CREPE (CREate Primers and Evaluate): A Computational Tool for Large-Scale Primer Design and Specificity Analysis
Abstract
1. Introduction
2. Methods
2.1. CREPE Pipeline Software Versions
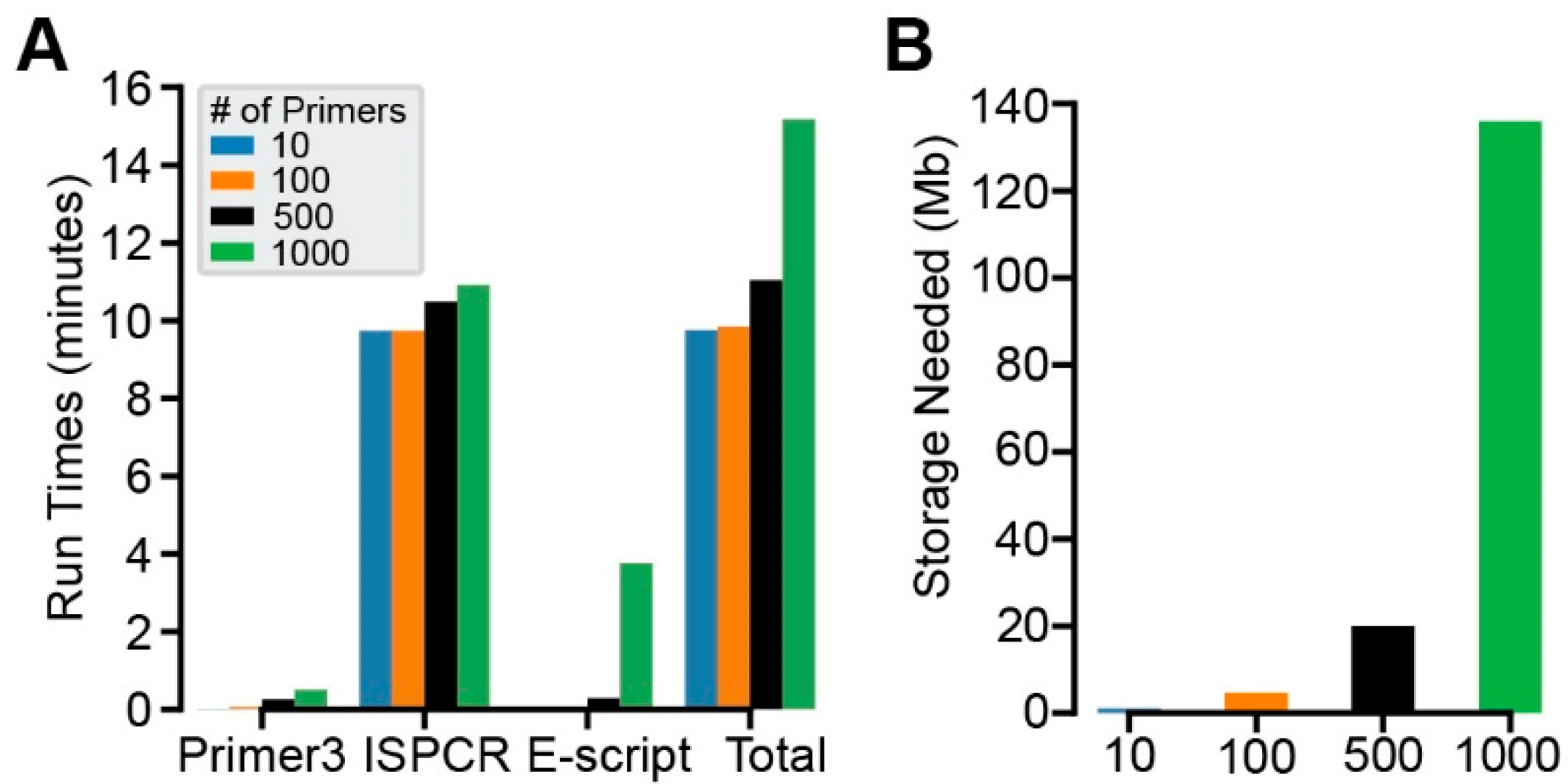
2.2. Run Time and Local Storage Testing
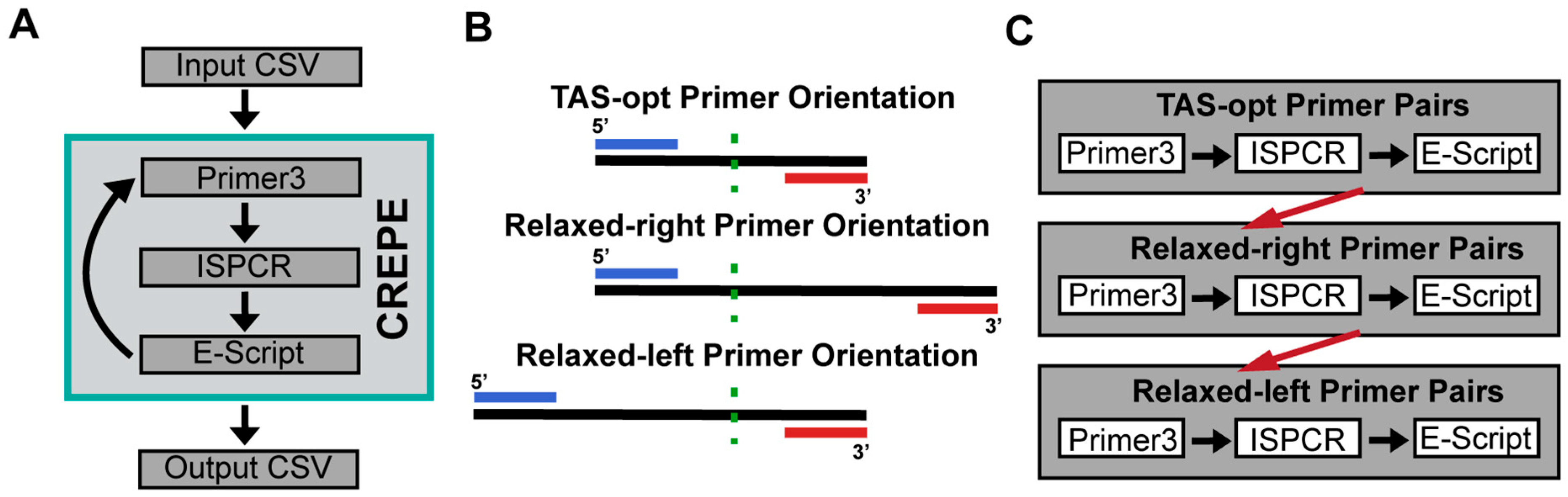
2.3. CREPE Pipeline Primer3 and ISPCR Overview
2.4. CREPE Pipeline Evaluation Script (Off-Target Assessment)
2.5. CREPE Pipeline Output File Format
2.6. Targeted Amplicon Sequencing
2.7. TAS Analysis
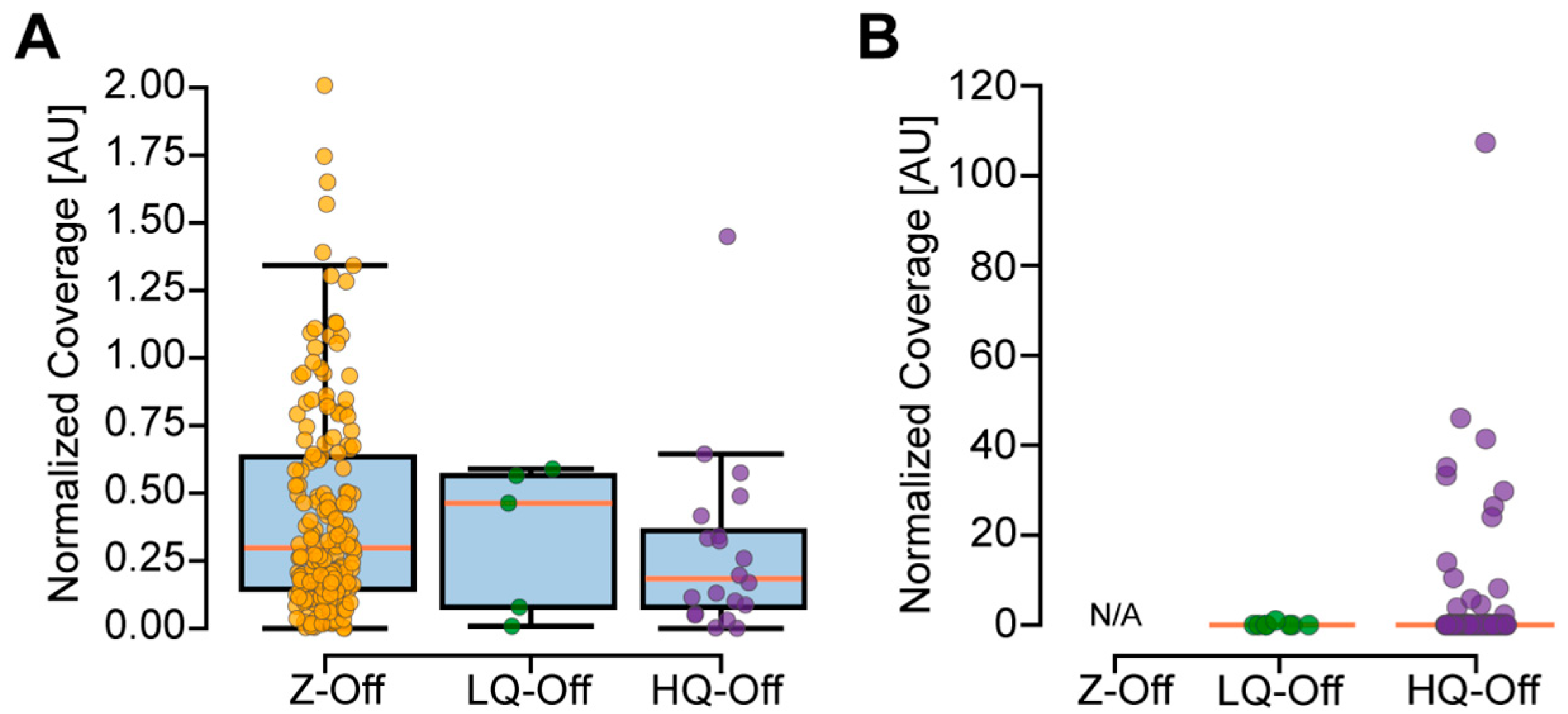
2.8. Measuring Off-Target Coverage
2.9. Data Analysis and Visualization
2.10. Human Subject Statement
2.11. DNA Sample Extraction
3. Results and Discussion
3.1. Overview of the Approach
3.2. Performance and Storage Needs
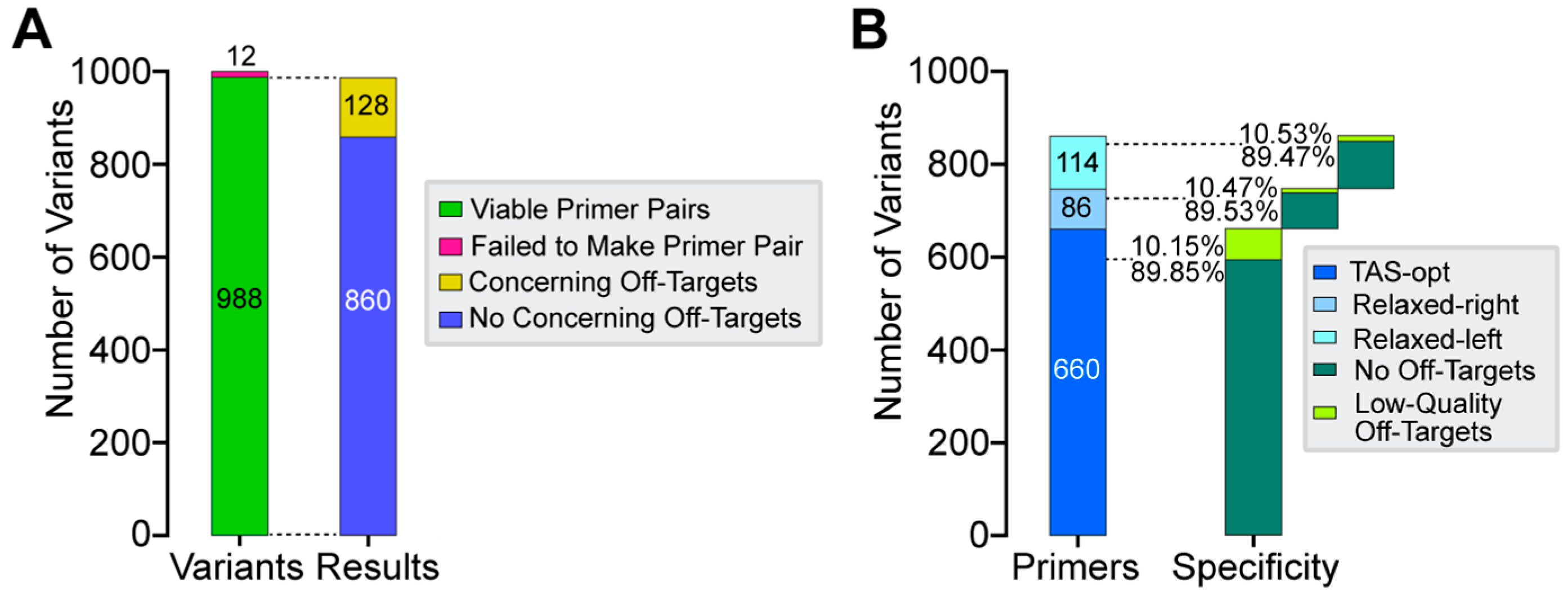
3.3. In Silico Performance of CREPE on 1000 ClinVar Variants
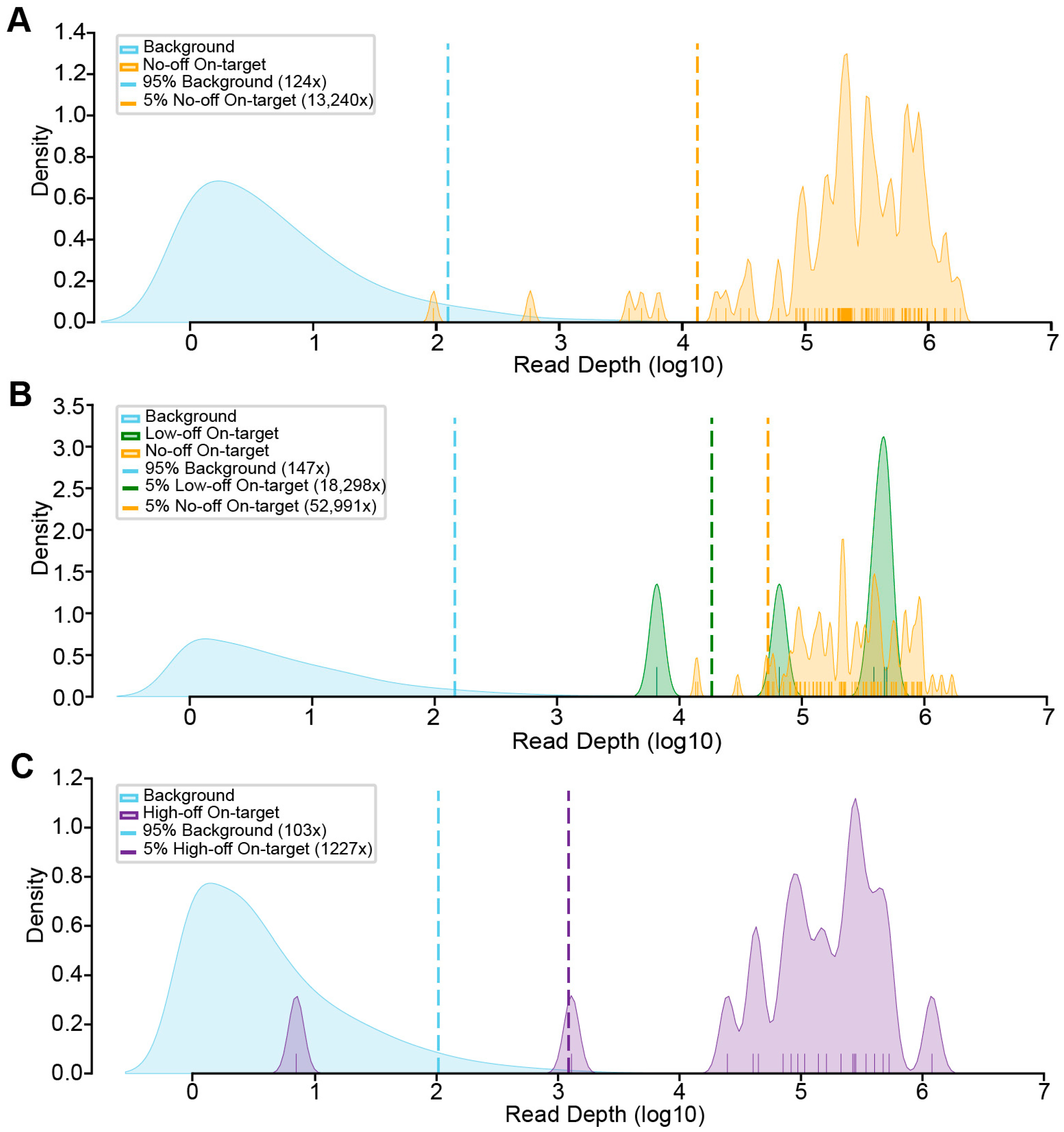
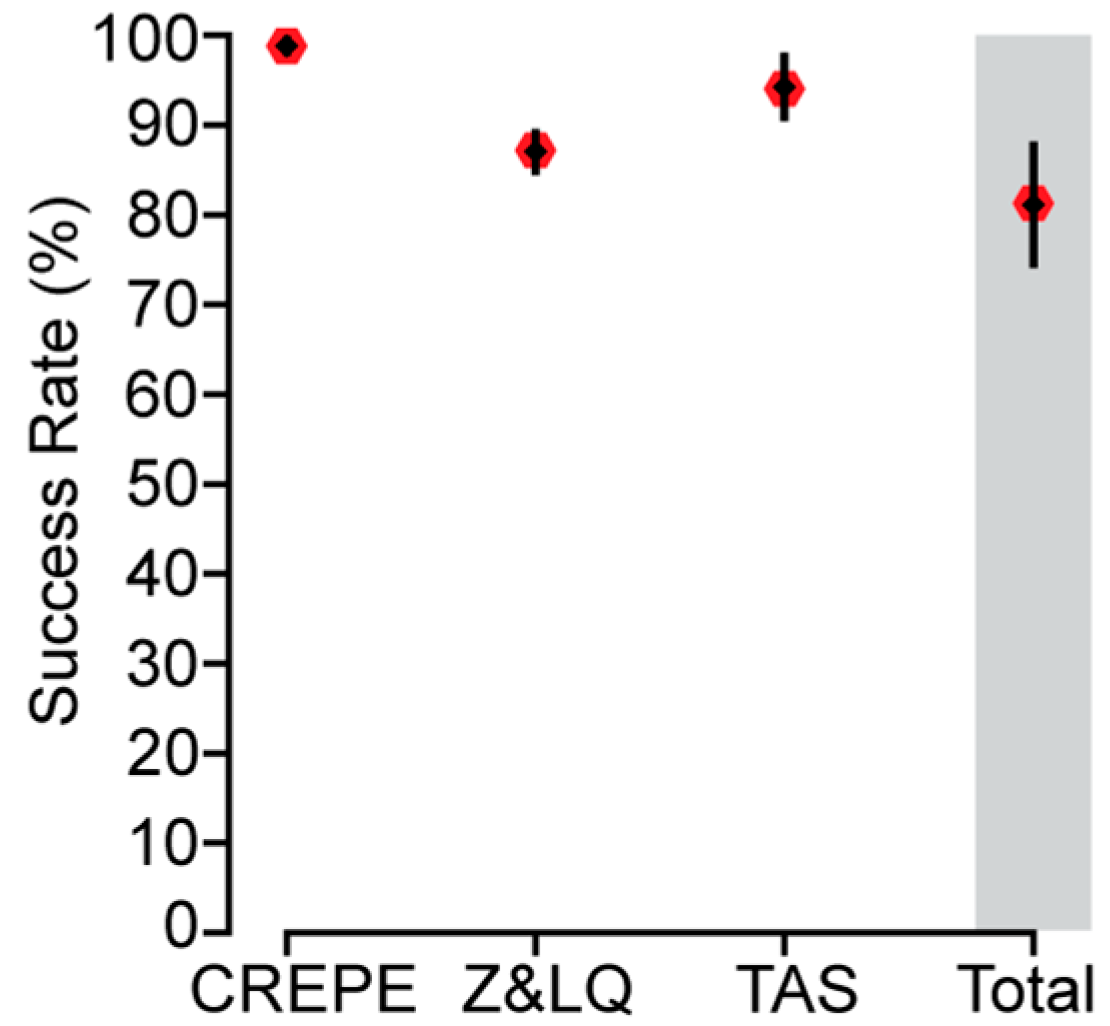
3.4. Experimental Evaluation of CREPE Performance
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mullis, K.B. The Unusual Origin of the Polymerase Chain-Reaction. Sci. Am. 1990, 262, 56–65. [Google Scholar] [CrossRef]
- Breuss, M.W.; Antaki, D.; George, R.D.; Kleiber, M.; James, K.N.; Ball, L.L.; Hong, O.; Mitra, I.; Yang, X.; Wirth, S.A.; et al. Autism risk in offspring can be assessed through quantification of male sperm mosaicism. Nat. Med. 2020, 26, 143–150. [Google Scholar] [CrossRef]
- Doan, R.N.; Miller, M.B.; Kim, S.N.; Rodin, R.E.; Ganz, J.; Bizzotto, S.; Morillo, K.S.; Huang, A.Y.; Digumarthy, R.; Zemmel, Z.; et al. MIPP-Seq: Ultra-sensitive rapid detection and validation of low-frequency mosaic mutations. BMC Med. Genom. 2021, 14, 47. [Google Scholar] [CrossRef]
- Xu, X.; Yang, X.; Wu, Q.; Liu, A.; Yang, X.; Ye, A.Y.; Huang, A.Y.; Li, J.; Wang, M.; Yu, Z.; et al. Amplicon Resequencing Identified Parental Mosaicism for Approximately 10% of “de novo” SCN1A Mutations in Children with Dravet Syndrome. Hum. Mutat. 2015, 36, 861–872. [Google Scholar] [CrossRef]
- Yang, X.; Breuss, M.W.; Xu, X.; Antaki, D.; James, K.N.; Stanley, V.; Ball, L.L.; George, R.D.; Wirth, S.A.; Cao, B.; et al. Developmental and temporal characteristics of clonal sperm mosaicism. Cell 2021, 184, 4772–4783. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef]
- You, F.M.; Huo, N.; Gu, Y.Q.; Luo, M.C.; Ma, Y.; Hane, D.; Lazo, G.R.; Dvorak, J.; Anderson, O.D. BatchPrimer3: A high throughput web application for PCR and sequencing primer design. BMC Bioinform. 2008, 9, 253. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.R. Target Enrichment Approaches for Next-Generation Sequencing Applications in Oncology. Diagnostics 2022, 12, 1539. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Perez, G.; Barber, G.P.; Benet-Pages, A.; Casper, J.; Clawson, H.; Diekhans, M.; Fischer, C.; Gonzalez, J.N.; Hinrichs, A.S.; Lee, C.M.; et al. The UCSC Genome Browser database: 2025 update. Nucleic Acids Res. 2025, 53, D1243–D1249. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Kent, W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef]
- Kalendar, R. A Guide to Using FASTPCR Software for PCR, in Silico PCR, and Oligonucleotide Analysis. Methods Mol. Biol. 2022, 2392, 223–243. [Google Scholar] [CrossRef] [PubMed]
- Hysom, D.A.; Naraghi-Arani, P.; Elsheikh, M.; Carrillo, A.C.; Williams, P.L.; Gardner, S.N. Skip the alignment: Degenerate, multiplex primer and probe design using K-mer matching instead of alignments. PLoS ONE 2012, 7, e34560. [Google Scholar] [CrossRef]
- Kechin, A.; Borobova, V.; Boyarskikh, U.; Khrapov, E.; Subbotin, S.; Filipenko, M. NGS-PrimerPlex: High-throughput primer design for multiplex polymerase chain reactions. PLoS Comput. Biol. 2020, 16, e1008468. [Google Scholar] [CrossRef] [PubMed]
- Andreas Heger, J.M.; Kevin, J. Pysam: Htslib Interface for Python. Available online: https://github.com/pysam-developers/pysam (accessed on 7 September 2025).
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- The Pandas Development Team. Pandas-dev/Pandas: Pandas. Available online: https://doi.org/10.5281/zenodo.3509134 (accessed on 9 September 2025).
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Broad Institute. Picard Toolkit. Available online: https://broadinstitute.github.io/picard/ (accessed on 7 September 2025).
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Seabold, S.; Josef, P. Statsmodels: Econometric and statistical modeling with python. In Proceedings of the Python in Science Conference 2010, Austin, TX, USA, 28 June–3 July 2010. [Google Scholar]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Waskom, M.L. Seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef] [PubMed]
| Primer Dataset | TAS-opt | Relaxed-Right | Relaxed-Left | Total |
|---|---|---|---|---|
| No-Off | 76 | 6 | 13 | 95 |
| Low-Off | 71 | 8 | 17 | 96 |
| High-Off | 16 | 2 | 3 | 21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pitsch, J.W.; Wirth, S.A.; Costantino, N.T.; Mejia, J.; Doss, R.M.; Warren, A.V.A.; Ustanik, J.; Yang, X.; Breuss, M.W. CREPE (CREate Primers and Evaluate): A Computational Tool for Large-Scale Primer Design and Specificity Analysis. Genes 2025, 16, 1062. https://doi.org/10.3390/genes16091062
Pitsch JW, Wirth SA, Costantino NT, Mejia J, Doss RM, Warren AVA, Ustanik J, Yang X, Breuss MW. CREPE (CREate Primers and Evaluate): A Computational Tool for Large-Scale Primer Design and Specificity Analysis. Genes. 2025; 16(9):1062. https://doi.org/10.3390/genes16091062
Chicago/Turabian StylePitsch, Jonathan W., Sara A. Wirth, Nicole T. Costantino, Josh Mejia, Rose M. Doss, Ava V. A. Warren, Jack Ustanik, Xiaoxu Yang, and Martin W. Breuss. 2025. "CREPE (CREate Primers and Evaluate): A Computational Tool for Large-Scale Primer Design and Specificity Analysis" Genes 16, no. 9: 1062. https://doi.org/10.3390/genes16091062
APA StylePitsch, J. W., Wirth, S. A., Costantino, N. T., Mejia, J., Doss, R. M., Warren, A. V. A., Ustanik, J., Yang, X., & Breuss, M. W. (2025). CREPE (CREate Primers and Evaluate): A Computational Tool for Large-Scale Primer Design and Specificity Analysis. Genes, 16(9), 1062. https://doi.org/10.3390/genes16091062








