Identification of Prognostic Genes and Establishment of a Risk Score Model Related to Pancreatic Adenocarcinoma and Brown Adipose Tissue Based on Transcriptomics and Experimental Validation
Abstract
1. Introduction
2. Methods
2.1. Data Acquisition
2.2. Discernment and Related Functional Analysis of Candidate Genes
2.3. Identification of Prognostic Genes
2.4. Construction and Evaluation of RS Models
2.5. Independent Prognostic Analysis and Construction of Nomogram
2.6. Function Analyses of HRG and LRG
2.7. Immune Microenvironment Analysis
2.8. Somatic Mutation Analysis
2.9. Drug Sensitivity Analysis
2.10. Localization and Immunohistochemistry of Prognostic Genes
2.11. Preprocessing, Dimensionality Reduction, Clustering, and Cell Subpopulation Annotation of Single-Cell Data
2.12. Identification of Key Cell Types
2.13. Cell Communication and Pseudotime Analysis
2.14. The Reverse Transcription Quantitative PCR (RT-qPCR)
2.15. Statistical Analysis
3. Results
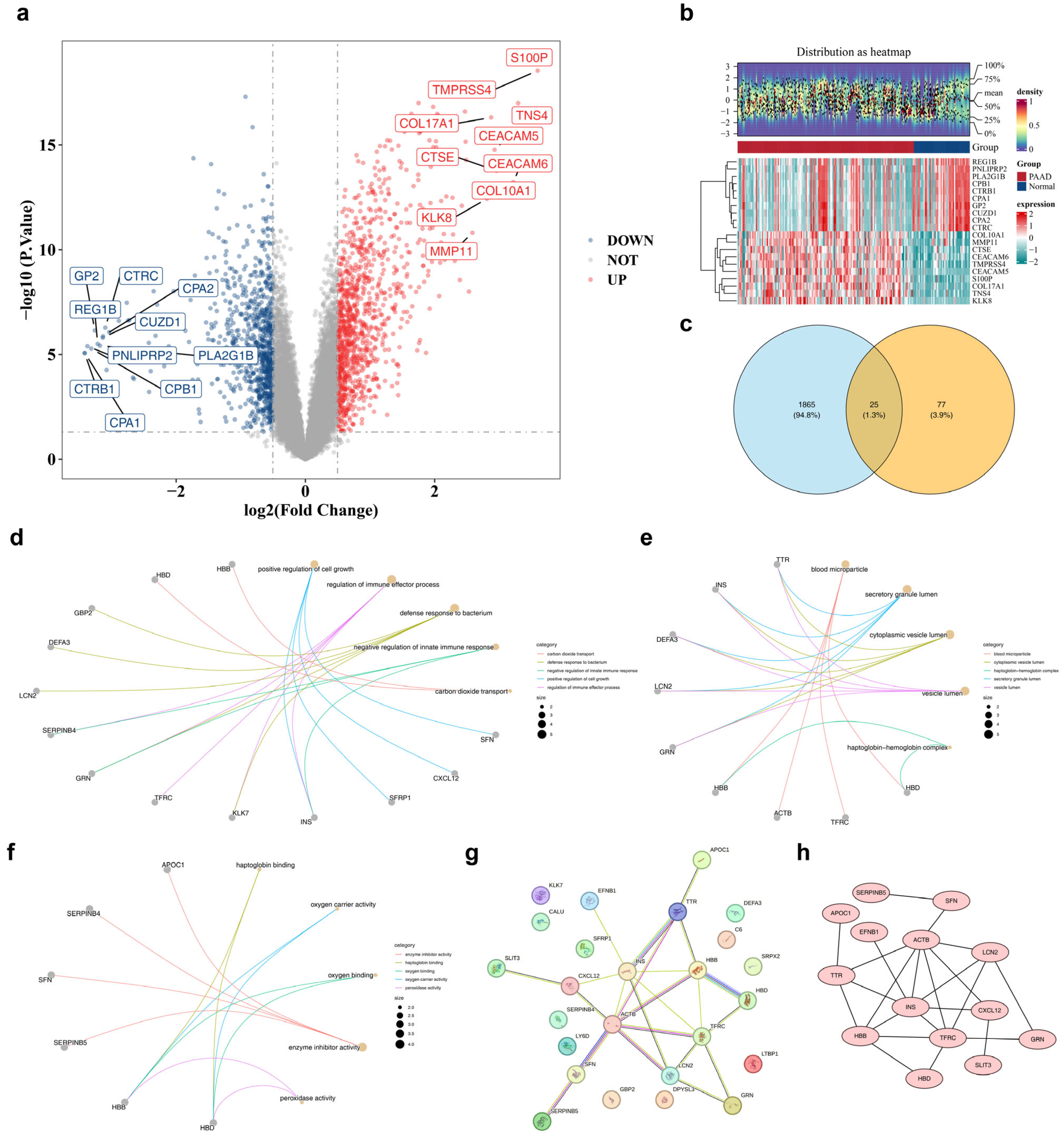
3.1. Discernment of 25 Candidate Genes and Exploration of Their Biological Functions
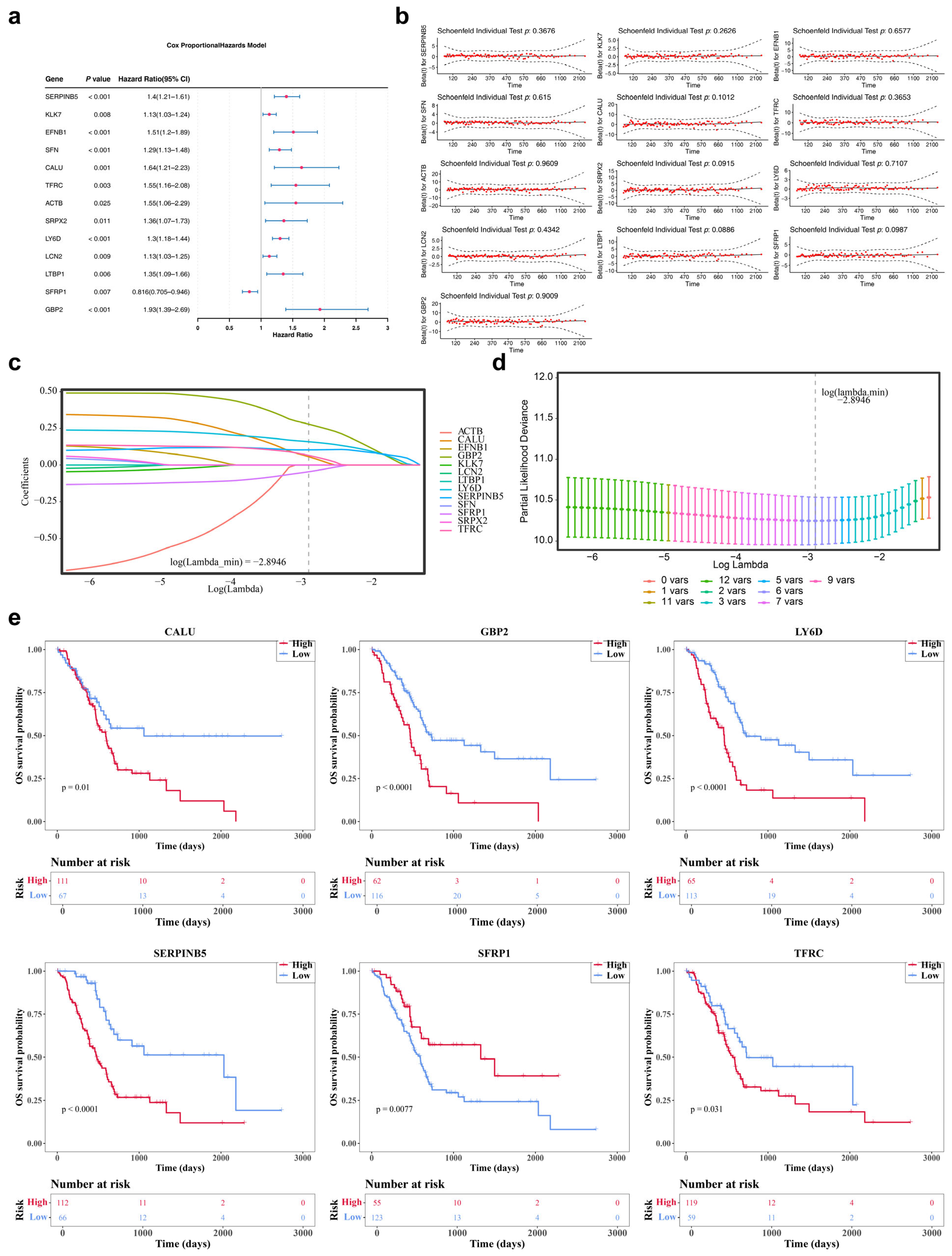
3.2. Acquisition of 6 Prognostic Genes: SERPINB5, CALU, TFRC, LY6D, SFRP1 and GBP2
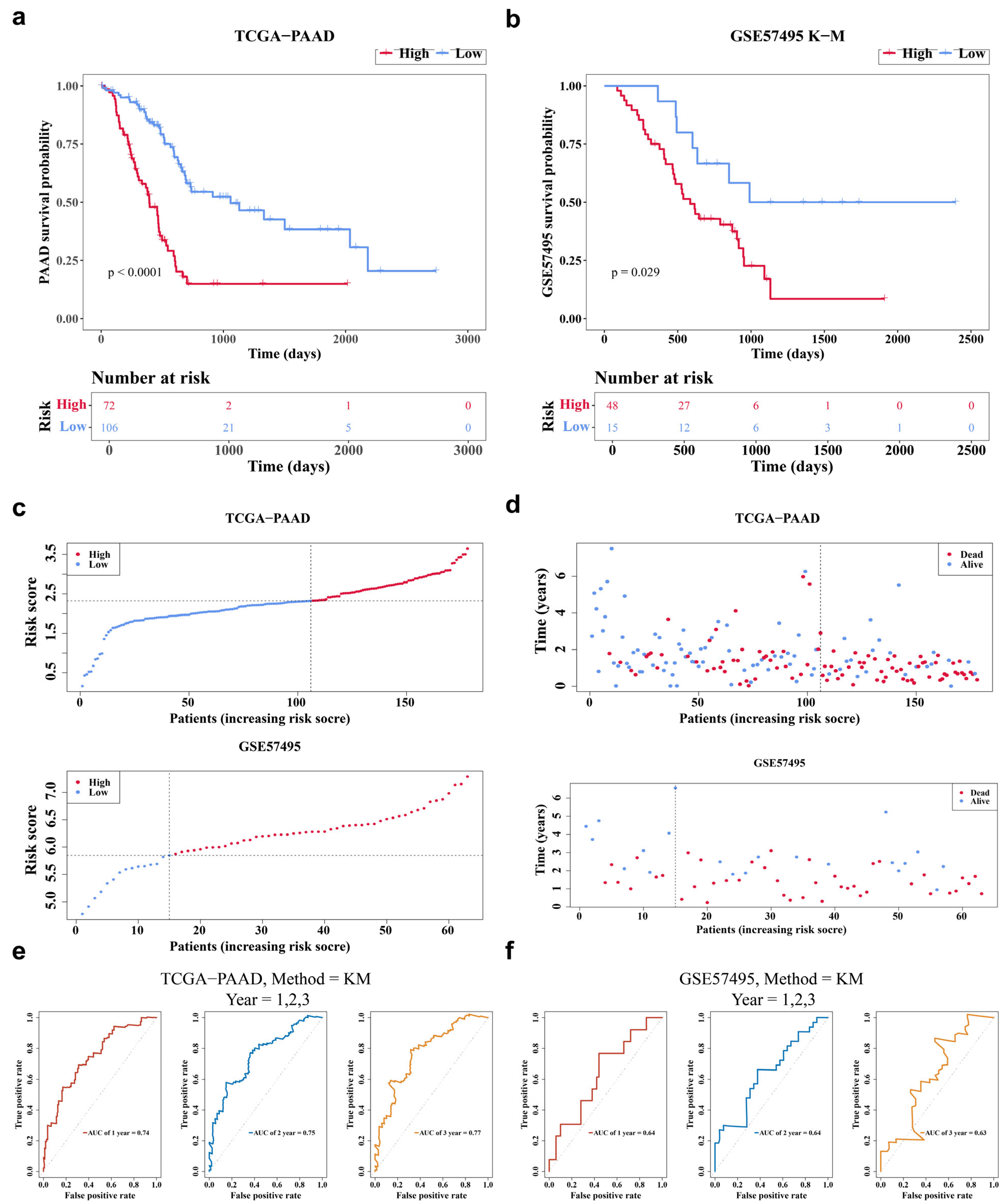
3.3. The RS Model Demonstrated a Favorable Predictive Performance
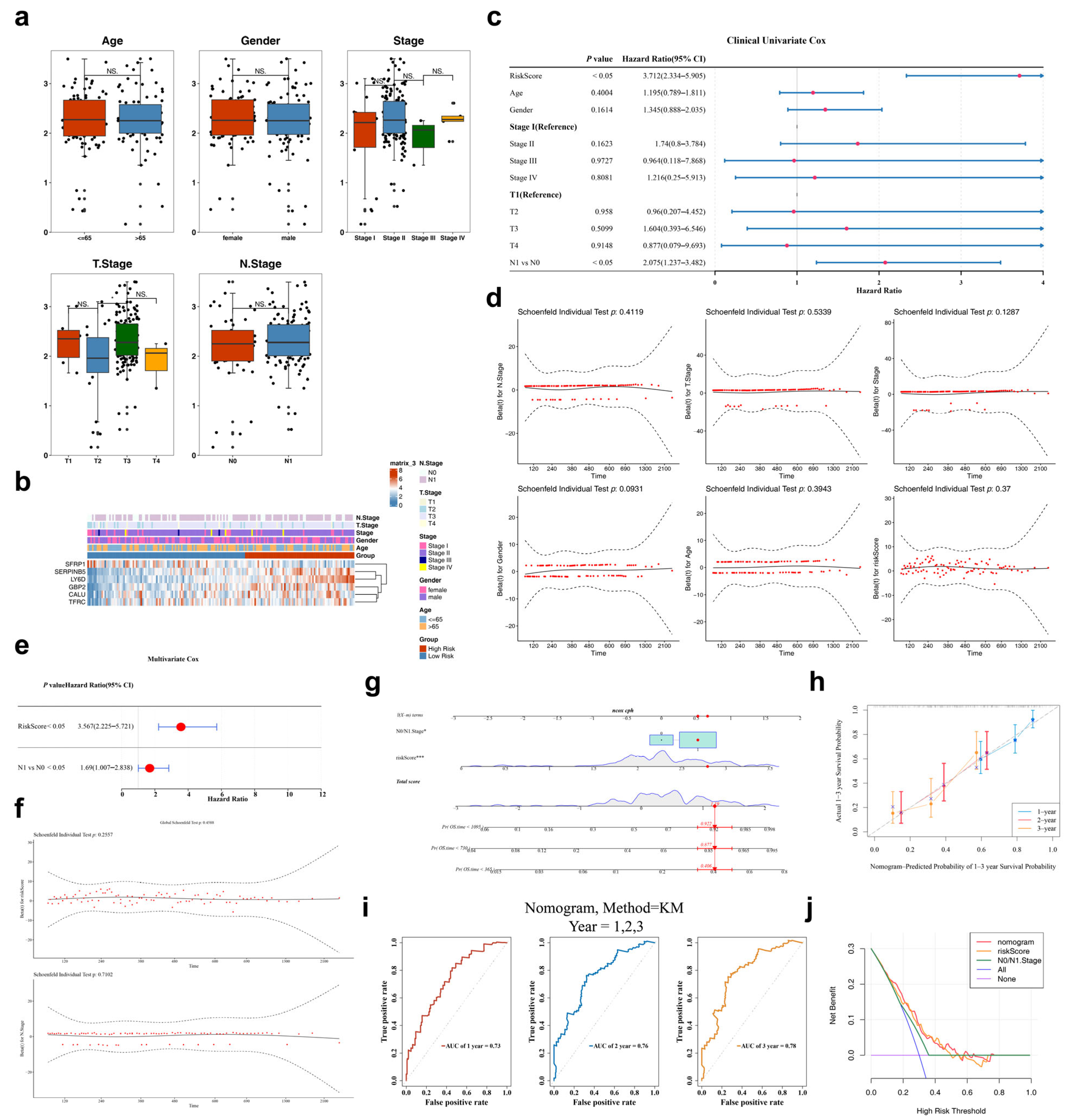
3.4. Independent Prognostic Factors: RS and N0/N1 Stage
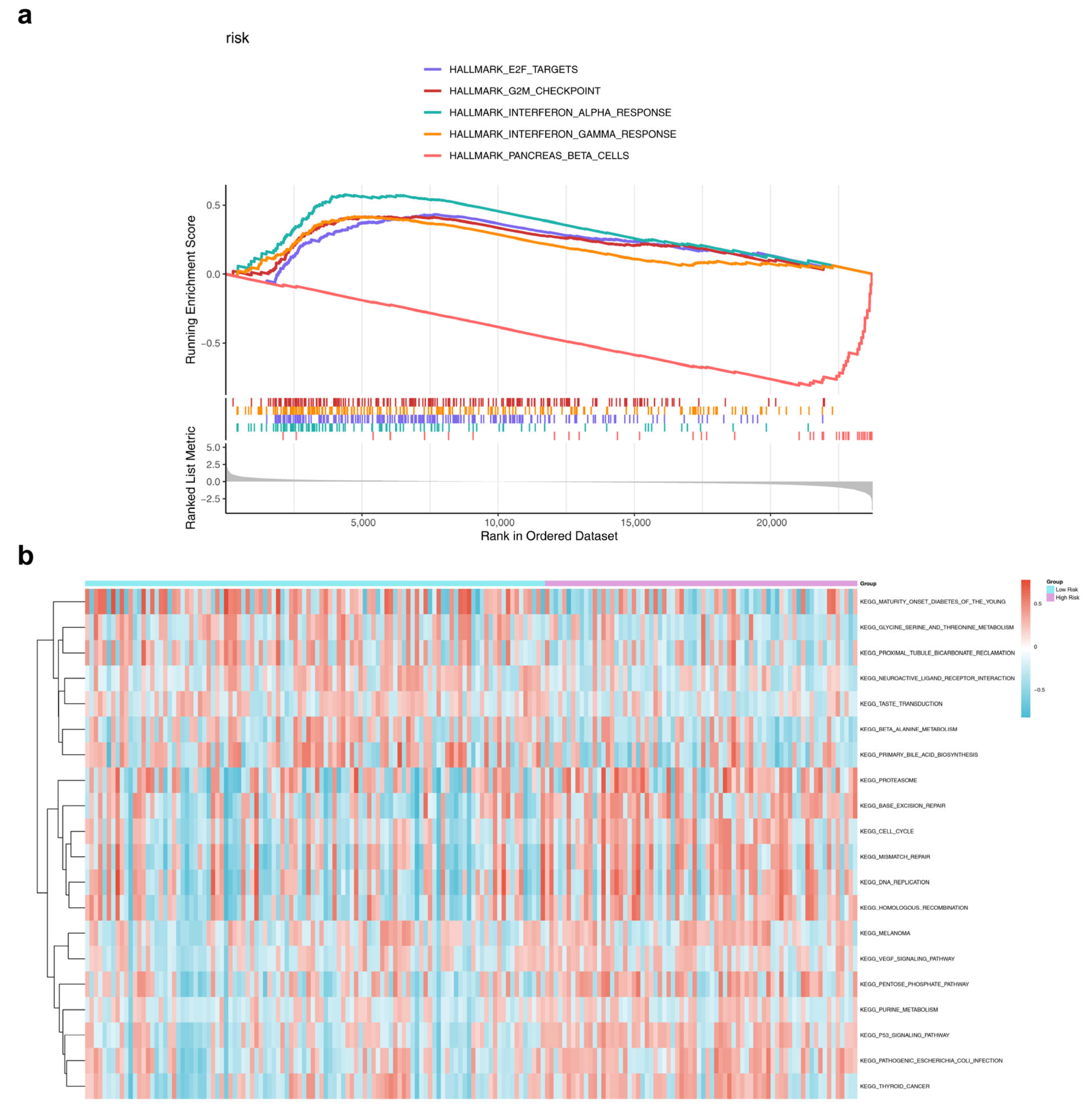
3.5. Differences in Enrichment Pathways Between HRG and LRG
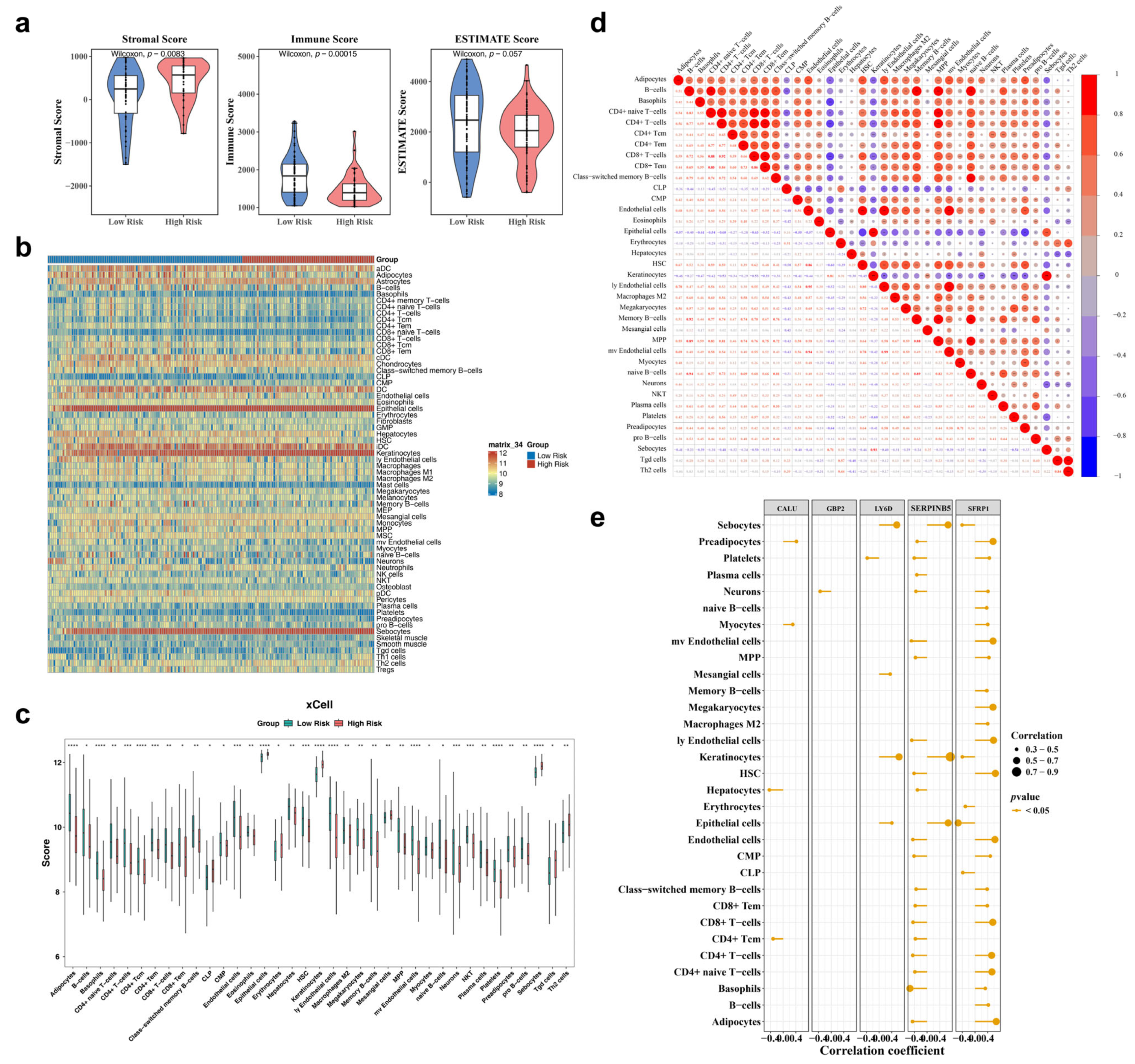
3.6. Estimation of Tumor Immune Microenvironment
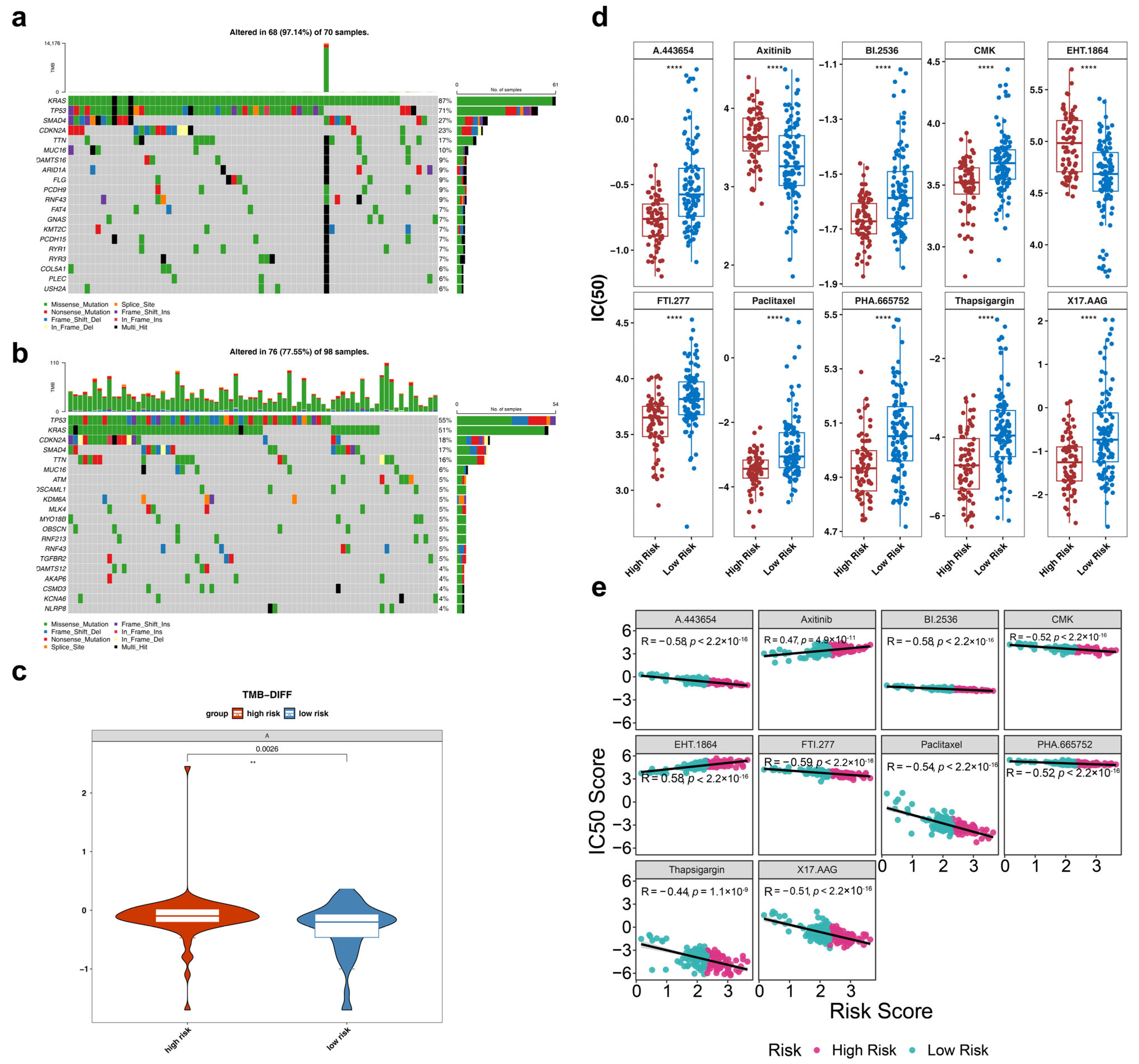
3.7. Somatic Mutation Analysis Between HRG and LRG
3.8. Chemotherapy Sensitivity Analysis Between HRG and LRG
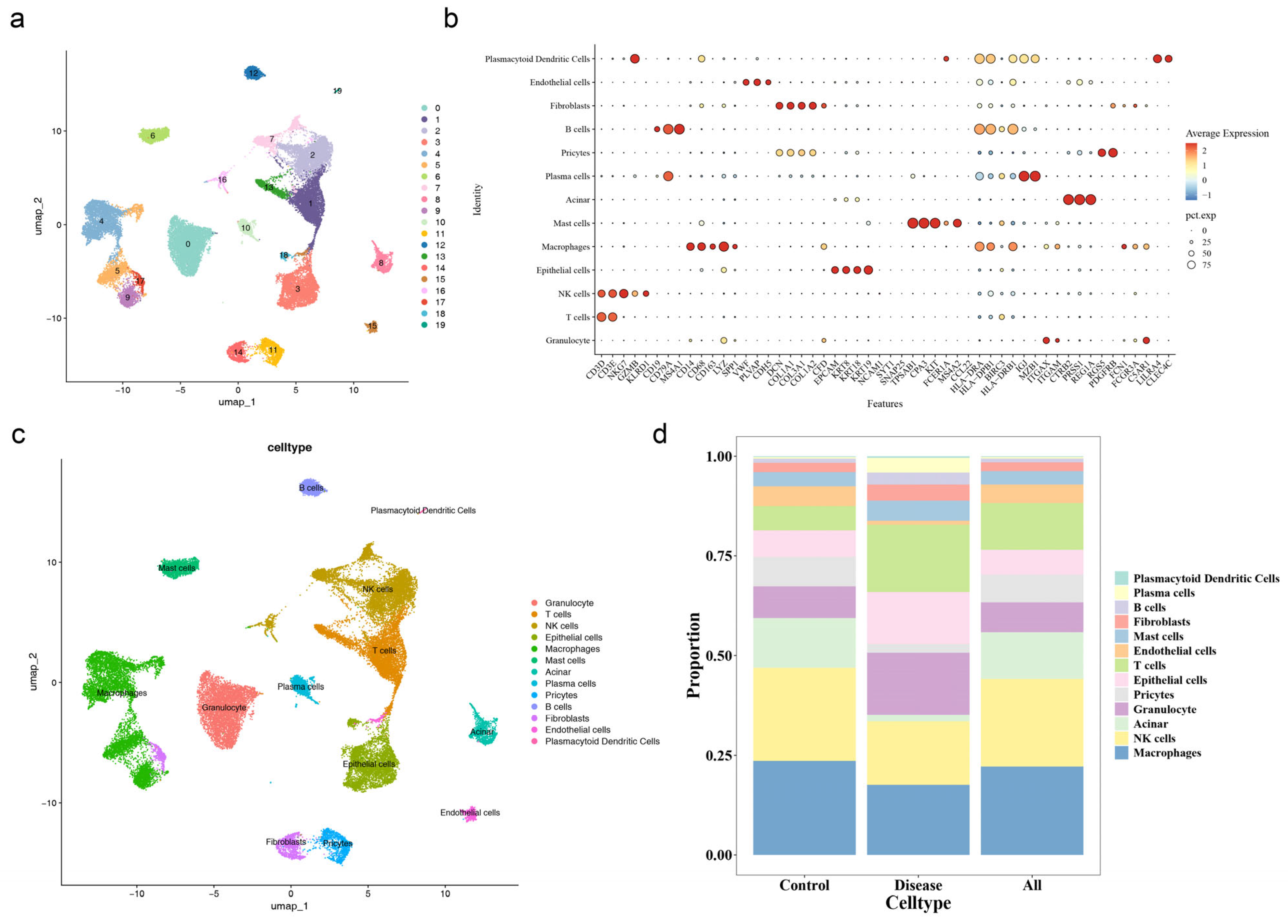
3.9. Quality Control and Annotation of Single-Cell Sequencing Data
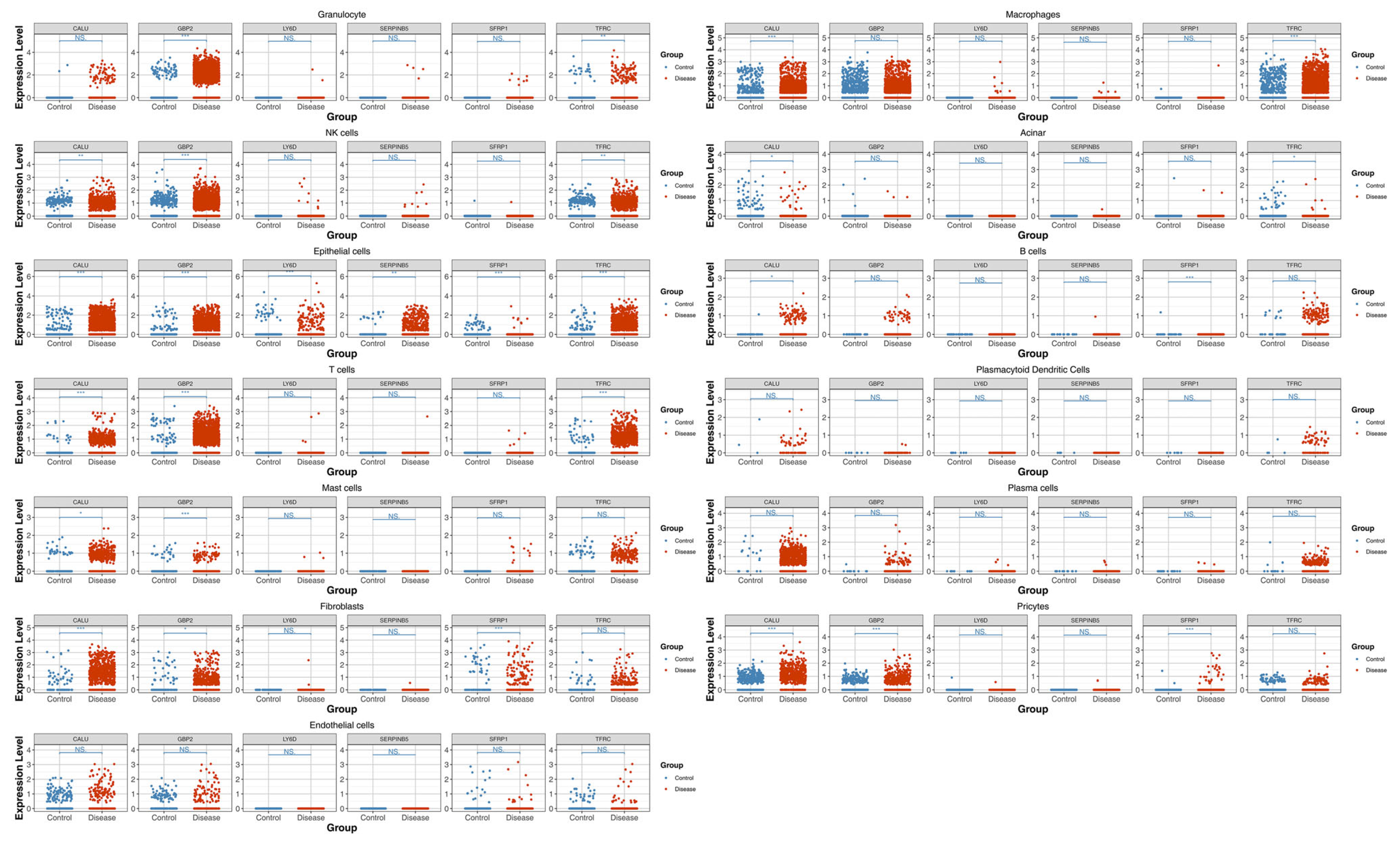
3.10. Epithelial Cells as the Key Cell Type
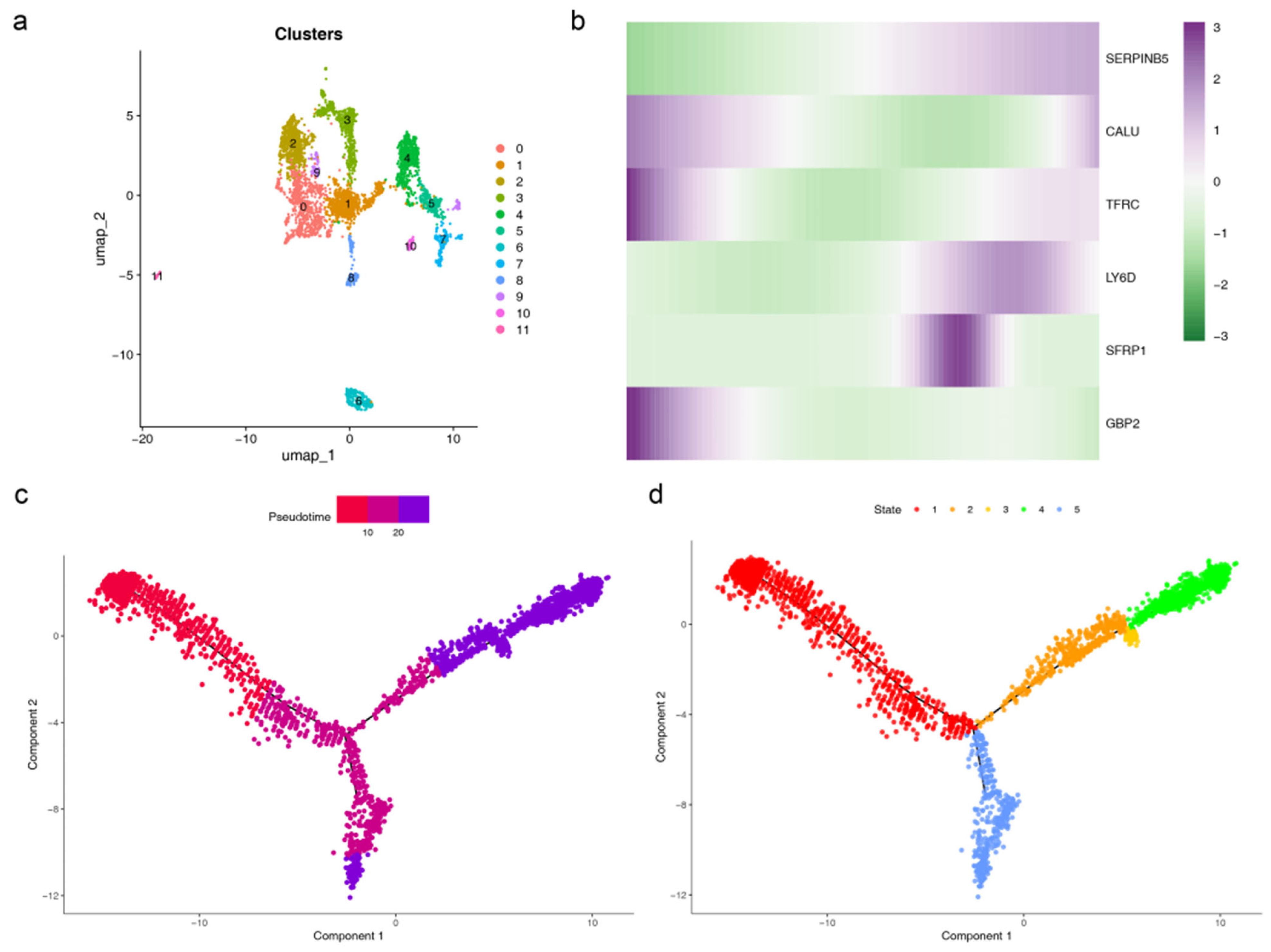
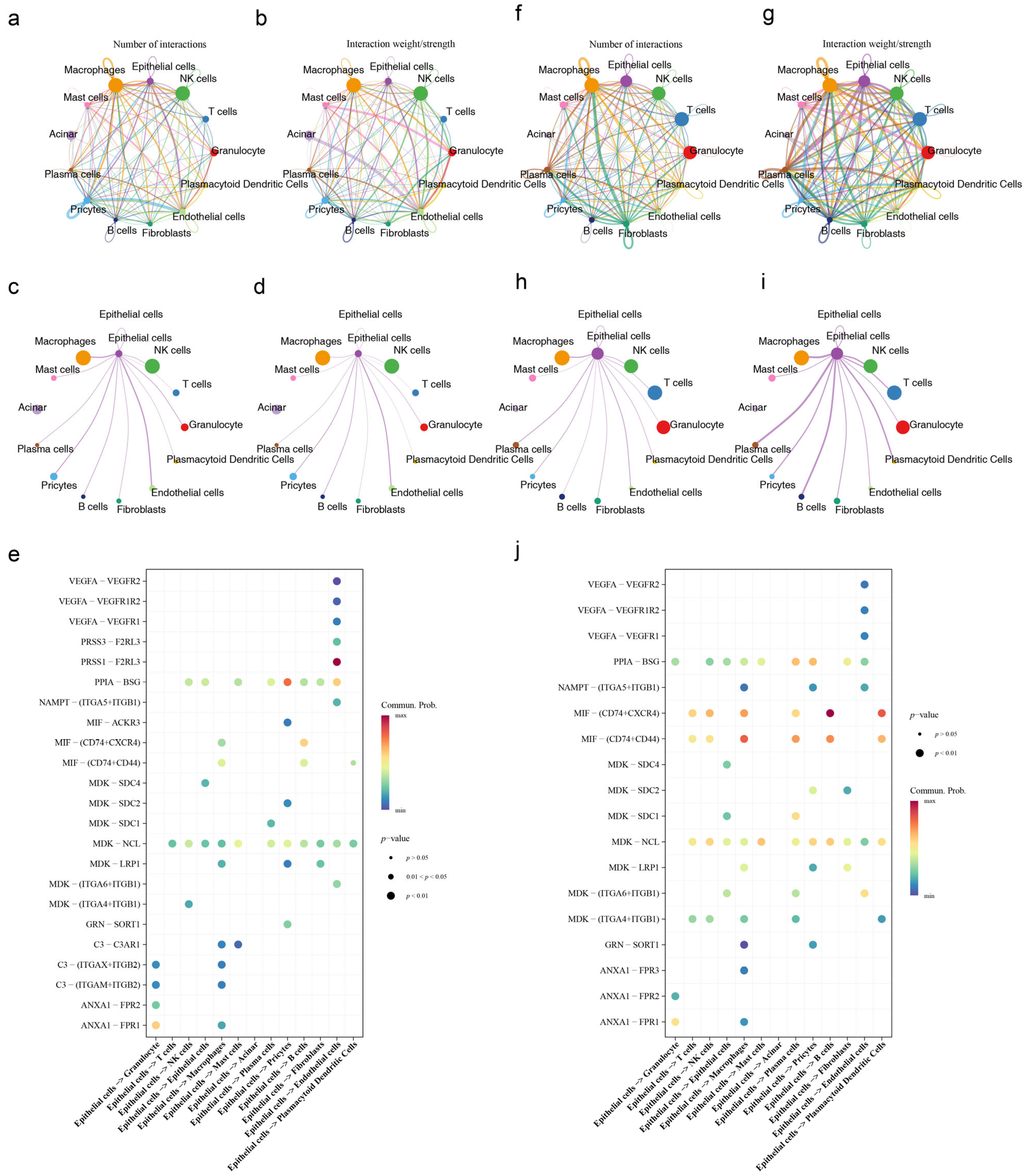
3.11. Cell Communication and Pseudotime Analysis
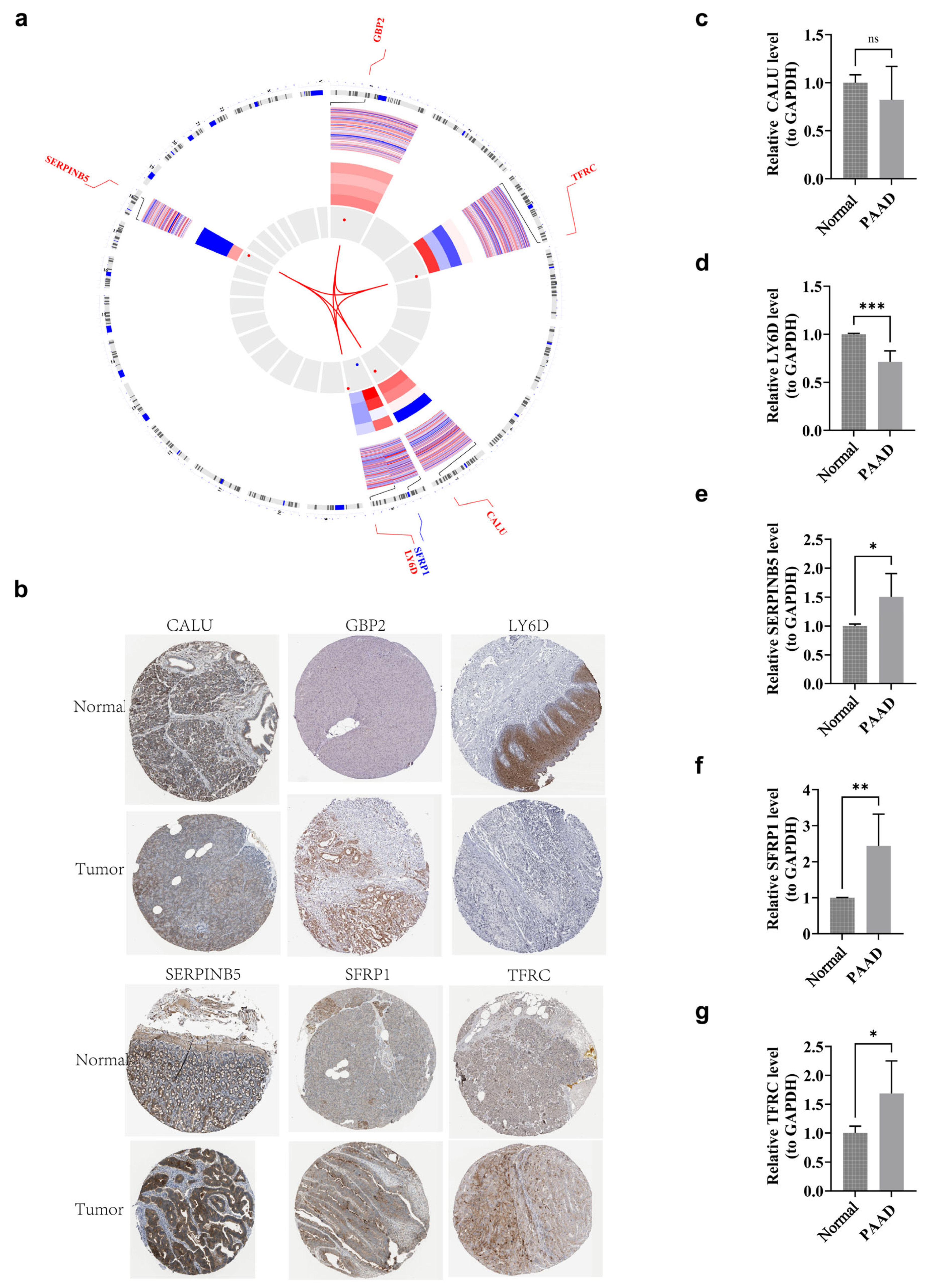
3.12. Localization Analysis and Clinical Trial Validation of Prognostic Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviation | Full Form |
| PAAD | Pancreatic Adenocarcinoma |
| BA | Brown Adipocyte |
| PH | Proportional Hazards |
| DCA | Decision Curve Analysis |
| RS | Risk Score |
| UCP1 | Uncoupling Protein 1 |
| TCGA | The Cancer Genome Atlas |
| GEO | Gene Expression Omnibus |
| BARGs | Brown Adipocyte-Related Genes |
| DEGs | Differentially Expressed Genes |
| GO | Gene Ontology |
| MF | Molecular Functions |
| CC | Cellular Components |
| BP | Biological Processes |
| PPI | Protein-Protein Interaction |
| HR | Hazard Ratio |
| K-M | Kaplan-Meier |
| HRG | High Risk Group |
| LRG | Low Risk Group |
| ROC | Receiver Operating Characteristic |
| AUC | Area Under Curve |
| GSEA | Gene Set Enrichment Analysis |
| GSVA | Gene Set Variation Analysis |
| MSigDB | Molecular Signatures Database |
| ssGSEA | Single Sample GSEA |
| TMB | Tumor Mutation Burden |
| IC50 | Half-Maximal Inhibitory Concentration |
| GDSC | Genomics of Drug Sensitivity in Cancer |
| HPA | Human Protein Atlas |
| RT-qPCR | Reverse Transcription Quantitative PCR |
References
- Kordes, M.; Larsson, L.; Engstrand, L.; Lohr, J.M. Pancreatic cancer cachexia: Three dimensions of a complex syndrome. Br. J. Cancer 2021, 124, 1623–1636. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Xiang, H.; Pan, Y.; Shang, D.; Guo, J.; Gao, G.; Xiao, G.G. Pancreatitis initiated pancreatic ductal adenocarcinoma: Pathophysiology explaining clinical evidence. Pharmacol. Res. 2021, 168, 105595. [Google Scholar] [CrossRef] [PubMed]
- Stoffel, E.M.; Brand, R.E.; Goggins, M. Pancreatic Cancer: Changing Epidemiology and New Approaches to Risk Assessment, Early Detection, and Prevention. Gastroenterology 2023, 164, 752–765. [Google Scholar] [CrossRef] [PubMed]
- Versteijne, E.; van Dam, J.L.; Suker, M.; Janssen, Q.P.; Groothuis, K.; Akkermans-Vogelaar, J.M.; Besselink, M.G.; Bonsing, B.A.; Buijsen, J.; Busch, O.R.; et al. Neoadjuvant Chemoradiotherapy Versus Upfront Surgery for Resectable and Borderline Resectable Pancreatic Cancer: Long-Term Results of the Dutch Randomized PREOPANC Trial. J. Clin. Oncol. 2022, 40, 1220–1230. [Google Scholar] [CrossRef]
- Masiak-Segit, W.; Rawicz-Pruszynski, K.; Skorzewska, M.; Polkowski, W.P. Surgical treatment of pancreatic cancer. Pol. Przegl. Chir. 2018, 90, 45–53. [Google Scholar] [CrossRef]
- Evans, D.B. What Makes a Pancreatic Cancer Resectable? Am. Soc. Clin. Oncol. Educ. Book 2018, 38, 300–305. [Google Scholar] [CrossRef]
- He, F.; Chen, Z.; Deng, W.; Zhan, T.; Huang, X.; Zheng, Y.; Yang, H. Development and validation of a novel ferroptosis-related gene signature for predicting prognosis and immune microenvironment in head and neck squamous cell carcinoma. Int. Immunopharmacol. 2021, 98, 107789. [Google Scholar] [CrossRef]
- Gao, Z.; Chen, J.; Zhou, Y.; Deng, P.; Sun, L.; Qi, J.; Zhang, P. A Novel Metabolism-Related Gene Signature for Predicting the Prognosis of HBV-Infected Hepatocellular Carcinoma. J. Oncol. 2022, 2022, 2391265. [Google Scholar] [CrossRef]
- Okamatsu-Ogura, Y.; Kuroda, M.; Tsutsumi, R.; Tsubota, A.; Saito, M.; Kimura, K.; Sakaue, H. UCP1-dependent and UCP1-independent metabolic changes induced by acute cold exposure in brown adipose tissue of mice. Metabolism 2020, 113, 154396. [Google Scholar] [CrossRef]
- Liu, X.; Tang, J.; Zhang, R.; Zhan, S.; Zhong, T.; Guo, J.; Wang, Y.; Cao, J.; Li, L.; Zhang, H.; et al. Cold exposure induces lipid dynamics and thermogenesis in brown adipose tissue of goats. BMC Genom. 2022, 23, 528. [Google Scholar] [CrossRef]
- Saito, M.; Matsushita, M.; Yoneshiro, T.; Okamatsu-Ogura, Y. Brown Adipose Tissue, Diet-Induced Thermogenesis, and Thermogenic Food Ingredients: From Mice to Men. Front. Endocrinol. 2020, 11, 222. [Google Scholar] [CrossRef] [PubMed]
- U-Din, M.; Rebelos, E.; Saari, T.; Niemi, T.; Kuellmer, K.; Eskola, O.; Fromme, T.; Rajander, J.; Taittonen, M.; Klingenspor, M.; et al. Thermogenic Capacity of Human Supraclavicular Brown Fat and Cold-Stimulated Brain Glucose Metabolism. Metabolites 2023, 13, 387. [Google Scholar] [CrossRef] [PubMed]
- Scheel, A.K.; Espelage, L.; Chadt, A. Many Ways to Rome: Exercise, Cold Exposure and Diet-Do They All Affect BAT Activation and WAT Browning in the Same Manner? Int. J. Mol. Sci. 2022, 23, 4759. [Google Scholar] [CrossRef] [PubMed]
- Mouisel, E.; Bodon, A.; Noll, C.; Cassant-Sourdy, S.; Marques, M.A.; Flores-Flores, R.; Riant, E.; Bergoglio, C.; Vezin, P.; Caspar-Bauguil, S.; et al. Cold-induced thermogenesis requires neutral-lipase-mediated intracellular lipolysis in brown adipocytes. Cell Metab. 2025, 37, 429–440.e5. [Google Scholar] [CrossRef]
- Suzuki, T.; Otsuka, M.; Seimiya, T.; Iwata, T.; Kishikawa, T.; Koike, K. The biological role of metabolic reprogramming in pancreatic cancer. MedComm (2020) 2020, 1, 302–310. [Google Scholar] [CrossRef]
- Kepple, J.D.; Barra, J.M.; Young, M.E.; Hunter, C.S.; Tse, H.M. Islet transplantation into brown adipose tissue can delay immune rejection. JCI Insight 2022, 7, e152800. [Google Scholar] [CrossRef]
- Bertola, A.; Gallerand, A.; Ivanov, S. Immune cell involvement in brown adipose tissue functions. Discov. Immunol. 2022, 1, kyac007. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Liu, Y.; Ouyang, Q.; Li, Q. A novel brown adipocytes-related gene signature predicts and validates prognosis and immune infiltration of clear cell renal cell carcinoma. Am. J. Cancer Res. 2024, 14, 4286–4305. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Gustavsson, E.K.; Zhang, D.; Reynolds, R.H.; Garcia-Ruiz, S.; Ryten, M. ggtranscript: An R package for the visualization and interpretation of transcript isoforms using ggplot2. Bioinformatics 2022, 38, 3844–3846. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Hübschmann, D. Make Interactive Complex Heatmaps in R. Bioinformatics 2022, 38, 1460–1462. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Gao, W.; Zhang, Q.; Cheng, X.; Liu, Y.; Qi, Z.; Li, T. Ferroptosis and Autophagy-Related Genes in the Pathogenesis of Ischemic Cardiomyopathy. Front. Cardiovasc. Med. 2022, 9, 906753. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Ramsay, I.S.; Ma, S.; Fisher, M.; Loewy, R.L.; Ragland, J.D.; Niendam, T.; Carter, C.S.; Vinogradov, S. Model selection and prediction of outcomes in recent onset schizophrenia patients who undergo cognitive training. Schizophr. Res. Cogn. 2018, 11, 1–5. [Google Scholar] [CrossRef]
- Tay, J.K.; Narasimhan, B.; Hastie, T. Elastic Net Regularization Paths for All Generalized Linear Models. J. Stat. Softw. 2023, 106, 1. [Google Scholar] [CrossRef]
- Liu, T.T.; Li, R.; Huo, C.; Li, J.P.; Yao, J.; Ji, X.L.; Qu, Y.Q. Identification of CDK2-Related Immune Forecast Model and ceRNA in Lung Adenocarcinoma, a Pan-Cancer Analysis. Front. Cell Dev. Biol. 2021, 9, 682002. [Google Scholar] [CrossRef]
- Heagerty, P.J.; Lumley, T.; Pepe, M.S. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 2000, 56, 337–344. [Google Scholar] [CrossRef]
- Sachs, M.C. plotROC: A Tool for Plotting ROC Curves. J. Stat. Softw. 2017, 79, 2. [Google Scholar] [CrossRef]
- Zou, Y.; Xie, J.; Zheng, S.; Liu, W.; Tang, Y.; Tian, W.; Deng, X.; Wu, L.; Zhang, Y.; Wong, C.W.; et al. Leveraging diverse cell-death patterns to predict the prognosis and drug sensitivity of triple-negative breast cancer patients after surgery. Int. J. Surg. 2022, 107, 106936. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Petinrin, O.O.; Chen, N.; Toseef, M.; Liu, F.; Zhu, Z.; Qi, F.; Wong, K.C. Identification and evaluation of candidate COVID-19 critical genes and medicinal drugs related to plasma cells. BMC Infect. Dis. 2024, 24, 1099. [Google Scholar] [CrossRef]
- Garcia-Barrios, G.; Crossa, J.; Cruz-Izquierdo, S.; Aguilar-Rincon, V.H.; Sandoval-Islas, J.S.; Corona-Torres, T.; Lozano-Ramirez, N.; Dreisigacker, S.; He, X.; Singh, P.K.; et al. Genomic Prediction of Resistance to Tan Spot, Spot Blotch and Septoria Nodorum Blotch in Synthetic Hexaploid Wheat. Int. J. Mol. Sci. 2023, 24, 10506. [Google Scholar] [CrossRef]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Geeleher, P.; Cox, N.; Huang, R.S. pRRophetic: An R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE 2014, 9, e107468. [Google Scholar] [CrossRef]
- Zhang, H.; Meltzer, P.; Davis, S. RCircos: An R package for Circos 2D track plots. BMC Bioinform. 2013, 14, 244. [Google Scholar] [CrossRef]
- Chen, L.; Ying, X.; Wang, H.; Xie, J.; Tang, Q.; Liu, W. Identification and Validation of Senescence-Related Signature by Combining Single Cell and Bulk Transcriptome Data Analysis to Predict the Prognosis and Identify the Key Gene CAV1 in Pancreatic Cancer. J. Inflamm. Res. 2024, 17, 9391–9406. [Google Scholar] [CrossRef]
- Chen, L.; Ying, X.; Ma, C.; Tang, Q.; Chen, S. Single-cell and multi-omics analysis identifies TRIM9 as a key ubiquitination regulator in pancreatic cancer. Front. Immunol. 2025, 16, 1631708. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.P.; An, K.; Ito, Y.; Kharbikar, B.N.; Sheng, R.; Paredes, B.; Murray, E.; Pham, K.; Bruck, M.; Zhou, X.; et al. Implantation of engineered adipocytes suppresses tumor progression in cancer models. Nat. Biotechnol. 2025, 43, 1979–1995. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.T.; Hsieh, M.J.; Lin, C.W.; Su, S.C.; Miao, N.F.; Yang, S.F.; Huang, H.C.; Lai, F.C.; Liu, Y.F. Combinations of SERPINB5 gene polymorphisms and environmental factors are associated with oral cancer risks. PLoS ONE 2017, 12, e0163369. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Wei, C.; Cheng, J.; Ding, R.; Li, Y.; Wang, Y.; Yang, Y.; Wang, J. BTG2 and SerpinB5, a novel gene pair to evaluate the prognosis of lung adenocarcinoma. Front. Immunol. 2023, 14, 1098700. [Google Scholar] [CrossRef]
- Liu, B.X.; Xie, Y.; Zhang, J.; Zeng, S.; Li, J.; Tao, Q.; Yang, J.; Chen, Y.; Zeng, C. SERPINB5 promotes colorectal cancer invasion and migration by promoting EMT and angiogenesis via the TNF-alpha/NF-kappaB pathway. Int. Immunopharmacol. 2024, 131, 111759. [Google Scholar] [CrossRef]
- Tian, Y.; Tang, R.; Xie, M.; Zhan, Y.; Guo, Y.; Zeng, X.; Wang, S.; Kuang, H.; Yang, S.; Gao, Z.; et al. SERPINB2 increases endothelial inflammation through augmented fatty acid oxidation to promote choroidal neovascularization. Exp. Eye Res. 2025, 259, 110524. [Google Scholar] [CrossRef]
- Omran, F.; Christian, M. Inflammatory Signaling and Brown Fat Activity. Front. Endocrinol. 2020, 11, 156. [Google Scholar] [CrossRef]
- Liu, D.; Li, Y.; Shang, Y.; Wang, W.; Chen, S.Z. Effect of brown adipose tissue/cells on the growth of mouse hepatocellular carcinoma in vitro and in vivo. Oncol. Lett. 2019, 17, 3203–3210. [Google Scholar] [CrossRef]
- Ning, J.; Liu, M.; Shen, J.; Wang, D.; Gao, L.; Li, H.; Cao, J. Expression signature and prognostic value of CREC gene family in human colorectal cancer. BMC Cancer 2023, 23, 878. [Google Scholar] [CrossRef]
- Li, Y.; Sun, S.; Zhang, H.; Jing, Y.; Ji, X.; Wan, Q.; Liu, Y. CALU promotes lung adenocarcinoma progression by enhancing cell proliferation, migration and invasion. Respir. Res. 2024, 25, 267. [Google Scholar] [CrossRef]
- Nasri Nasrabadi, P.; Nayeri, Z.; Gharib, E.; Salmanipour, R.; Masoomi, F.; Mahjoubi, F.; Zomorodipour, A. Establishment of a CALU, AURKA, and MCM2 gene panel for discrimination of metastasis from primary colon and lung cancers. PLoS ONE 2020, 15, e0233717. [Google Scholar]
- Seki, T.; Yang, Y.; Sun, X.; Lim, S.; Xie, S.; Guo, Z.; Xiong, W.; Kuroda, M.; Sakaue, H.; Hosaka, K.; et al. Brown-fat-mediated tumour suppression by cold-altered global metabolism. Nature 2022, 608, 421–428. [Google Scholar] [PubMed]
- Shen, B.; Zheng, P.; Qian, N.; Chen, Q.; Zhou, X.; Hu, J.; Chen, J.; Teng, J. Calumenin-1 Interacts with Climp63 to Cooperatively Determine the Luminal Width and Distribution of Endoplasmic Reticulum Sheets. iScience 2019, 22, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Hu, Y.; Wu, Z.; Li, Y.; Kong, M.; Kang, Z.; Zuoyuan, B.; Yang, Z. TFRC upregulation promotes ferroptosis in CVB3 infection via nucleus recruitment of Sp1. Cell Death Dis. 2022, 13, 592. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.Y.; Pathak, J.L.; Lin, H.Y.; Guo, W.Q.; Chen, W.J.; Luo, G.; Wang, L.J.; Sun, X.F.; Ding, Y.; Li, J.; et al. Inflammation Triggers Chondrocyte Ferroptosis in TMJOA via HIF-1alpha/TFRC. J. Dent. Res. 2024, 103, 712–722. [Google Scholar]
- Wang, C.; Liang, X.; Tao, C.; Yao, X.; Wang, Y.; Wang, Y.; Li, K. Induction of copper and iron in acute cold-stimulated brown adipose tissues. Biochem. Biophys. Res. Commun. 2017, 488, 496–500. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, Y.; Li, X.; Zhou, J.; Yang, W.; Wang, X.; Jiao, S.; Zuo, W.; You, Z.; Ying, W.; et al. O-GlcNAcylation regulates the stability of transferrin receptor (TFRC) to control the ferroptosis in hepatocellular carcinoma cells. Redox Biol. 2024, 73, 103182. [Google Scholar]
- Brener, L.; Horwitz, R.; Cama, E.; Vu, H.M.K.; Jin, D.; Wu, K.O.E.; Rance, J.; Broady, T.; Treloar, C.; Mao, L.; et al. Understanding stigma and attitudes towards hepatitis B among university students in Australia of Chinese and Vietnamese background. BMC Public Health 2024, 24, 2801. [Google Scholar] [CrossRef]
- Lee, J.; Kim, H.; Kang, Y.W.; Kim, Y.; Park, M.Y.; Song, J.H.; Jo, Y.; Dao, T.; Ryu, D.; Lee, J.; et al. LY6D is crucial for lipid accumulation and inflammation in nonalcoholic fatty liver disease. Exp. Mol. Med. 2023, 55, 1479–1491. [Google Scholar] [CrossRef]
- Wang, J.; Sheng, N.; Li, Y.; Fan, Y.; Nan, X.; Fu, R. Ly6D facilitates chemoresistance in laryngeal squamous cell carcinoma through miR-509/beta-catenin signaling pathway. Am. J. Cancer Res. 2023, 13, 2155–2171. [Google Scholar]
- Semba, T.; Sato, R.; Kasuga, A.; Suina, K.; Shibata, T.; Kohno, T.; Suzuki, M.; Saya, H.; Arima, Y. Lung Adenocarcinoma Mouse Models Based on Orthotopic Transplantation of Syngeneic Tumor-Initiating Cells Expressing EpCAM, SCA-1, and Ly6d. Cancers 2020, 12, 3805. [Google Scholar] [CrossRef]
- Wang, J.; Fan, J.; Gao, W.; Wu, Y.; Zhao, Q.; Chen, B.; Ding, Y.; Wen, S.; Nan, X.; Wang, B. LY6D as a Chemoresistance Marker Gene and Therapeutic Target for Laryngeal Squamous Cell Carcinoma. Stem Cells Dev. 2020, 29, 774–785. [Google Scholar] [CrossRef] [PubMed]
- Okimura, S.; Nishida, N.; Takahashi, H.; Yokoyama, Y.; Yamamoto, H.; Hamabe, A.; Ogino, T.; Miyoshi, N.; Takahashi, H.; Uemura, M.; et al. Lymphocyte Antigen 6 Family Member D (LY6D) Affects Stem Cell Phenotype and Progression of Pancreatic Adenocarcinoma. Anticancer Res. 2024, 44, 4737–4749. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Wu, L.; Zhong, S.; Shan, H.; Luo, J.L. The role of GPI-anchored LY6/uPAR family proteins in connecting membrane microdomains with immune regulation and diseases. Crit. Rev. Oncol. Hematol. 2025, 216, 104971. [Google Scholar] [CrossRef] [PubMed]
- Villarroya, F.; Cereijo, R.; Villarroya, J.; Gavalda-Navarro, A.; Giralt, M. Toward an Understanding of How Immune Cells Control Brown and Beige Adipobiology. Cell Metab. 2018, 27, 954–961. [Google Scholar] [CrossRef]
- Gregory, K.J.; Mason, H.; Casaubon, J.; Schneider, S.S. SFRP1 decreases WNT-Mediated M2 macrophage marker expression in breast tissue. Cancer Immunol. Immunother. 2024, 73, 86. [Google Scholar] [CrossRef]
- Pehlivan, M.; Caliskan, C.; Yuce, Z.; Sercan, H.O. sFRP1 Expression Regulates Wnt Signaling in Chronic Myeloid Leukemia K562 Cells. Anticancer Agents Med. Chem. 2022, 22, 1354–1362. [Google Scholar] [CrossRef]
- Carson, C.; Macias-Velasco, J.F.; Gunawardana, S.; Miranda, M.A.; Oyama, S.; St Pierre, C.L.; Schmidt, H.; Wayhart, J.P.; Lawson, H.A. Brown Adipose Expansion and Remission of Glycemic Dysfunction in Obese SM/J Mice. Cell Rep. 2020, 33, 108237. [Google Scholar] [CrossRef]
- Wang, Y.; Pan, J.; An, F.; Chen, K.; Chen, J.; Nie, H.; Zhu, Y.; Qian, Z.; Zhan, Q. GBP2 is a prognostic biomarker and associated with immunotherapeutic responses in gastric cancer. BMC Cancer 2023, 23, 925. [Google Scholar] [CrossRef]
- Discov, C. Uridine Utilization Drives Pancreatic Cancer in Low-Nutrient Conditions. Cancer Discov. 2023, 13, OF5. [Google Scholar] [CrossRef]
- Li, X.; Liu, J.; Zeng, M.; Yang, K.; Zhang, S.; Liu, Y.; Yin, X.; Zhao, C.; Wang, W.; Xiao, L. GBP2 promotes M1 macrophage polarization by activating the notch1 signaling pathway in diabetic nephropathy. Front. Immunol. 2023, 14, 1127612. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhou, Y.; Zhang, Y.; Fang, S.; Zhang, M.; Li, H.; Xu, F.; Liu, L.; Liu, J.; Zhao, Q.; et al. Subtyping of microsatellite stability colorectal cancer reveals guanylate binding protein 2 (GBP2) as a potential immunotherapeutic target. J. Immunother. Cancer 2022, 10, e004302. [Google Scholar] [CrossRef] [PubMed]
- Okamatsu-Ogura, Y.; Fukano, K.; Tsubota, A.; Nio-Kobayashi, J.; Nakamura, K.; Morimatsu, M.; Sakaue, H.; Saito, M.; Kimura, K. Cell-cycle arrest in mature adipocytes impairs BAT development but not WAT browning, and reduces adaptive thermogenesis in mice. Sci. Rep. 2017, 7, 6648. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Challa, S.; Jones, A.; Kraus, W.L. PARPs and ADP-ribosylation in RNA biology: From RNA expression and processing to protein translation and proteostasis. Genes Dev. 2020, 34, 302–320. [Google Scholar] [CrossRef]
- Aravindhan, S.; Younus, L.A.; Hadi Lafta, M.; Markov, A.; Ivanovna Enina, Y.; Yushchenko, N.A.; Thangavelu, L.; Mostafavi, S.M.; Pokrovskii, M.V.; Ahmadi, M. P53 long noncoding RNA regulatory network in cancer development. Cell Biol. Int. 2021, 45, 1583–1598. [Google Scholar] [CrossRef]
- Donadelli, M.; Dalla Pozza, E.; Scupoli, M.T.; Costanzo, C.; Scarpa, A.; Palmieri, M. Intracellular zinc increase inhibits p53(-/-) pancreatic adenocarcinoma cell growth by ROS/AIF-mediated apoptosis. Biochim. Biophys. Acta 2009, 1793, 273–280. [Google Scholar] [CrossRef]
- Fiorini, C.; Menegazzi, M.; Padroni, C.; Dando, I.; Dalla Pozza, E.; Gregorelli, A.; Costanzo, C.; Palmieri, M.; Donadelli, M. Autophagy induced by p53-reactivating molecules protects pancreatic cancer cells from apoptosis. Apoptosis 2013, 18, 337–346. [Google Scholar] [CrossRef]
- Tabuchi, C.; Sul, H.S. Signaling Pathways Regulating Thermogenesis. Front. Endocrinol. 2021, 12, 595020. [Google Scholar] [CrossRef]
- Prattico, F.; Garajova, I. Focus on Pancreatic Cancer Microenvironment. Curr. Oncol. 2024, 31, 4241–4260. [Google Scholar] [CrossRef]
- Eil, R.; Vodnala, S.K.; Clever, D.; Klebanoff, C.A.; Sukumar, M.; Pan, J.H.; Palmer, D.C.; Gros, A.; Yamamoto, T.N.; Patel, S.J.; et al. Ionic immune suppression within the tumour microenvironment limits T cell effector function. Nature 2016, 537, 539–543. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Kang, B.; Wang, W.; Guo, X.; Bai, T.; Lv, C.; Shen, Y. Identification of Prognostic Genes and Establishment of a Risk Score Model Related to Pancreatic Adenocarcinoma and Brown Adipose Tissue Based on Transcriptomics and Experimental Validation. Genes 2026, 17, 48. https://doi.org/10.3390/genes17010048
Kang B, Wang W, Guo X, Bai T, Lv C, Shen Y. Identification of Prognostic Genes and Establishment of a Risk Score Model Related to Pancreatic Adenocarcinoma and Brown Adipose Tissue Based on Transcriptomics and Experimental Validation. Genes. 2026; 17(1):48. https://doi.org/10.3390/genes17010048
Chicago/Turabian StyleKang, Bin, Weina Wang, Xin Guo, Tong Bai, Chengyu Lv, and Yunzhi Shen. 2026. "Identification of Prognostic Genes and Establishment of a Risk Score Model Related to Pancreatic Adenocarcinoma and Brown Adipose Tissue Based on Transcriptomics and Experimental Validation" Genes 17, no. 1: 48. https://doi.org/10.3390/genes17010048
APA StyleKang, B., Wang, W., Guo, X., Bai, T., Lv, C., & Shen, Y. (2026). Identification of Prognostic Genes and Establishment of a Risk Score Model Related to Pancreatic Adenocarcinoma and Brown Adipose Tissue Based on Transcriptomics and Experimental Validation. Genes, 17(1), 48. https://doi.org/10.3390/genes17010048






