 2010, 23, 367–375. [Google Scholar] [CrossRef]
- Wang, B.; Howel, P.; Bruheim, S.; Ju, J.; Owen, L.B.; Fodstad, O.; Xi, Y. Systematic evaluation of three microRNA profiling platforms: Microarray, beads array, and quantitative real-time PCR array. PLOS ONE 2011, 6, e17167. [Google Scholar] [CrossRef] [PubMed]
- Camarillo, C.; Swerdel, M.; Hart, R.P. Comparison of microarray and quantitative real-time PCR methods for measuring microRNA levels in msc cultures. Methods Mol. Biol. 2011, 698, 419–429. [Google Scholar] [PubMed]
- Karaayvaz, M.; Zhai, H.; Ju, J. Mir-129 promotes apoptosis and enhances chemosensitivity to 5-fluorouracil in colorectal cancer. Cell Death Dis. 2013, 4, e659. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.W.; Wu, C.W.; Hu, L.Y.; Li, S.C.; Liao, Y.L.; Lai, C.H.; Kao, H.W.; Fang, W.L.; Huang, K.H.; Chan, W.C.; et al. Epigenetic regulation of mir-34b and mir-129 expression in gastric cancer. Int. J. Cancer 2011, 129, 2600–2610. [Google Scholar] [CrossRef]
- Dyrskjot, L.; Ostenfeld, M.S.; Bramsen, J.B.; Silahtaroglu, A.N.; Lamy, P.; Ramanathan, R.; Fristrup, N.; Jensen, J.L.; Andersen, C.L.; Zieger, K.; et al. Genomic profiling of microRNAs in bladder cancer: Mir-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009, 69, 4851–4860. [Google Scholar] [CrossRef]
- Bandres, E.; Agirre, X.; Bitarte, N.; Ramirez, N.; Zarate, R.; Roman-Gomez, J.; Prosper, F.; Garcia-Foncillas, J. Epigenetic regulation of microRNA expression in colorectal cancer. Int. J. Cancer 2009, 125, 2737–2743. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Qian, J.; Li, C.; Kwok, L.; Cheng, F.; Liu, P.; Perdomo, C.; Kotton, D.; Vaziri, C.; Anderlind, C.; et al. Mir-129 regulates cell proliferation by downregulating cdk6 expression. Cell Cycle 2010, 9, 1809–1818. [Google Scholar] [CrossRef] [PubMed]
- Pignatelli, M.; Luna-Medina, R.; Perez-Rendon, A.; Santos, A.; Perez-Castillo, A. The transcription factor early growth response factor-1 (egr-1) promotes apoptosis of neuroblastoma cells. Biochem. J. 2003, 373, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Pu, J.; Jiang, G.; Weng, M.; He, J.; Mei, H.; Hou, X.; Tong, Q. Abnormal expression of early growth response 1 in gastric cancer: Association with tumor invasion, metastasis and heparanase transcription. Pathol. Int. 2010, 60, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Gitenay, D.; Baron, V.T. Is egr1 a potential target for prostate cancer therapy? Future Oncol. 2009, 5, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Edfeldt, K.; Bjorklund, P.; Akerstrom, G.; Westin, G.; Hellman, P.; Stalberg, P. Different gene expression profiles in metastasizing midgut carcinoid tumors. Endoc.-Relat. Cancer 2011, 18, 479–489. [Google Scholar] [CrossRef]
- Silverman, E.S.; Collins, T. Pathways of egr-1-mediated gene transcription in vascular biology. Am. J. Pathol. 1999, 154, 665–670. [Google Scholar] [CrossRef] [PubMed]
- Thiel, G.; Cibelli, G. Regulation of life and death by the zinc finger transcription factor egr-1. J. Cell. Physiol. 2002, 193, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Karhoff, D.; Sauer, S.; Schrader, J.; Arnold, R.; Fendrich, V.; Bartsch, D.K.; Horsch, D. Rap1/b-raf signaling is activated in neuroendocrine tumors of the digestive tract and raf kinase inhibition constitutes a putative therapeutic target. Neuroendocrinology 2007, 85, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Ramos, J.W. The regulation of extracellular signal-regulated kinase (erk) in mammalian cells. Int. J. Biochem. Cell Biol. 2008, 40, 2707–2719. [Google Scholar] [CrossRef] [PubMed]
- Zwang, Y.; Sas-Chen, A.; Drier, Y.; Shay, T.; Avraham, R.; Lauriola, M.; Shema, E.; Lidor-Nili, E.; Jacob-Hirsch, J.; Amariglio, N.; et al. Two phases of mitogenic signaling unveil roles for p53 and egr1 in elimination of inconsistent growth signals. Mol. Cell 2011, 42, 524–535. [Google Scholar] [CrossRef] [PubMed]
- Welin, S.; Fjallskog, M.L.; Saras, J.; Eriksson, B.; Janson, E.T. Expression of tyrosine kinase receptors in malignant midgut carcinoid tumors. Neuroendocrinology 2006, 84, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Rickman, O.B.; Vohra, P.K.; Sanyal, B.; Vrana, J.A.; Aubry, M.C.; Wigle, D.A.; Thomas, C.F., Jr. Analysis of erbb receptors in pulmonary carcinoid tumors. Clin. Cancer Res. 2009, 15, 3315–3324. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.H.; Su, Z.Y.; Chae, J.I.; Kim, D.J.; Zhu, F.; Ma, W.Y.; Bode, A.M.; Yang, C.S.; Dong, Z. Epigallocatechin gallate suppresses lung cancer cell growth through ras-gtpase-activating protein sh3 domain-binding protein 1. Cancer Prev. Res. 2010, 3, 670–679. [Google Scholar] [CrossRef]
- Zekri, L.; Chebli, K.; Tourriere, H.; Nielsen, F.C.; Hansen, T.V.; Rami, A.; Tazi, J. Control of fetal growth and neonatal survival by the rasgap-associated endoribonuclease g3bp. Mol. Cell. Biol. 2005, 25, 8703–8716. [Google Scholar] [CrossRef] [PubMed]
- Taniuchi, K.; Nishimori, I.; Hollingsworth, M.A. The N-terminal domain of g3bp enhances cell motility and invasion by posttranscriptional regulation of bart. Mol. Cancer Res. 2011, 9, 856–866. [Google Scholar] [CrossRef] [PubMed]
- French, J.; Stirling, R.; Walsh, M.; Kennedy, H.D. The expression of ras-gtpase activating protein sh3 domain-binding proteins, g3bps, in human breast cancers. Histochem. J. 2002, 34, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Wan, P.T.; Garnett, M.J.; Roe, S.M.; Lee, S.; Niculescu-Duvaz, D.; Good, V.M.; Jones, C.M.; Marshall, C.J.; Springer, C.J.; Barford, D.; et al. Mechanism of activation of the raf-erk signaling pathway by oncogenic mutations of b-raf. Cell 2004, 116, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Tannapfel, A.; Vomschloss, S.; Karhoff, D.; Markwarth, A.; Hengge, U.R.; Wittekind, C.; Arnold, R.; Horsch, D. Braf gene mutations are rare events in gastroenteropancreatic neuroendocrine tumors. Am. J. Clin. Pathol. 2005, 123, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Roush, S.; Slack, F.J. The let-7 family of micrornas. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Nimmo, R.A.; Slack, F.J. An elegant mirror: MicroRNAs in stem cells, developmental timing and cancer. Chromosoma 2009, 118, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Qian, Z.R.; Wang, E.L.; Sultana, R.; Kudo, E.; Nakasono, M.; Hayashi, T.; Kakiuchi, S.; Sano, T. Frequent overexpression of hmga1 and 2 in gastroenteropancreatic neuroendocrine tumours and its relationship to let-7 downregulation. Br. J. Cancer 2009, 100, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Lorenz, P.; Gross, G.; Ibrahim, S.; Kunz, M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008, 18, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; He, L.; Zhao, X.; Miao, Y.; Gu, Y.; Guo, C.; Xue, Z.; Dou, W.; Hu, F.; Wu, K.; et al. MicroRNA let-7f inhibits tumor invasion and metastasis by targeting myh9 in human gastric cancer. PLOS ONE 2011, 6, e18409. [Google Scholar] [CrossRef] [PubMed]
- King, C.E.; Cuatrecasas, M.; Castells, A.; Sepulveda, A.R.; Lee, J.S.; Rustgi, A.K. Lin28b promotes colon cancer progression and metastasis. Cancer Res. 2011, 71, 4260–4268. [Google Scholar] [CrossRef] [PubMed]
- Heinzelmann, J.; Henning, B.; Sanjmyatav, J.; Posorski, N.; Steiner, T.; Wunderlich, H.; Gajda, M.R.; Junker, K. Specific miRNA signatures are associated with metastasis and poor prognosis in clear cell renal cell carcinoma. World J. Urol. 2011, 29, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.R.; Asa, S.L.; Siomi, H.; Siomi, M.C.; Yoshimoto, K.; Yamada, S.; Wang, E.L.; Rahman, M.M.; Inoue, H.; Itakura, M.; et al. Overexpression of hmga2 relates to reduction of the let-7 and its relationship to clinicopathological features in pituitary adenomas. Modern Pathol. 2009, 22, 431–441. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Døssing, K.B.V.; Binderup, T.; Kaczkowski, B.; Jacobsen, A.; Rossing, M.; Winther, O.; Federspiel, B.; Knigge, U.; Kjær, A.; Friis-Hansen, L. Down-Regulation of miR-129-5p and the let-7 Family in Neuroendocrine Tumors and Metastases Leads to Up-Regulation of Their Targets Egr1, G3bp1, Hmga2 and Bach1. Genes 2015, 6, 1-21. https://doi.org/10.3390/genes6010001
Døssing KBV, Binderup T, Kaczkowski B, Jacobsen A, Rossing M, Winther O, Federspiel B, Knigge U, Kjær A, Friis-Hansen L. Down-Regulation of miR-129-5p and the let-7 Family in Neuroendocrine Tumors and Metastases Leads to Up-Regulation of Their Targets Egr1, G3bp1, Hmga2 and Bach1. Genes. 2015; 6(1):1-21. https://doi.org/10.3390/genes6010001
Chicago/Turabian StyleDøssing, Kristina B. V., Tina Binderup, Bogumil Kaczkowski, Anders Jacobsen, Maria Rossing, Ole Winther, Birgitte Federspiel, Ulrich Knigge, Andreas Kjær, and Lennart Friis-Hansen. 2015. "Down-Regulation of miR-129-5p and the let-7 Family in Neuroendocrine Tumors and Metastases Leads to Up-Regulation of Their Targets Egr1, G3bp1, Hmga2 and Bach1" Genes 6, no. 1: 1-21. https://doi.org/10.3390/genes6010001
APA StyleDøssing, K. B. V., Binderup, T., Kaczkowski, B., Jacobsen, A., Rossing, M., Winther, O., Federspiel, B., Knigge, U., Kjær, A., & Friis-Hansen, L. (2015). Down-Regulation of miR-129-5p and the let-7 Family in Neuroendocrine Tumors and Metastases Leads to Up-Regulation of Their Targets Egr1, G3bp1, Hmga2 and Bach1. Genes, 6(1), 1-21. https://doi.org/10.3390/genes6010001






