Implication of an Aldehyde Dehydrogenase Gene and a Phosphinothricin N-Acetyltransferase Gene in the Diversity of Pseudomonas cichorii Virulence
Abstract
:1. Introduction
2. Results and Discussion
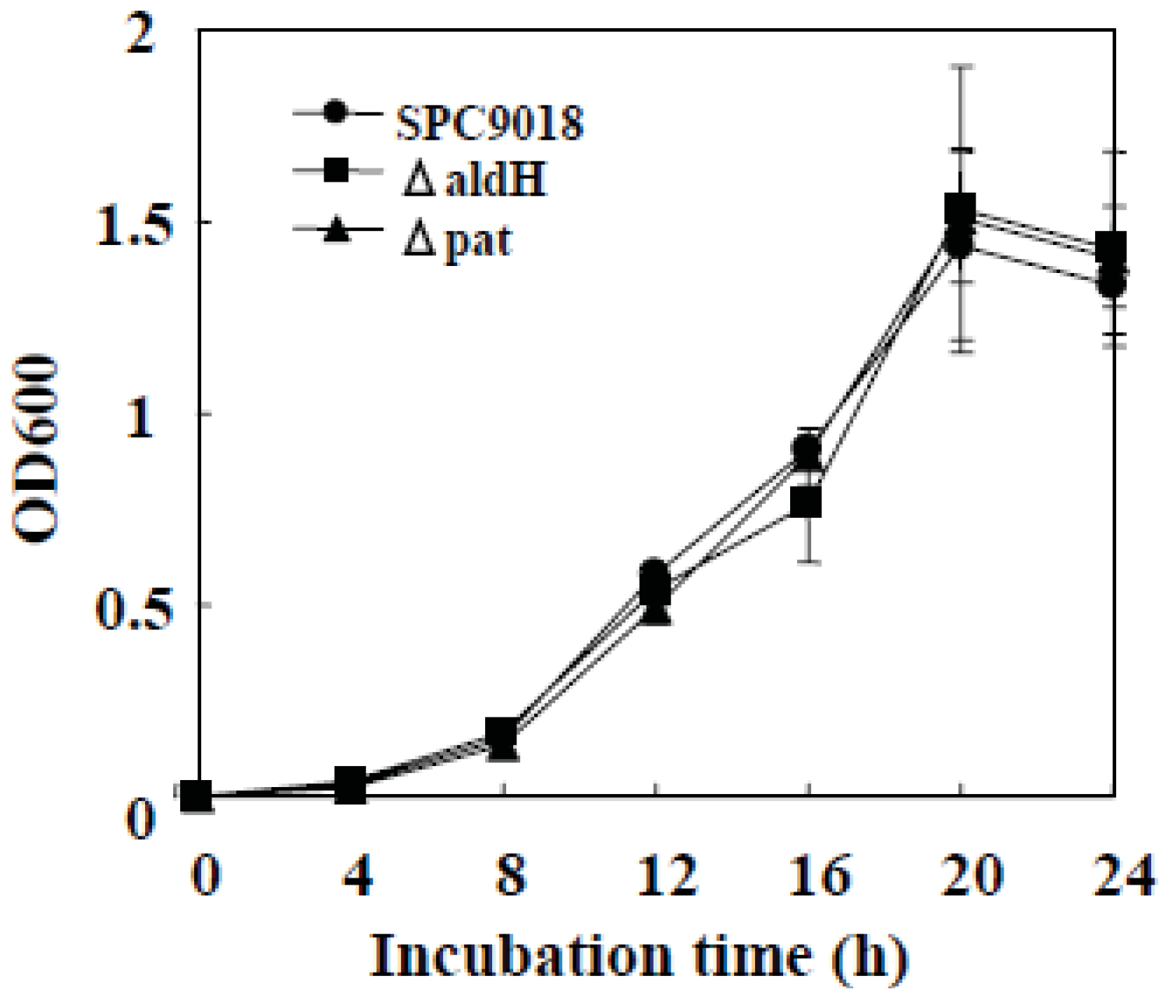
2.1. In Vitro Growth of the aldH-Deficient Mutant and the pat-Deficient Mutant of P. cichorii
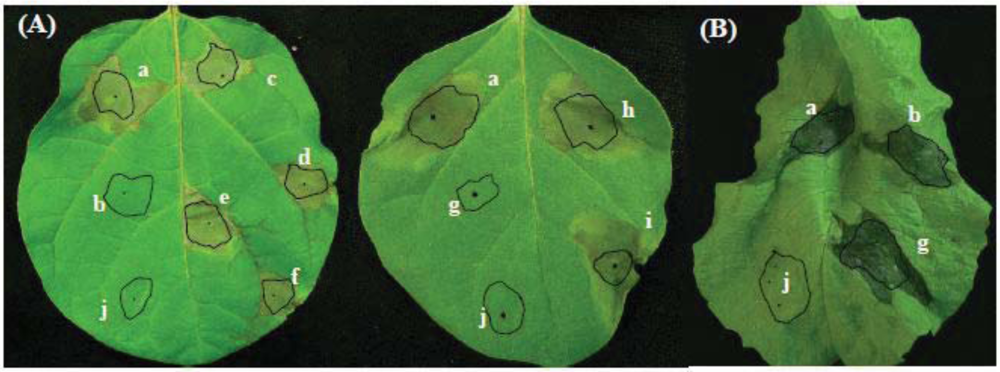
2.2. Involvement of aldH and pat in Virulence of P. cichorii on Eggplant but not on Lettuce
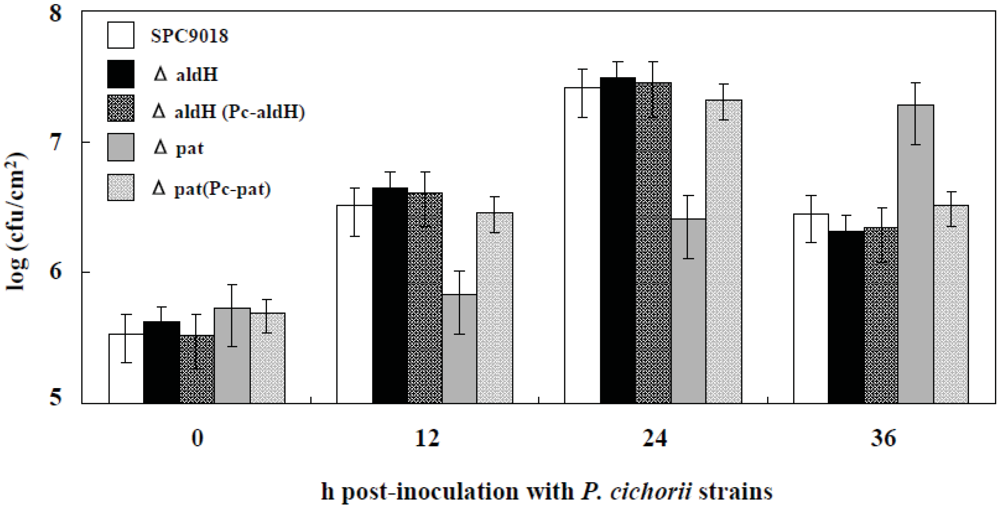
2.3. Involvement of pat, but not aldH, in Bacterial Growth in Planta
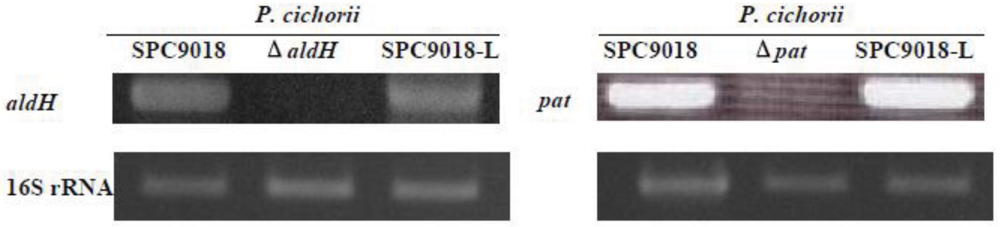
2.4. Expression of aldH and pat is not Regulated by HrpL
2.5. Phylogenetic Diversity and Functional Conservation of aldH among Pseudomonads
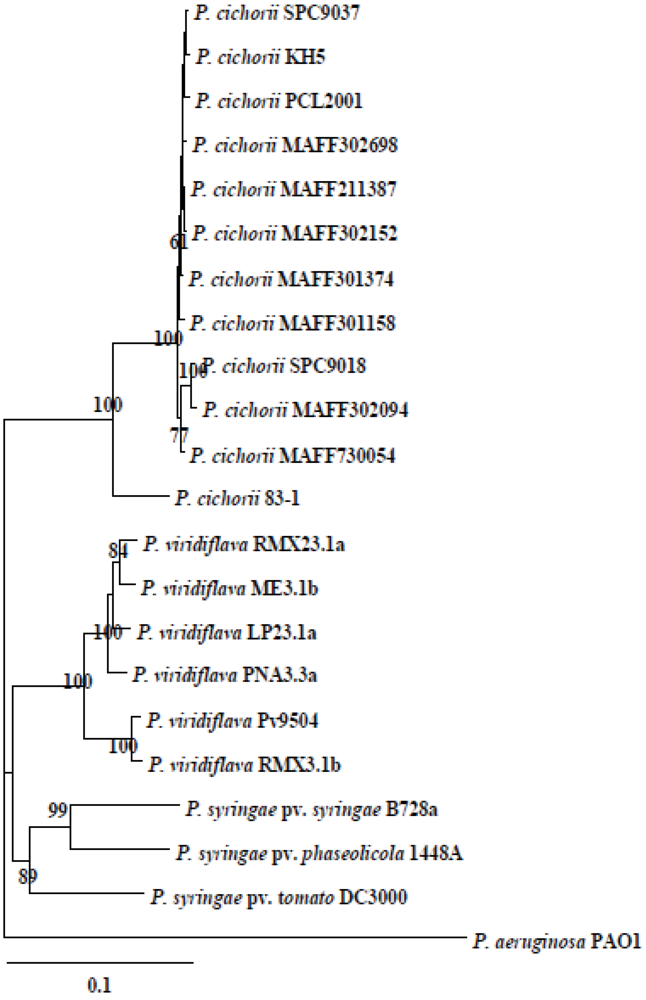
2.6. Functional Conservation of pat
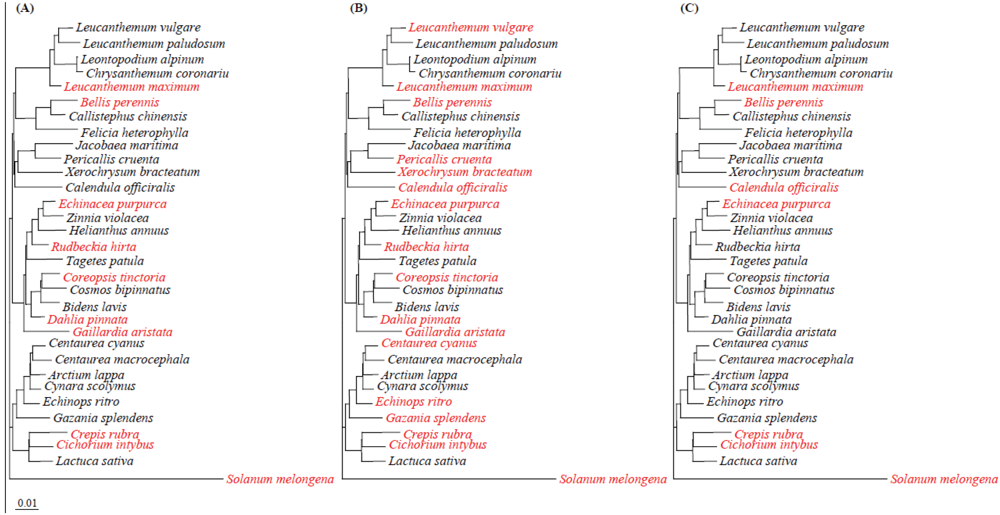
2.7. Implication of hrp, aldH and pat in SPC9018 Virulence on Asteraceae Plants
3. Experimental Section
3.1. Bacterial Strains, Plasmids and Culture Conditions
3.2. DNA Manipulations
3.3. Creation of aldH- and pat-Deleted Mutants
3.4. Creation of the hrcC-Mutant
3.5. Complementation of ΔaldH with aldH Originating from Pseudomonads
3.6. Complementation of Δpat with pat Originating from SPC9018 and Pv9504
3.7. Expression Analysis of aldH and pat in P. cichorii Strains by RT-PCR
3.8. Sequencing of aldH from P. cichorii Strains
3.9. Sequencing of ndhF and rbcL from Asteraceae Plants
3.10. Data Analysis
3.11. Virulence Assays
3.12. Bacterial Population in Planta
3.13. Nucleotide Sequence Accession Numbers
4. Conclusions
Acknowledgments
References
- Grogan, R.G.; Misaghi, I.J.; Kimble, K.A.; Greathead, A.S.; Ririe, D.; Bardin, R. Varnish spot, a destructive disease of lettuce in California caused by Pseudomonas cichorii. Phytopathology 1977, 67, 957–960. [Google Scholar]
- Hikichi, Y.; Saito, A.; Suzuki, K. Infection sites of Pseudomonas cichorii into head leaf of lettuce. Ann. Phytopathol. Soc. Jpn. 1996, 62, 125–129. [Google Scholar] [CrossRef]
- Hikichi, Y.; Saito, A.; Suzuki, K. Relationship between population dynamics of Pseudomonas cichorii on lettuce and disease incidence of bacterial rot of lettuce. Ann. Phytopathol. Soc. Jpn. 1996, 62, 141–146. [Google Scholar] [CrossRef]
- Cottyn, B.; Heylen, K.; Heyrman, J.; Vanhouteghem, K.; Pauwelyn, E.; ABleyaert, P.; van Vaerenbergh, J.; Hofte, M.; de Vos, P.; Maes, M. Pseudomonas cichorii as the causal agent of midrib rot, anemerging disease of greenhouse-grown butterhead lettuce in Flanders. Syst. Appl. Microbiol. 2009, 32, 211–225. [Google Scholar] [CrossRef]
- Kiba, A.; Sangawa, Y.; Ohnishi, K.; Yao, N.; Park, P.; Nakayashiki, H.; Tosa, Y.; Mayama, S.; Hikichi, Y. Induction of apoptotic cell death leads to the development of bacterial rot caused by Pseudomonas cichorii. Mol. Plant Microbe Interact. 2006, 19, 112–122. [Google Scholar] [CrossRef]
- Hojo, H.; Koyanagi, M.; Tanaka, M.; Kajihara, S.; Ohnishi, K.; Kiba, A.; Hikichi, Y. The hrp genes of Pseudomonas cichorii are essential for pathogenicity on aubergine plants but not on lettuce. Microbiology 2008, 154, 2920–2928. [Google Scholar] [CrossRef]
- Kiba, A.; Takata, O.; Ohnishi, K.; Hikichi, Y. Comparative analysis of induction pattern of programmed cell death and defense-related responses during hypersensitive cell death and development of bacterial necrotic leaf spots in aubergine plants. Planta 2006, 224, 981–994. [Google Scholar]
- Alfano, J.R.; Collmer, A. The type III (Hrp) secretion pathway of plant pathogenic bacteria: Trafficking harpins, avr proteins, and death. J. Bacteriol. 1997, 179, 5655–5662. [Google Scholar]
- Bogdanove, A.J.; Beer, S.V.; Bonas, U.; Boucher, C.A.; Collmer, A.; Coplin, D.L.; Cornelis, G.R.; Huang, H.C.; Hutcheson, S.W.; Panopoulos, N.J.; et al. Unified nomenclature for broadly conserved hrp genes of phytopathogenic bacteria. Mol. Microbiol. 1996, 20, 681–683. [Google Scholar]
- Araki, H.; Tian, D.; Goss, E.M.; Jakob, K.; Halldorsdottir, S.S.; Kreitman, M.; Bergelson, J. Presence/absence polymorphism for alternative pathogenicity islands in Pseudomonas viridiflava, a pathogen of Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 5887–5892. [Google Scholar]
- Buell, C.R.; Joardar, V.; Lindeberg, M.; Selengut, J.; Paulsen, I.T.; Lindeberg, M.; Selengut, J.; Paulsen, I.T.; Gwinn, M.L.; Dodson, R.J.; et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 2003, 100, 10181–10186. [Google Scholar]
- Wang, C.; Zhang, H.B.; Wang, L.H.: ; Zhang, L.H. Succinic semialdehyde couples stress response to quorum-sensing signal decay in Agrobacterium tumefaciens. Mol. Microbiol. 2006, 62, 45–56. [Google Scholar] [CrossRef]
- Parsot, C.; Mekalanos, J.J. Expression of the Vibrio cholerae gene encoding aldehyde dehydrogenase is under control of ToxR, the cholera toxin transcriptional activator. J. Bacteriol. 1991, 173, 2842–2851. [Google Scholar]
- Liang, X.; Pham, X.Q.; Olson, M.V.; Lory, S. Identification of a genomic island present in the majority of pathogenic isolates of Pseudomonas aeruginosa. J. Bacteriol. 2001, 183, 843–853. [Google Scholar] [CrossRef]
- Lutz, A.K.; Knapp, E.J.; Maliga, P. Expression of bar in the plastid genome confers herbicide resistance. Plant Physiol. 2001, 125, 1585–1590. [Google Scholar] [CrossRef]
- Thompson, J.C.; Movva, N.R.; Tizard, R.; Crameri, R.; Davies, E.J.; Lauwereys, M.; Botterman, J. Characterization of the herbicide-resistance gene bar from Streptomyces hygroscopics. EMBO J. 1987, 6, 2519–2523. [Google Scholar]
- Yun, C.S.; Hasegawa, H.; Nanamiya, H. Novel bacterial N-acetyltransferase gene for herbicide detoxification in land plants and selection marker in plant transformation. Biosci. Biotechnol. Biochem. 2009, 73, 1000–1006. [Google Scholar] [CrossRef]
- Schechter, L.M.; Roberts, K.A.; Jamir, Y.; Alfano, J.R.; Collmer, A. Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 2004, 186, 543–555. [Google Scholar]
- Xiao, Y.; Hutcheson, S.W. A single promoter sequence recognized by a newly identified alternate sigma factor directs expression of pathogenicity and host range determinants in Pseudomonas syringae. J. Bacteriol. 1994, 176, 3089–3091. [Google Scholar]
- Innes, R.W.; Bent, A.F.; Kunkel, B.N.; Bisgrove, S.R.; Staskawicz, B.J. Molecular analysis of avirulence gene avrRpt2 and identification of a putative regulatory sequence common to all known Pseudomonas syringae avirulence genes. J. Bacteriol. 1993, 175, 4859–4869. [Google Scholar]
- Shen, H.; Keen, N.T. Characterization of the promoter of avirulence gene D from Pseudomonas syringae pv. tomato. J. Bacteriol. 1993, 175, 5916–5924. [Google Scholar]
- ClustalW. Available online: http://clustalw.ddbj.nig.ac.jp/top-j.html/ (accessed on 28 October 2011).
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Hacker, J.; Carniel, E. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2001, 2, 376–381. [Google Scholar]
- Ochman, H. Lateral and oblique gene transfer. Curr. Opin. Genet. Dev. 2001, 11, 616–619. [Google Scholar]
- Gogarten, J.P.; Doolittle, W.F.; Lawrence, J.G. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 2002, 19, 2226–2238. [Google Scholar]
- Lan, R.; Reeves, P.R. When does a clone deserve a name? A perspective on bacterial species based on population genetics. Trends Microbiol. 2001, 9, 419–424. [Google Scholar]
- Nakamura, Y.; Itoh, T.; Matsuda, H.; Gojobori, T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat. Genet. 2004, 36, 760–766. [Google Scholar]
- Hacker, J.; Kaper, J.B. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 2000, 54, 641–679. [Google Scholar]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Uematsu, T.; Takatsu, A.; Ohata, K. A medium for the selective isolation of Pseudomonas cichorii. Ann. Phytopathol. Soc. Jpn. 1982, 48, 425–432. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed; Cold Spring Habor Laboratory Press: Cold Spring Habor, NY, USA, 1989. [Google Scholar]
- Tsuge, S.; Furutani, A.; Fukunaka, R.; Kubo, Y.; Horino, O. Growth complementation of hrpXo mutants of Xanthomonas oryzae pv. oryzae by virulent strains in rice cultivars resistant and susceptible to the parental strain. J. Gen. Plant Pathol. 2001, 67, 51–57. [Google Scholar] [CrossRef]
- Gay, P.; Le Coq, D.; Steinmetz, M.; Berkelman, T.; Kado, C.I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 1985, 164, 918–921. [Google Scholar]
- Kanda, A.; Yasukohchi, M.; Ohnishi, K.; Kiba, A.; Okuno, T.; Hikichi, Y. Ectopic expression of Ralstonia solanacearum effector protein PopA early in invasion results in loss of virulence. Mol. Plant Microbe Interact. 2003, 16, 447–455. [Google Scholar] [CrossRef]
- Staskawicz, B.; Dahlbeck, D.; Keen, N.; Napoli, C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 1987, 169, 5789–5794. [Google Scholar]
- de Feyter, R.; Gabriel, D.W. Use of cloned DNA methylase genes to increase the frequency of transfer of foreign genes into Xanthomonas campestris pv. malvacearum. J. Bacteriol. 1991, 173, 6421–6427. [Google Scholar]
- Sukchawalit, R.; Vattanaviboon, P.; Sallabhan, R.; Mongkolsuk, S. Construction and characterization of regulated L-arabinose-inducible broad host range expression vectors in Xanthomonas. FEMS Microbiol. Lett. 1999, 181, 217–223. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 1111–1120. [Google Scholar]
Supplementary Material
1. Construction of Plasmids for aldH and pat-Deletion Mutants

2. Construction of Plasmids for the hrcC-Mutant
3. Construction of Plasmids for Complementation of ΔaldH with aldH Originating from Pseudomonads
4. Construction of Plasmids for Complementation of Δpat with pat Originating from SPC9018 and Pv9504
| Bacteria | DDBJ Accession Number | Reference or Source |
|---|---|---|
| P. cichorii | ||
| SPC9018 | AB433910 | [6] |
| MAFF730054 | AB530808 | This study |
| SPC9037 | AB530809 | This study |
| PCL2001 | AB530810 | This study |
| MAFF211387 | AB530811 | This study |
| MAFF301158 | AB530812 | This study |
| MAFF301374 | AB530813 | This study |
| MAFF302094 | AB530814 | This study |
| MAFF302698 | AB530815 | This study |
| KH5 | AB530816 | This study |
| MAFF302152 | AB530817 | This study |
| P. viridiflava | ||
| Pv9504 | AB530818 | [6] |
| Plant Species | P. cichorii Strains | DDBJ Accession Number | |||||
|---|---|---|---|---|---|---|---|
| SPC9018 | SPC9018-hrcC | ΔaldH | Δpat | ndhF | rbcL | ||
| Bidens laevis | V | V | V | V | AB530917 | AB530951 | |
| Bellis perennis | V | NV | NV | NV | AB530918 | AB530952 | |
| Calendula officinalis | V | V | NV | NV | AB530919 | AB530953 | |
| Callistephus chinensis | V | V | V | V | AB530920 | AB530954 | |
| Centaurea cyanus | V | V | NV | V | AB530921 | AB530955 | |
| Centaurea macrocephala | V | V | V | V | AB530922 | AB530956 | |
| Leucanthemum vulgare | V | V | NV | V | AB530923 | AB530957 | |
| Leucanthemum maximum | V | NV | NV | NV | AB530924 | AB530958 | |
| Coreopsis tinctoria | V | NV | NV | V | AB530925 | AB530959 | |
| Cosmos bipinnatus | V | V | V | V | AB530926 | AB530960 | |
| Crepis rubra | V | NV | NV | NV | AB530927 | AB530961 | |
| Dahlia pinnata | V | NV | NV | V | AB530928 | AB530962 | |
| Echinacea purpurea | V | NV | NV | NV | AB530929 | AB530963 | |
| Echinops ritro | V | V | NV | V | AB530930 | AB530964 | |
| Felicia heterophylla | V | V | V | V | AB530931 | AB530965 | |
| Gaillardia aristata | V | NV | NV | V | AB530932 | AB530966 | |
| Gazania splendens | V | V | NV | V | AB530933 | AB530967 | |
| Helianthus annuus | V | V | V | V | AB530934 | AB530968 | |
| Xerochrysum bracteatum | V | V | NV | V | AB530935 | AB530969 | |
| Leucanthemum paludosum | V | V | V | V | AB530937 | AB530971 | |
| Leontopodium alpinum | V | V | V | V | AB530938 | AB530972 | |
| Rudbeckia hirta | V | NV | NV | V | AB530939 | AB530973 | |
| Jacobaea maritima | V | V | V | V | AB530940 | AB530974 | |
| Pericallis cruenta | V | V | NV | V | AB530941 | AB530975 | |
| Tagetes patula | V | V | V | V | AB530942 | AB530976 | |
| Zinnia violacea | V | V | V | V | AB530943 | AB530977 | |
| Arctium lappa | V | V | V | V | AB530944 | AB530978 | |
| Chrysanthemum coronarium | V | V | V | V | AB530945 | AB530979 | |
| Cichorium intybus | V | NV | NV | NV | AB530946 | AB530980 | |
| Cynara scolymus | V | V | V | V | AB530947 | AB530981 | |
| Lactuca sativa | V | V | V | V | AB530948 | AB530982 | |
| Relevant Characteristics | Ref. or Source | |
|---|---|---|
| E. coli | ||
| DH5α | recA1 endA1 gyrA96 thi-1 hsdR17supE44, Δ(lac) U169 (φ80lacΔM15) | Takara |
| DH5α-aldH | Transformant of DH5α with pUC118-aldH, Apr | This study |
| P. cichorii | ||
| SPC9018 | Wild-type | [5] |
| ΔaldH | aldH-deleted mutant of SPC9018, Kmr | This study |
| ΔaldH (Pc-aldH) | Transformant of ΔaldH with pPc-aldH, Kmr, Cmr | This study |
| ΔaldH (Pv-aldH) | Transformant of ΔaldH with pPv-aldH, Kmr, Tcr | This study |
| ΔaldH (Pst-aldH) | Transformant of ΔaldH with pPst-aldH, Kmr, Cmr | This study |
| ΔaldH (Pa-aldH) | Transformant of ΔaldH with pPa-aldH, Kmr, Cmr | This study |
| Δpat | pat-deleted mutant of SPC9018, Kmr | This study |
| Δpat (Pc-pat) | Transformant of Δpat with pPc-pat, Kmr, Cmr | This study |
| Δpat (Pv-pat) | Transformant of Δpat with pPv-pat, Kmr, Cmr | This study |
| SPC9018-hrcC | hrcC-deleted mutant of SPC9018, Kmr | This study |
| SPC9018-L | hrpL-deleted mutant of SPC9018, Kmr | [6] |
| P. viridiflava | ||
| Pv9504 | BS type | [6] |
| P. syringae pv. tomato | ||
| DC3000 | [11] | |
| P. aeruginosa | ||
| PAO1 | [13] | |
| Plasmid | ||
| pUC118 | Ampr | Takara |
| pHSG398 | Cmr | Takara |
| pLAFR3 | pLAFR1 containing HaeII fragment of pUC8 | [36] |
| pUFR043 | Cosmid derivative of pUFRO42, IncW, Mob’ lacZα, Gmr, Kmr | [37] |
| pBbad22K | Derivative of pBAD22, mob, rep, araC, Kmr | [38] |
| pUCK191 | pUC18 derivative containing Kmr from Tn903 | [33] |
| pUCD800 | pUCD5 derivative containing sacB, Kmr | [34] |
| phrpFoperon | A 3.6 kb PCR fragment containing the hrpF operon from SPC9018 genomic DNA | [6] |
| pUC118-cml | A 1.1 kb Sau3AI-digested pHSG398 in pUC118, Apr, Cmr | This study |
| pPc-aldH | A 2.0 kb PCR-fragment containing aldH of SPC9018 and SacI-digested 1.1 kb fragment of pUC118-cml in pUFR043 | This study |
| pPv-aldH | A 2.8 kb PCR-fragment containing aldH of Pv9504 in pLAFR3 | This study |
| pPst-aldH | A 2.0 kb PCR-fragment containing aldH of DC3000 and SacI-digested 1.1 kbp fragment of pUC118-cml in pUFR043 | This study |
| pPa-aldH | A 2.4 kb PCR-fragment containing aldH of PAO1 and KpnI-digested 1.1 kb fragment of pUC118-cml in pBbad22K | This study |
| pPc-pat | A 1.5 kb PCR-fragment containing pat of SPC9018 and SacI-digested 1.1 kb fragment of pUC118-cml in pUFR043 | This study |
| pPv-pat | A 1.2 kb PCR-fragment containing pat of Pv9504 and SacI-digested 1.1 kb fragment of pUC118-cml in pUFR043 | This study |
| Name | Sequence a |
|---|---|
| Kpn-D4-1-Fw | 5'-GGGGTACCACAGTTTTGTCCCTAAACCCG-3' |
| Bam-D4-1-Rv | 5'-CGGGATCCGGCGTCCACAAAAAAGAGCG-3' |
| Bam-D4-2-Fw | 5'-CGGGATCCTCACATCGGTATCTCCTGTTG-3' |
| Sal-D4-2-Rv | 5'-GCGTCGACGCTATGATCATTCATCCTCAGC-3' |
| Bam-Km1 | 5'-CGGGATCCGGTACCCCCCCGCGCCTGATGC-3' |
| Bam-Km2 | 5'-CGGGATCCGGTACCCCCCCGCGCCTGATGC-3' |
| Sal-SacB1 | 5'-CGACGCGTCGACGGATCCTTTTTAACCCATC-3' |
| Sal-SacB2 | 5'-CGACGTCGACTGCAGTTCACTTACACCGC-3' |
| delta5-1-FW-Kpn | 5'-GGGGTACCTGCCATCTGATGCTTTGAAAG-3' |
| delta5-1-RV-Bam | 5'-CGGGATCCTCACATCGCTTCGAGATCGTCTTCAG-3' |
| delata5-2-Fw-Bam | 5'-CGGGATCCTTCGCCAGCGTTGAAAAAAGGG-3' |
| delata5-2-Rv-Sal | 5'-GCGTCGACGCGATTCGTTCCTGCCGCTATC-3' |
| Kpn-Pc-aldH-Fw | 5'-GGGGTACCCCCGCATCAAACCGGTCATGG-3' |
| Kpn-Pc-aldH-Rv | 5'-GGGGTACCGTCAGACGATAGGCTGGTC-3' |
| Bam-PV4-Fw | 5'-CGGGATCCAAGCTATGATTAATCATCCAC-3' |
| Bam-PV4-Rv | 5'-CGGGATCCTCAAGCGATCGGCTGATCACTC-3' |
| Kpn-Pst-Fw | 5'-GGGGTACCCCCGCATCAAGCCGGTGATGG-3' |
| Kpn-Pst-Rv | 5'-GGGGTACCTCACGCGACAGGCTGATC-3' |
| Pa-Fw | 5'-GCTACGCGCCTGCTGCTACGGGC-3' |
| Pa-Rv | 5'-GACCGCCTACGCCGCTGCCGCAG-3' |
| Kpn-PV-ORF5-RV | 5'-GGGGTACCTCAGACAGCCTCCGATACGTG-3' |
| Bam-PV-ORF5-FW | 5'-CGGGATCCGGTGGCATCACAACTGCGTATC-3' |
| SEMI-Back | 5'-CTCACCGTTGACCAGACGC-3' |
| SEMI-Front | 5'-GTCCAGCACTTGCTGGAGC-3' |
| ORF5-RT-Fw | 5'-GGGGCCAACTCGCCGGTTAC-3' |
| aldH-Fw1 | 5'-GCGATTCGTTCCTGCCGCTATC-3' |
| aldH-Rv | 5'-CCGCTCTTTTTTGTGGACGCCGG-3' |
| 16S-rRNA-Rv | 5'-AAATTCCACCACCCTCTGC-3' |
| 16S-rRNA-FwndhF-11FW | 5'-GCCTAGGTCGGATTAGCTAG-3'5'-GGGYTGGGACTTCTTCTTTTYCC-3' |
| ndhF-22RV | 5'-CCSCCKACYSATTTAATAACC-3' |
| 1-1 | 5'-ATGTCACCACAAACAGAGACTAAAGC-3' |
| NN3-2 | 5'-GCAGCAGCTAGTTCCGGGCTCCA-3' |
| ndhF-11RV | 5'-TAGGYGAATACAACCAACTATC-3' |
| ndhF-22FW | 5'-TTGCYTGTTTTTGGTCNAAAGATG-3' |
| 1-2FW | 5'-CAGTACTTCCATGTTGG-3' |
| 1-2RV | 5'-TATCCAACAAGAGTTTCC-3' |
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tanaka, M.; Wali, U.M.; Nakayashiki, H.; Fukuda, T.; Mizumoto, H.; Ohnishi, K.; Kiba, A.; Hikichi, Y. Implication of an Aldehyde Dehydrogenase Gene and a Phosphinothricin N-Acetyltransferase Gene in the Diversity of Pseudomonas cichorii Virulence. Genes 2012, 3, 62-80. https://doi.org/10.3390/genes3010062
Tanaka M, Wali UM, Nakayashiki H, Fukuda T, Mizumoto H, Ohnishi K, Kiba A, Hikichi Y. Implication of an Aldehyde Dehydrogenase Gene and a Phosphinothricin N-Acetyltransferase Gene in the Diversity of Pseudomonas cichorii Virulence. Genes. 2012; 3(1):62-80. https://doi.org/10.3390/genes3010062
Chicago/Turabian StyleTanaka, Masayuki, Ullah Md Wali, Hitoshi Nakayashiki, Tatsuya Fukuda, Hiroyuki Mizumoto, Kouhei Ohnishi, Akinori Kiba, and Yasufumi Hikichi. 2012. "Implication of an Aldehyde Dehydrogenase Gene and a Phosphinothricin N-Acetyltransferase Gene in the Diversity of Pseudomonas cichorii Virulence" Genes 3, no. 1: 62-80. https://doi.org/10.3390/genes3010062
APA StyleTanaka, M., Wali, U. M., Nakayashiki, H., Fukuda, T., Mizumoto, H., Ohnishi, K., Kiba, A., & Hikichi, Y. (2012). Implication of an Aldehyde Dehydrogenase Gene and a Phosphinothricin N-Acetyltransferase Gene in the Diversity of Pseudomonas cichorii Virulence. Genes, 3(1), 62-80. https://doi.org/10.3390/genes3010062




