Novel Biallelic INTS1 Variants May Expand the Phenotypic Spectrum of INTS1-Related Disorders—Case Report and Literature Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Whole-Exome Sequencing
2.2. Sanger Sequencing
2.3. Three-Dimensional Modeling of the INTS1 Protein Mutation Site
3. Results
3.1. Clinical Report
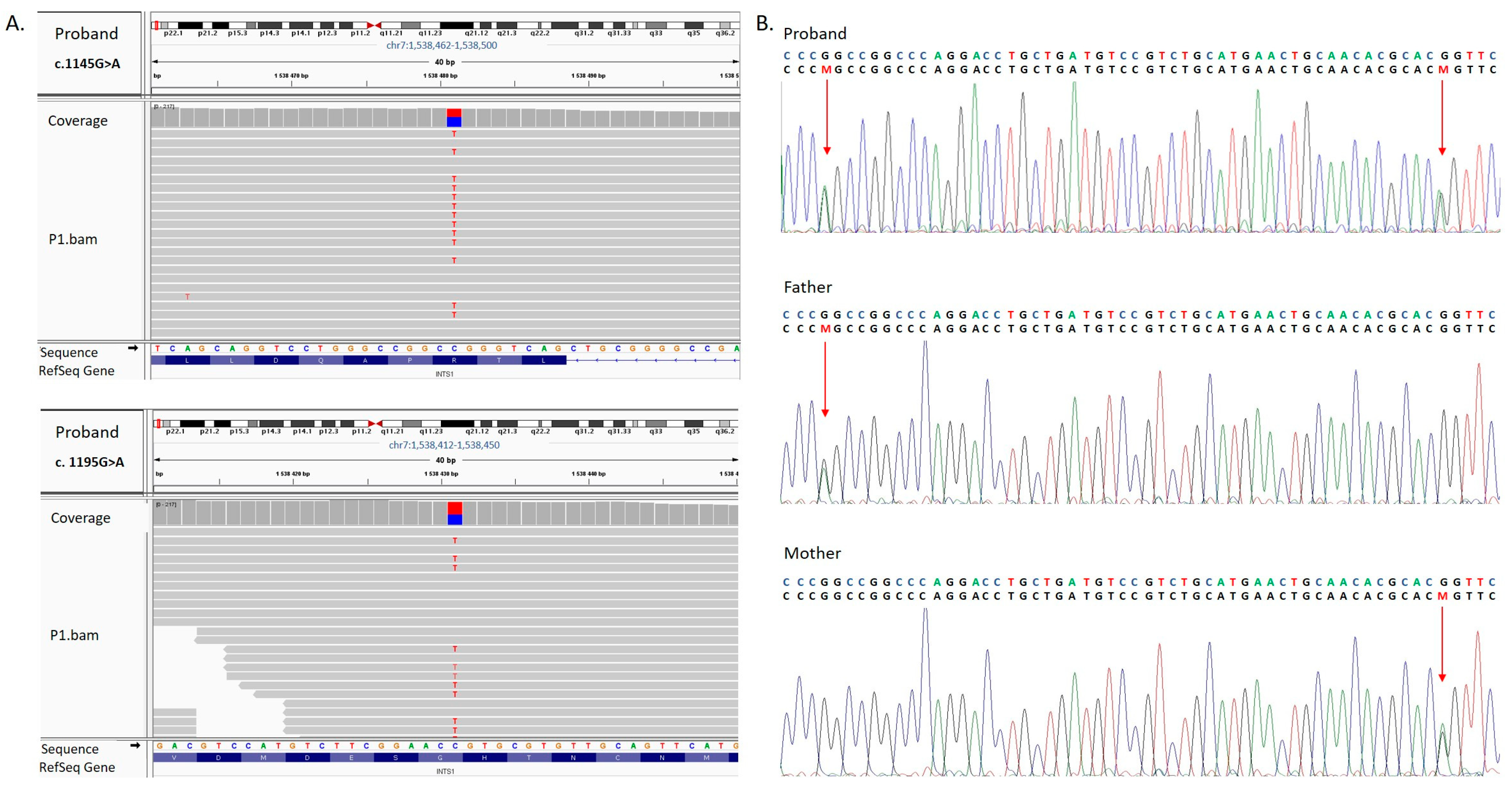
3.2. Molecular Findings and In Silico Assessment
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Manickam, K.; McClain, M.R.; Demmer, L.A.; Biswas, S.; Kearney, H.M.; Malinowski, J.; Massingham, L.J.; Miller, D.; Yu, T.W.; Hisama, F.M. Exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability: An evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2021, 23, 2029–2037. [Google Scholar] [CrossRef] [PubMed]
- Malinowski, J.; Miller, D.T.; Demmer, L.A.; Gannon, J.; Pereira, E.M.; Schroeder, M.C.; Scheuner, M.T.; Tsai, A.C.-H.; Hickey, S.E.; Shen, J. Systematic Evidence-Based Review: Outcomes from Exome and Genome Sequencing for Pediatric Patients with Congenital Anomalies or Intellectual Disability. Genet. Med. 2020, 22, 986–1004. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.; Tang, X.; Weng, W.; Xu, W.; Song, X.; Yang, Y.; Sun, H.; Ye, H.; Zhang, H.; Yu, G.; et al. Diagnostic Utility of Trio-Exome Sequencing for Children with Neurodevelopmental Disorders. JAMA Netw. Open 2025, 8, e251807. [Google Scholar] [CrossRef] [PubMed]
- Oegema, R.; Baillat, D.; Schot, R.; van Unen, L.M.; Brooks, A.; Kia, S.K.; Hoogeboom, A.J.M.; Xia, Z.; Li, W.; Cesaroni, M.; et al. Human Mutations in Integrator Complex Subunits Link Transcriptome Integrity to Brain Development. PLoS Genet. 2017, 13, e1006809. [Google Scholar] [CrossRef] [PubMed]
- Baillat, D.; Hakimi, M.-A.; Näär, A.M.; Shilatifard, A.; Cooch, N.; Shiekhattar, R. Integrator, a Multiprotein Mediator of Small Nuclear RNA Processing, Associates with the C-Terminal Repeat of RNA Polymerase II. Cell 2005, 123, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wagner, E.J. snRNA 3′ End Formation: The Dawn of the Integrator Complex. Biochem. Soc. Trans. 2010, 38, 1082–1087. [Google Scholar] [CrossRef] [PubMed]
- Gardini, A.; Baillat, D.; Cesaroni, M.; Hu, D.; Marinis, J.M.; Wagner, E.J.; Lazar, M.A.; Shilatifard, A.; Shiekhattar, R. Integrator Regulates Transcriptional Initiation and Pause Release via snRNA Processing. Mol. Cell 2014, 56, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Stadelmayer, B.; Micas, G.; Gamot, A.; Martin, P.; Malirat, N.; Koval, S.; Raffel, R.; Sobhian, B.; Severac, D.; Rialle, S.; et al. Integrator Complex Regulates NELF-Mediated RNA Polymerase II Pausing. Nat. Commun. 2014, 5, 5531. [Google Scholar] [CrossRef] [PubMed]
- Krall, M.; Htun, S.; Schnur, R.E.; Brooks, A.S.; Baker, L.; Campomanes, A.; Lamont, R.E.; Gripp, K.W.; Care4Rare Canada Consortium; Schneidman-Duhovny, D.; et al. Biallelic Sequence Variants in INTS1 in Patients with Developmental Delays, Cataracts, and Craniofacial Anomalies. Eur. J. Hum. Genet. 2019, 27, 582–593. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Yang, F.; Tang, J.; Xu, X.; Yang, L.; Yang, X.-A.; Wu, D. Biallelic INTS1 Mutations Cause a Rare Neurodevelopmental Disorder in Two Chinese Siblings. J. Mol. Neurosci. 2020, 70, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Confino, S.; Wexler, Y.; Medvetzky, A.; Elazary, Y.; Ben-Moshe, Z.; Reiter, J.; Dor, T.; Edvardson, S.; Prag, G.; Harel, T.; et al. A Deleterious Variant of INTS1 Leads to Disrupted Sleep–Wake Cycles. Dis. Model. Mech. 2024, 17, dmm050746. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Jin, Q.; Wang, X.; Qi, Y.; Liu, W.; Ren, Y.; Zhao, D.; Chen, F.X.; Cheng, J.; Chen, X.; et al. Structural basis of INTAC-regulated transcription. Protein Cell. 2023, 14, 698–702. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Qi, Y.; Hu, S.; Cao, X.; Xu, C.; Yin, Z.; Chen, X.; Li, Y.; Liu, W.; Li, J.; et al. Identification of Integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase. Science 2020, 370, eabb5872. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.V.; Ascher, D.B.; Blundell, T.L. MCSM: Predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 2014, 30, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Fenton, T.R.; Kim, J.H. A systematic review and meta-analysis to revise the Fenton growth chart for preterm infants. BMCadhą Pediatrics. 2013, 13, 59. [Google Scholar] [CrossRef] [PubMed]
- Fianu, I.; Chen, Y.; Dienemann, C.; Dybkov, O.; Linden, A.; Urlaub, H.; Cramer, P. Structural basis of Integrator-mediated transcription regulation. Science 2021, 374, 883–887. [Google Scholar] [CrossRef]
- Pfleiderer, M.M.; Galej, W.P. Structure of the Catalytic Core of the Integrator Complex. Mol. Cell 2021, 81, 1246–1259.e8. [Google Scholar] [CrossRef] [PubMed]
- Chamova, T.; Zlatareva, D.; Raycheva, M.; Bichev, S.; Kalaydjieva, L.; Tournev, I. Cognitive Impairment and Brain Imaging Characteristics of Patients with Congenital Cataracts, Facial Dysmorphism, Neuropathy Syndrome. Behav. Neurol. 2015, 2015, 639539. [Google Scholar] [CrossRef] [PubMed]


| − | This Study | Previously Reported Cases | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family 1 | Family 2 | Family 3 | − | − | − | − | Family 4 | |||||||
| Features | Our Patient | P2 [10] a | P3 [10] | P4 [9] | P5 [9] | P6 [9] | P7 [9] | P8 [9] | P9 [4] | P10 [4] | P11 [4] | P12 [11] | P13 [11] | Total |
| Variant: NM_004429.4 | c.1145G>A, p. (Arg382Gln); c.1195G>A, p. (Gly399Ser) | c.1645A>G, p. Met549Val; c.5881C>T, p. Gln1961Ter | c.229C>T, p. (Arg77Cys); c.5398dupC, p. (Arg1800ProfsTer20) | c.5621C>T, p. (Pro1874Leu); c.4207G>A, p. (Val1403Met) | c.5290delC, p. (Leu1764CysfsTer16); c.6491T>C, p. (Leu2164Pro) | c.5351C>A,p. (Ser1784Ter), | c.5351C>A,p. (Ser1784Ter) | c.5351C>A,p. (Ser1784Ter) | c.5224G>A,p. (Glu1742Lys); c.560C>T, p. (Thr187Met) | |||||
| Zygosity | Het b | Het | Het | Het | Het | Hom c | Hom | Het | Hom | Hom | Hom | Hom | Hom | |
| Sex | F | F | M | M | M | M | F | M | F | M | M | M | F | |
| Age at last examination | 9 yr | 5 yr | 11 yr | 14 yr | 9 yr | 11 yr | 6 yr | 21 mo | 10 yr | 19 yr | 6 yr | 9 yr | 7 yr | |
| Short stature | + | + | + | + | + | + | + | + | + | + | + | + | + | 13/13 |
| Facial Dysmorphism | ||||||||||||||
| Frontal Bossing | + | + | − | − | − | + | + | − | + | + | + | − | − | 7/13 |
| High anterior hairline | + | + | − | − | − | − | − | − | + | + | + | − | − | 5/13 |
| Horizontal eyebrows | + | − | + | + | + | − | − | − | + | + | + | − | − | 7/13 |
| Hypertelorism | − | + | + | + | + | + | + | − | + | + | + | + | + | 11/13 |
| Broad nasal bridge | + | + | + | + | + | + | + | − | + | + | + | + | − | 11/13 |
| Bulbous nasal tip | + | + | + | + | + | − | − | + | + | + | + | − | − | 9/13 |
| Low-set ears | + | − | − | + | + | − | − | − | + | + | + | − | − | 6/13 |
| Dysplastic/Prominent ears | + | − | − | + | + | − | − | − | +/− | + | + | − | − | 5/13 |
| Deep and long philtrum | + | − | − | + | + | − | − | − | − | − | − | − | − | 3/13 |
| Downturned corners of mouth | − | + | − | + | + | + | + | − | + | + | + | − | − | 8/13 |
| Abnormal dentition | + | + | ND d | + | + | + | − | − | + | + | + | − | + | 8/12 |
| Neurological and Neurodevelopmental | ||||||||||||||
| Absent speech/severely impaired speech | − * | + | + | + | + | + | + | + | + | + | + | + | + | 12/13 |
| Cognitive impairment | + ** | + | + | + | + | + | + | + | + | + | + | + | + | 13/13 |
| Motor impairment | + | + | + | + | + | + | + | + | + | + | + | + | + | 13/13 |
| Seizure | − | − | − | − | − | − | − | − | + | − | − | + | + | 3/13 |
| Hypotonia | + | ND | ND | + | + | + | + | + | + | + | + | + | + | 11/11 |
| Autism | + | + | − | − | + | + | + | − | − | − | − | − | − | 5/13 |
| Ocular | ||||||||||||||
| Cataract | − | + | + | + | + | + | + | + | + | + | + | + | + | 12/13 |
| Strabismus | + | + | + | − | − | + | − | − | + | + | + | + | − | 9/13 |
| Abnormality of refraction | + | ND | ND | + | − | + | + | − | − | + | + | ND | ND | 6/9 |
| Skeletal | ||||||||||||||
| Scoliosis | + | ND | ND | − | + | − | − | − | − | − | + | − | + | 4/11 |
| Hip dysplasia | + | ND | ND | − | − | − | − | − | − | + | + | − | − | 3/11 |
| Pectus abnormality | + | + | − | + | + | − | − | − | + | + | + | − | − | 7/13 |
| Overlapping toes | + | ND | ND | + | + | − | − | − | + | + | + | − | − | 6/11 |
| Visceral anomalies | ||||||||||||||
| Renal malformation | + | ND | ND | − | + | − | − | + | − | + | − | − | − | 4/11 |
| Congenital heart disease | + | ND | ND | − | − | − | − | − | − | + | + | + | − | 4/11 |
| c.1145G>A p.(Arg382Gln) | c.1195G>A p.(Gly399ser) | |
|---|---|---|
| gDNA level | Chr7(GRCh38):g.1498845C>T | Chr7(GRCh38):g1498795C>T |
| dbSNP rs number | rs1782994531 | rs1439392344 |
| Exon | 9 | 9 |
| gnomAD Exomes (v4) | Total: 0.00000138; NFE: 0.0000009027 | Total: 0.0000248, NFE: 0.0000307 |
| gnomAD Genomes (v4) | - | Total: 0.0000131, NFE: 0.0000294 |
| ACMG variant classification | Uncertain significance | Uncertain significance |
| Criteria | PP4 Supporting PM2 Moderate | PP4 Supporting PM2 Moderate BP4 Moderate |
| mCSM: Protein stability change (ΔΔG) | Stabilizing (0.136 kcal/mol) | Destabilizing (−0.616 kcal/mol) |
| mCSM: Protein–DNA affinity change (ΔΔG) | Destabilizing (−0.912 kcal/mol) | Destabilizing (−0.082 kcal/mol) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wnuk-Kłosińska, A.; Sowińska-Seidler, A.; Piechota, M.; Jamsheer, A. Novel Biallelic INTS1 Variants May Expand the Phenotypic Spectrum of INTS1-Related Disorders—Case Report and Literature Review. Genes 2025, 16, 1081. https://doi.org/10.3390/genes16091081
Wnuk-Kłosińska A, Sowińska-Seidler A, Piechota M, Jamsheer A. Novel Biallelic INTS1 Variants May Expand the Phenotypic Spectrum of INTS1-Related Disorders—Case Report and Literature Review. Genes. 2025; 16(9):1081. https://doi.org/10.3390/genes16091081
Chicago/Turabian StyleWnuk-Kłosińska, Aleksandra, Anna Sowińska-Seidler, Michał Piechota, and Aleksander Jamsheer. 2025. "Novel Biallelic INTS1 Variants May Expand the Phenotypic Spectrum of INTS1-Related Disorders—Case Report and Literature Review" Genes 16, no. 9: 1081. https://doi.org/10.3390/genes16091081
APA StyleWnuk-Kłosińska, A., Sowińska-Seidler, A., Piechota, M., & Jamsheer, A. (2025). Novel Biallelic INTS1 Variants May Expand the Phenotypic Spectrum of INTS1-Related Disorders—Case Report and Literature Review. Genes, 16(9), 1081. https://doi.org/10.3390/genes16091081






