Novel Wx Gene Functional Markers for High-Resistant Starch Rice Breeding
Abstract
1. Introduction
2. Materialsand Methods
2.1. Plant Materials
2.2. Determination of Apparent Amylose and RS Contents
2.3. DNA Extraction from Rice Leaves
2.4. Sequence Comparison, Sequencing and Primer Design
2.5. PCR Amplification and Recovery of the Wx Allele Fragment
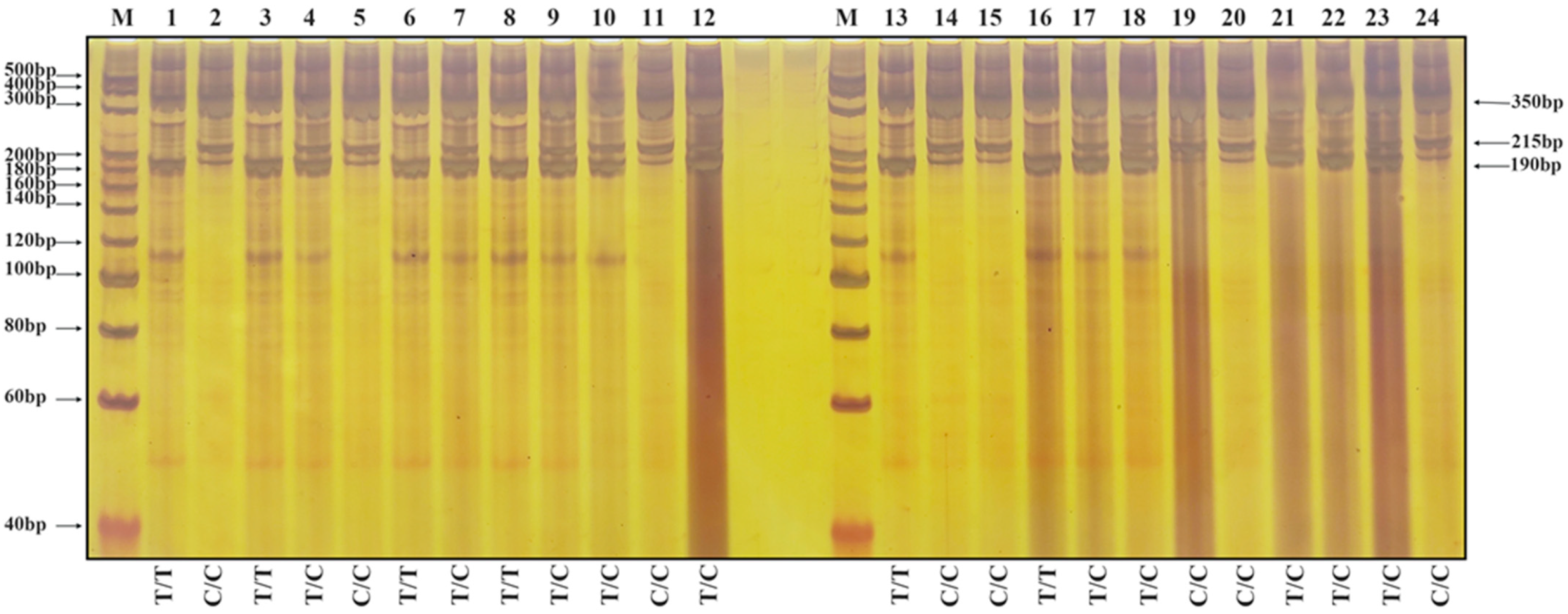
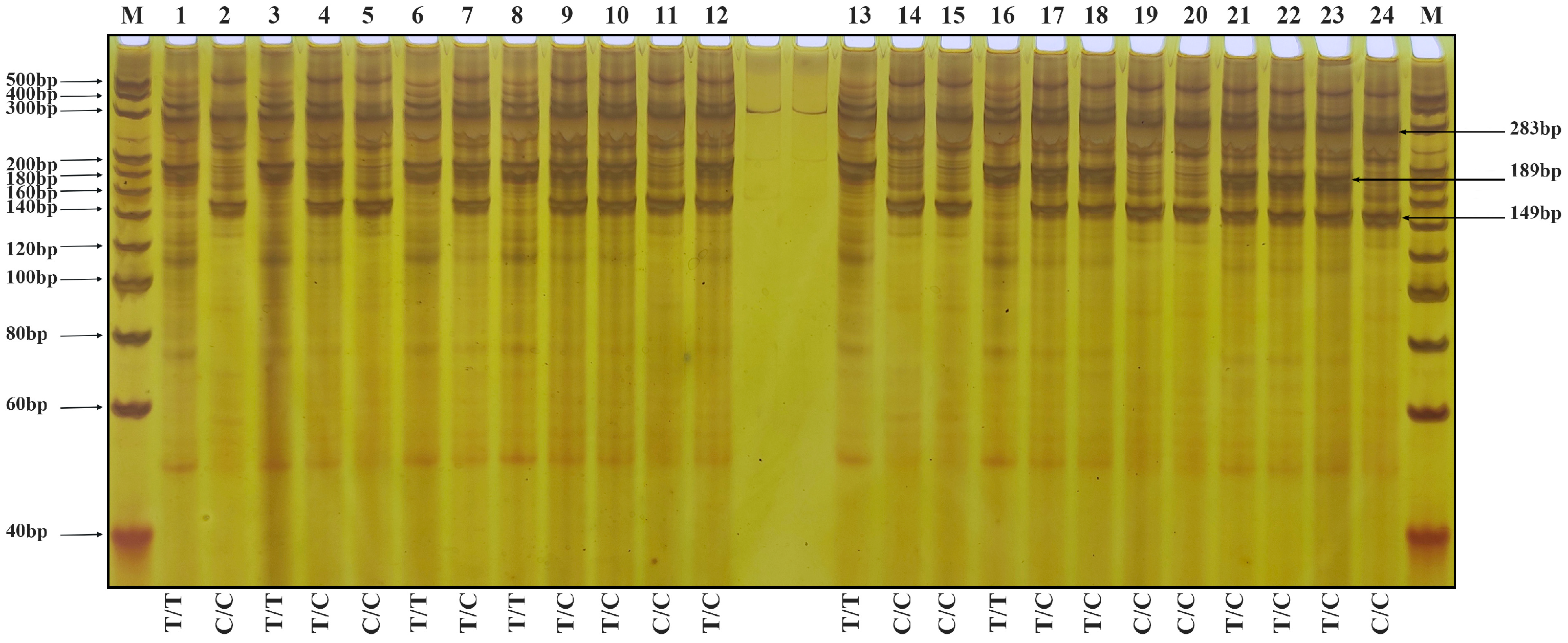
2.6. Genotype Identification
3. Results
3.1. AAC and RS Contents
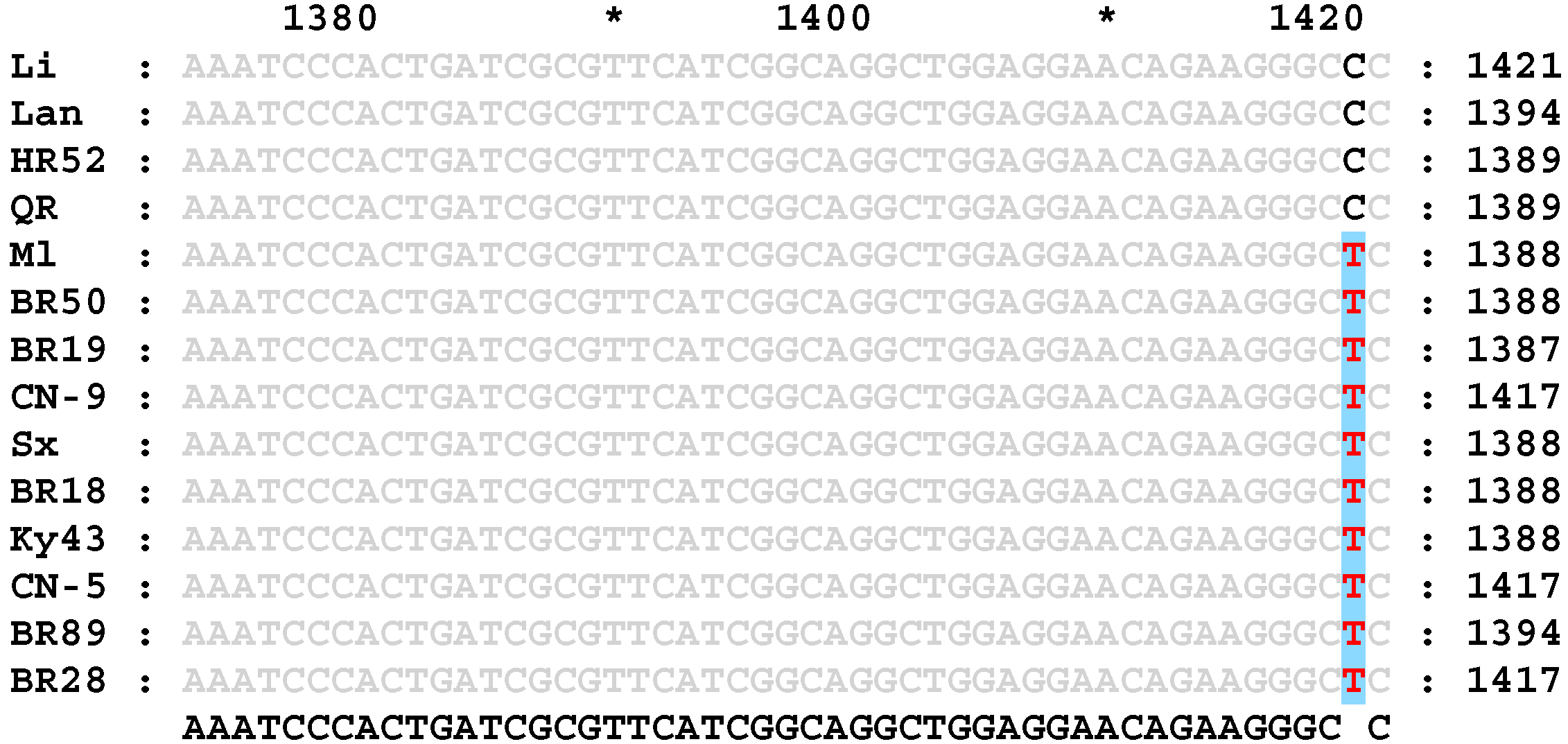
3.2. Identification and Primer Design of Specific SNPs in the Wx Gene of High-RS Rice Varieties
3.3. Validation of Molecular Markers
4. Discussion
4.1. Core Findings and Marker Utility
4.2. Interpretation of the Synonymous Mutation and Its Potential Mechanisms
4.3. Marker Development, Application, and Technical Considerations
4.4. Limitations, Generalizability, and Future Perspectives
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ARMS-PCR | Amplification refractory mutation system polymerase chain reaction |
| RS | Resistant starch |
| GBSSI | Granule-bound starch synthase I |
| GT | Gelatinization temperature |
| ECQ | Eating and cooking quality |
| FMs | Functional markers |
| MAS | Marker-assisted selection |
| AAC | Apparent amylose content |
| LD | Linkage Disequilibrium |
References
- Praphasanobol, P.; Purnama, P.R.; Junbuathong, S.; Chotechuen, S.; Moung-Ngam, P.; Kasettranan, W.; Paliyavuth, C.; Comai, L.; Pongpanich, M.; Buaboocha, T.; et al. Genome-Wide Association Study of Starch Properties in Local Thai Rice. Plants 2023, 12, 3290. [Google Scholar] [CrossRef]
- Yan, C.; Meng, H.; Pei, Y.; Sun, W.; Zhang, J. Breeding by Design for Functional Rice with Genome Editing Technologies. JoVE 2025, 215, 67336. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Liu, Y.; Deng, Z.; Liu, L.; Yu, T.; Ge, G.; Chen, B.; Wang, T. Creating of Novel Wx Allelic Variations Significantly Altering Wx Expression and Rice Eating and Cooking Quality. J. Plant Physiol. 2024, 303, 154384. [Google Scholar] [CrossRef]
- Zhu, M.; Liu, Y.; Jiao, G.; Yu, J.; Zhao, R.; Lu, A.; Zhou, W.; Cao, N.; Wu, J.; Hu, S.; et al. The Elite Eating Quality Alleles Wx(b) and ALK(b) Are Regulated by OsDOF18 and Coordinately Improve Head Rice Yield. Plant Biotechnol. J. 2024, 22, 1582–1595. [Google Scholar] [CrossRef]
- Fu, Y.; Hua, Y.; Luo, T.; Liu, C.; Zhang, B.; Zhang, X.; Liu, Y.; Zhu, Z.; Tao, Y.; Zhu, Z.; et al. Generating Waxy Rice Starch with Target Type of Amylopectin Fine Structure and Gelatinization Temperature by Waxy Gene Editing. Carbohydr. Polym. 2023, 306, 120595. [Google Scholar] [CrossRef]
- Kumar, S.; Li, Y.; Zheng, J.; Liu, J.; Xu, Q.; Zhang, Y.; Tang, H.; Qi, P.; Deng, M.; Ma, J.; et al. The Impact of GBSSI Inactivation on Starch Structure and Functionality in EMS-Induced Mutant Lines of Wheat. BMC Genom. 2025, 26, 501. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, X.; Song, J.; Xu, K.; Qin, T.; Zhang, X.; Song, Z.; He, Y.; Zhang, B.; Zhang, H.; et al. OsSPL14 and OsNF-YB9/YC8-12 Subunits Cooperate to Enhance Grain Appearance Quality by Promoting Waxy and PDIL1-1 Expression in Rice. Plant Commun. 2025, 6, 101348. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Niu, F.; Yan, P.; Wang, K.; Zhang, L.; Yan, Y.; Zhu, Y.; Dong, S.; Ma, F.; Lan, D.; et al. The Kinase OsSK41/OsGSK5 Negatively Regulates Amylose Content in Rice Endosperm by Affecting the Interaction between OsEBP89 and OsBP5. J. Integr. Plant Biol. 2023, 65, 1782–1793. [Google Scholar] [CrossRef]
- Li, J.; Zhang, C.; Luo, X.; Zhang, T.; Zhang, X.; Liu, P.; Yang, W.; Lei, Y.; Tang, S.; Kang, L.; et al. Fine Mapping of the Grain Chalkiness Quantitative Trait Locus qCGP6 Reveals the Involvement of Wx in Grain Chalkiness Formation. J. Exp. Bot. 2023, 74, 3544–3559. [Google Scholar] [CrossRef]
- Lu, Y.; Lv, D.; Zhou, L.; Yang, Y.; Hao, W.; Huang, L.; Fan, X.; Zhao, D.; Li, Q.; Zhang, C.; et al. Combined Effects of SSII-2RNAi and Different Wx Alleles on Rice Grain Transparency and Physicochemical Properties. Carbohydr. Polym. 2023, 308, 120651. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, Y.; Yu, S.; Deng, G.; Dai, G.; Bao, J. Combined Effects of BEIIb and SSIIa Alleles on Amylose Contents, Starch Fine Structures and Physicochemical Properties of Indica Rice. Foods 2022, 12, 119. [Google Scholar] [CrossRef]
- Ying, Y.; Hu, Y.; Liu, X.; Zhao, J.; Deng, B.; Zhang, Z.; Bao, J. Effects of Wx, SSIIa and FLO2 Alleles and Their Interactions on the Formation of Multi-Scale Structures of Rice Starch. Int. J. Biol. Macromol. 2025, 303, 140658. [Google Scholar] [CrossRef]
- Zhong, Q.; Jia, Q.; Yin, W.; Wang, Y.; Rao, Y.; Mao, Y. Advances in Cloning Functional Genes for Rice Yield Traits and Molecular Design Breeding in China. Front. Plant Sci. 2023, 14, 1206165. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Liu, H.; Jiang, W.; Hu, Y.; Zhou, Z.; An, X.; Miao, S.; Qin, Y.; Du, B.; Zhu, L.; et al. Large Scale Rice Germplasm Screening for Identification of Novel Brown Planthopper Resistance Sources. Mol. Breed. 2023, 43, 70. [Google Scholar] [CrossRef] [PubMed]
- Scheuermann, K.K.; Pereira, A. Development of a Molecular Marker for the Pi1 Gene Based on the Association of the SNAP Protocol with the Touch-up Gradient Amplification Method. J. Microbiol. Methods 2023, 214, 106845. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; He, N.; Yu, M.; Li, D.; Yang, D. Identification and Fine Mapping of a New Bacterial Blight Resistance Gene, Xa43(t), in Zhangpu Wild Rice (Oryza rufipogon). Plant Biol. 2023, 25, 433–439. [Google Scholar] [CrossRef]
- Thulasinathan, T.; Ayyenar, B.; Kambale, R.; Manickam, S.; Chellappan, G.; Shanmugavel, P.; Narayanan, M.B.; Swaminathan, M.; Muthurajan, R. Marker Assisted Introgression of Resistance Genes and Phenotypic Evaluation Enabled Identification of Durable and Broad-Spectrum Blast Resistance in Elite Rice Cultivar, CO 51. Genes 2023, 14, 719. [Google Scholar] [CrossRef]
- Chen, S.; Feng, A.; Wang, C.; Zhao, J.; Feng, J.; Chen, B.; Yang, J.; Wang, W.; Zhang, M.; Chen, K.; et al. Identification and Fine-Mapping of Xo2, a Novel Rice Bacterial Leaf Streak Resistance Gene. Theor. Appl. Genet. 2022, 135, 3195–3209. [Google Scholar] [CrossRef]
- Srivastava, A.; Pusuluri, M.; Balakrishnan, D.; Vattikuti, J.L.; Neelamraju, S.; Sundaram, R.M.; Mangrauthia, S.K.; Ram, T. Identification and Functional Characterization of Two Major Loci Associated with Resistance against Brown Planthoppers (Nilaparvata lugens (Stal)) Derived from Oryza nivara. Genes 2023, 14, 66. [Google Scholar] [CrossRef]
- Bhatia, S.K.; Vikal, Y.; Kaur, P.; Dhillon, G.S.; Kaur, G.; Neelam, K.; Malik, P.; Lore, J.S.; Khanna, R.; Singh, K. Introgression and Mapping of a Novel Bacterial Blight Resistance Gene Xa49(t) from Oryza rufipogon Acc. CR100098A into O. sativa. Phytopathology 2024, 114, 2412–2420. [Google Scholar] [CrossRef]
- Zhang, Y.; Lin, X.-F.; Li, L.; Piao, R.-H.; Wu, S.; Song, A.; Gao, M.; Jin, Y.-M. CRISPR/Cas9-Mediated Knockout of Bsr-D1 Enhances the Blast Resistance of Rice in Northeast China. Plant Cell Rep. 2024, 43, 100. [Google Scholar] [CrossRef]
- Bian, Z.; Cao, D.; Zou, Y.; Xie, D.; Zhuang, W.; Sun, Z.; Mou, N.; Sun, Y.; Zhang, C.; Li, Q.; et al. Genetic Dissection of Major Rice QTLs for Strong Culms and Fine Mapping of qWS5 for Breeding Application in Transplanted System. Rice 2024, 17, 43. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Shen, S.; Sun, M.; Corke, H. Analysis of Genotypic Diversity in the Starch Physicochemical Properties of Nonwaxy Rice: Apparent Amylose Content, Pasting Viscosity and Gel Texture. Starch Stärke 2006, 58, 259–267. [Google Scholar] [CrossRef]
- Chen, X.; Temnykh, S.; Xu, Y.; Cho, Y.G.; McCouch, S.R. Development of a Microsatellite Framework Map Providing Genome-Wide Coverage in Rice (Oryza sativa L.). Theor. Appl. Genet. 1997, 95, 553–567. [Google Scholar] [CrossRef]
- Ye, S.; Dhillon, S.; Ke, X.; Collins, A.R.; Day, I.N.M. An Efficient Procedure for Genotyping Single Nucleotide Polymorphisms. Nucleic Acids Res. 2001, 29, e88. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.A.; Daygon, V.D.; Resurreccion, A.P.; Cuevas, R.P.; Corpuz, H.M.; Fitzgerald, M.A. A Single Nucleotide Polymorphism in the Waxy Gene Explains a Significant Component of Gel Consistency. Theor. Appl. Genet. 2011, 123, 519–525. [Google Scholar] [CrossRef] [PubMed]


| Variety | AAC (%) | AAC Grade | RS (%) | RS Grade |
|---|---|---|---|---|
| Li | 14.86 ± 0.39 i | low | 0.48 ± 0.14 g | low |
| QR | 15.01 ± 0.28 hi | low | 0.44 ± 0.22 g | low |
| Lan | 15.14 ± 0.29 hi | low | 0.48 ± 0.18 g | low |
| HR52 | 15.67 ± 0.17 h | low | 0.57 ± 0.24 g | low |
| BR18 | 21.26 ± 0.42 g | medium | 7.78 ± 0.42 de | high |
| BR19 | 22.68 ± 0.47 f | medium | 8.33 ± 0.39 bcd | high |
| BR50 | 23.28 ± 0.53 ef | medium | 9.10 ± 0.47 b | high |
| BR28 | 23.77 ± 0.29 de | medium | 8.09 ± 0.68 cd | high |
| Sx | 23.85 ± 0.25 de | medium | 8.31 ± 0.62 bcd | high |
| Ky43 | 24.24 ± 0.33 cd | medium | 6.25 ± 0.56 f | high |
| BR89 | 24.85 ± 0.76 bc | medium | 8.94 ± 0.70 bc | high |
| CN-5 | 24.99 ± 0.38 b | medium | 6.99 ± 0.53 ef | high |
| CN-9 | 25.35 ± 0.49 ab | high | 10.66 ± 0.88 a | high |
| ML | 25.78 ± 0.43 a | high | 6.68 ± 0.68 f | high |
| Marker Name | Primer Name | Primer Sequence |
|---|---|---|
| WL3 | GCAGATCAAGGTTGCAGACA | |
| WR | TGGCAATAAGCCACACACAT | |
| T-Wx9-RS1 | W9-IF1 | GCATGGACGTGAGTACT |
| W9-IR1 | ACTTGGCGGTGGATGTACTTGTCCTTGATG | |
| W9-OF1 | GGAGGAGGAAGATCAACTGGATGAA | |
| W9-OR1 | TTGCCTGAAATTGTTACTCATTCTTGCC | |
| T-Wx9-RS2 | W9-IF1 | GCATGGACGTGAGTACT |
| W9-IR1 | ACTTGGCGGTGGATGTACTTGTCCTTGATG | |
| W9-OF2 | CCCGTACTACGCCGAGGAGCTCATCT | |
| W9-OR2 | TGCCTGAAATTGTTCACTCATTCTTGCCTT |
| Number | Source | Genotype | Resistant Starch (g/100 g) | Number | Source | Genotype | Resistant Starch (g/100 g) |
|---|---|---|---|---|---|---|---|
| 2021p1637 | Lan/CN-5 | − | 0.50 ± 0.16 | 2021p1657 | QR/Ky43 | − | 0.52 ± 0.21 |
| 2021p1638 | Lan/CN-5 | − | 0.52 ± 0.09 | 2021p1658 | QR/Ky43 | − | 0.59 ± 0.23 |
| 2021p1639 | Lan/CN-5 | − | 0.49 ± 0.15 | 2021p1659 | QR/Ky43 | − | 0.62 ± 0.14 |
| 2021p1640 | Lan/CN-9 | − | 1.76 ± 0.24 | 2021p1660 | Lan/CN-9 | + | 3.96 ± 1.66 |
| 2021p1641 | Lan/CN-9 | − | 1.30 ± 0.35 | 2021p1661 | Lan/CN-9 | + | 3.74 ± 1.65 |
| 2021p1642 | Lan/Sx | − | 1.31 ± 0.33 | 2021p1662 | Li/BR50 | + | 4.52 ± 0.18 |
| 2021p1643 | Lan/Sx | − | 0.51 ± 0.24 | 2021p1663 | QR/BR50 | + | 7.24 ± 0.94 |
| 2021p1644 | QR/BR50 | − | 1.99 ± 0.65 | 2021p1664 | QR/BR50 | + | 6.47 ± 0.96 |
| 2021p1645 | QR/BR50 | − | 0.91 ± 0.10 | 2021p1665 | QR/BR50 | + | 11.10 ± 1.70 |
| 2021p1646 | QR/BR50 | − | 0.41 ± 0.04 | 2021p1666 | QR/BR50 | + | 7.14 ± 1.38 |
| 2021p1647 | QR/BR50 | − | 1.04 ± 0.07 | 2021p1667 | QR/BR50 | + | 3.10 ± 0.44 |
| 2021p1648 | QR/BR50 | − | 0.77 ± 0.17 | 2021p1668 | Lan/Sx | H | 4.00 ± 1.49 |
| 2021p1649 | QR/BR50 | − | 0.78 ± 0.11 | 2021p1669 | Lan/Sx | H | 3.48 ± 1.19 |
| 2021p1650 | QR/BR50 | − | 0.68 ± 0.10 | 2021p1670 | QR/BR50 | H | 1.53 ± 0.45 |
| 2021p1651 | QR/BR50 | − | 0.68 ± 0.14 | 2021p1671 | QR/BR50 | H | 1.42 ± 0.40 |
| 2021p1652 | QR/Sx | − | 1.12 ± 0.18 | 2021p1672 | QR/BR50 | H | 3.04 ± 0.73 |
| 2021p1653 | QR/Sx | − | 1.45 ± 0.44 | 2021p1673 | QR/BR50 | H | 3.35 ± 0.33 |
| 2021p1654 | QR/Sx | − | 1.06 ± 0.17 | 2021p1674 | QR/BR50 | H | 2.84 ± 0.56 |
| 2021p1655 | QR/Ky43 | − | 0.73 ± 0.28 | 2021p1675 | QR/BR50 | H | 3.91 ± 0.94 |
| 2021p1656 | QR/Ky43 | − | 0.59 ± 0.18 | 2021p1676 | QR/BR50 | H | 3.44 ± 0.96 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Ouyang, J.; Zhu, Z.; Guan, Y.; Huang, Q.; Huang, T.; Zang, S.; Pan, C. Novel Wx Gene Functional Markers for High-Resistant Starch Rice Breeding. Genes 2026, 17, 89. https://doi.org/10.3390/genes17010089
Ouyang J, Zhu Z, Guan Y, Huang Q, Huang T, Zang S, Pan C. Novel Wx Gene Functional Markers for High-Resistant Starch Rice Breeding. Genes. 2026; 17(1):89. https://doi.org/10.3390/genes17010089
Chicago/Turabian StyleOuyang, Jie, Zichao Zhu, Yusheng Guan, Qianlong Huang, Tao Huang, Shun Zang, and Chuxiang Pan. 2026. "Novel Wx Gene Functional Markers for High-Resistant Starch Rice Breeding" Genes 17, no. 1: 89. https://doi.org/10.3390/genes17010089
APA StyleOuyang, J., Zhu, Z., Guan, Y., Huang, Q., Huang, T., Zang, S., & Pan, C. (2026). Novel Wx Gene Functional Markers for High-Resistant Starch Rice Breeding. Genes, 17(1), 89. https://doi.org/10.3390/genes17010089





