Identifying Key Genes of Proanthocyanidin Intervention in Fluoride-Induced Liver Injury: Integrated Molecular Docking and Experimental Validation
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Rearing and Treatment
2.2. Main Reagents and Instruments
2.3. Determination of Serum ALT and AST
2.4. Western Blot
2.5. Statistical Analysis
2.6. Prediction of Potential Targets of PC
2.7. Prediction of Potential Targets for Fluoride-Induced Liver Injury
2.8. Construction of PPI Network and Screening of Key Targets
2.9. GO and KEGG Enrichment Analysis
2.10. Construction of the miRNA Regulatory Network for Key Targets
2.11. Molecular Docking
3. Results
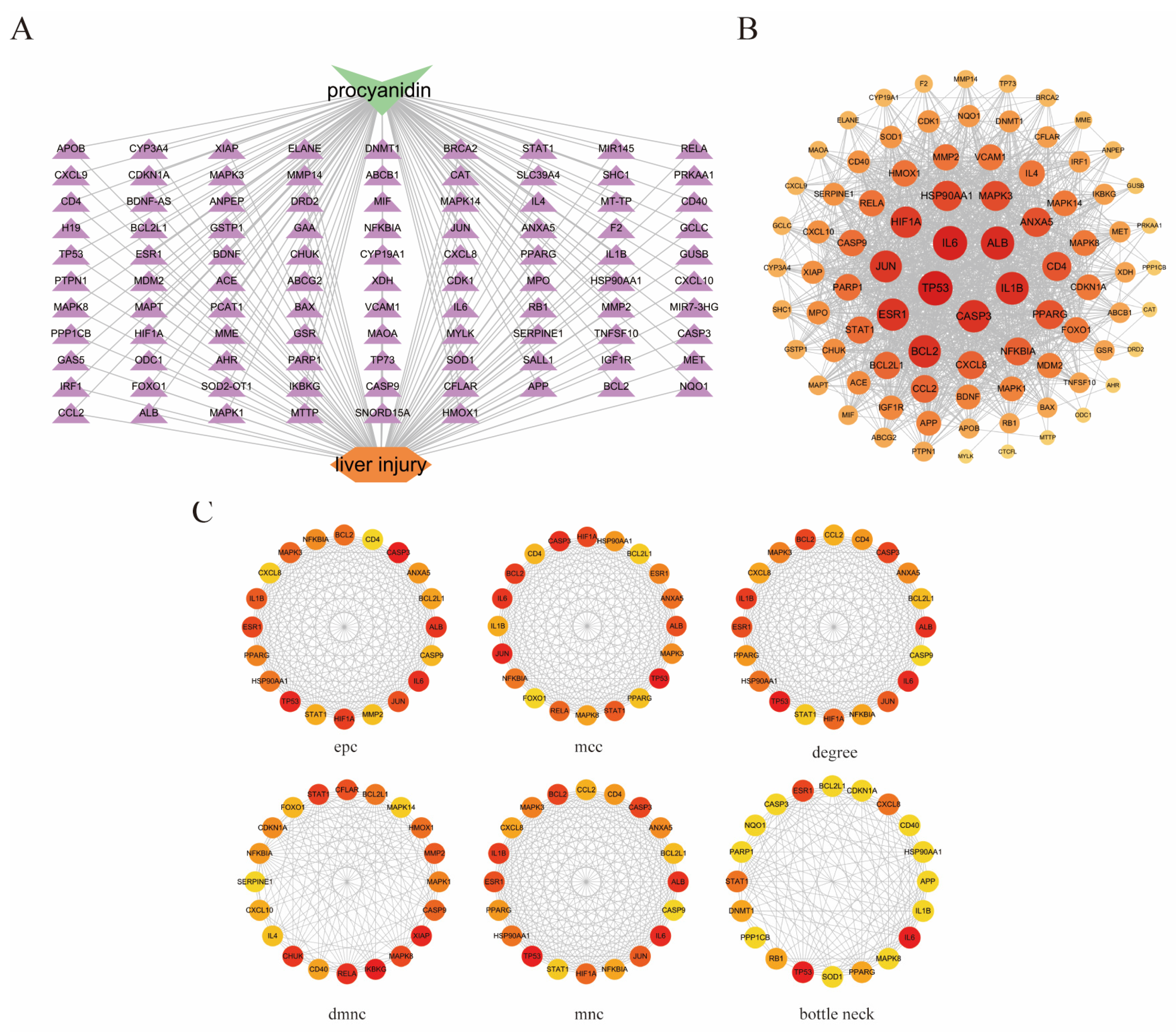
3.1. Prediction of Potential Targets for PC and Fluoride-Induced Liver Injury
3.2. Common Targets of Proanthocyanidins and Fluoride-Induced Liver Injury
3.3. Construction of PPI Network and Screening of Key Targets
3.4. GO and KEGG Pathway Enrichment Analysis
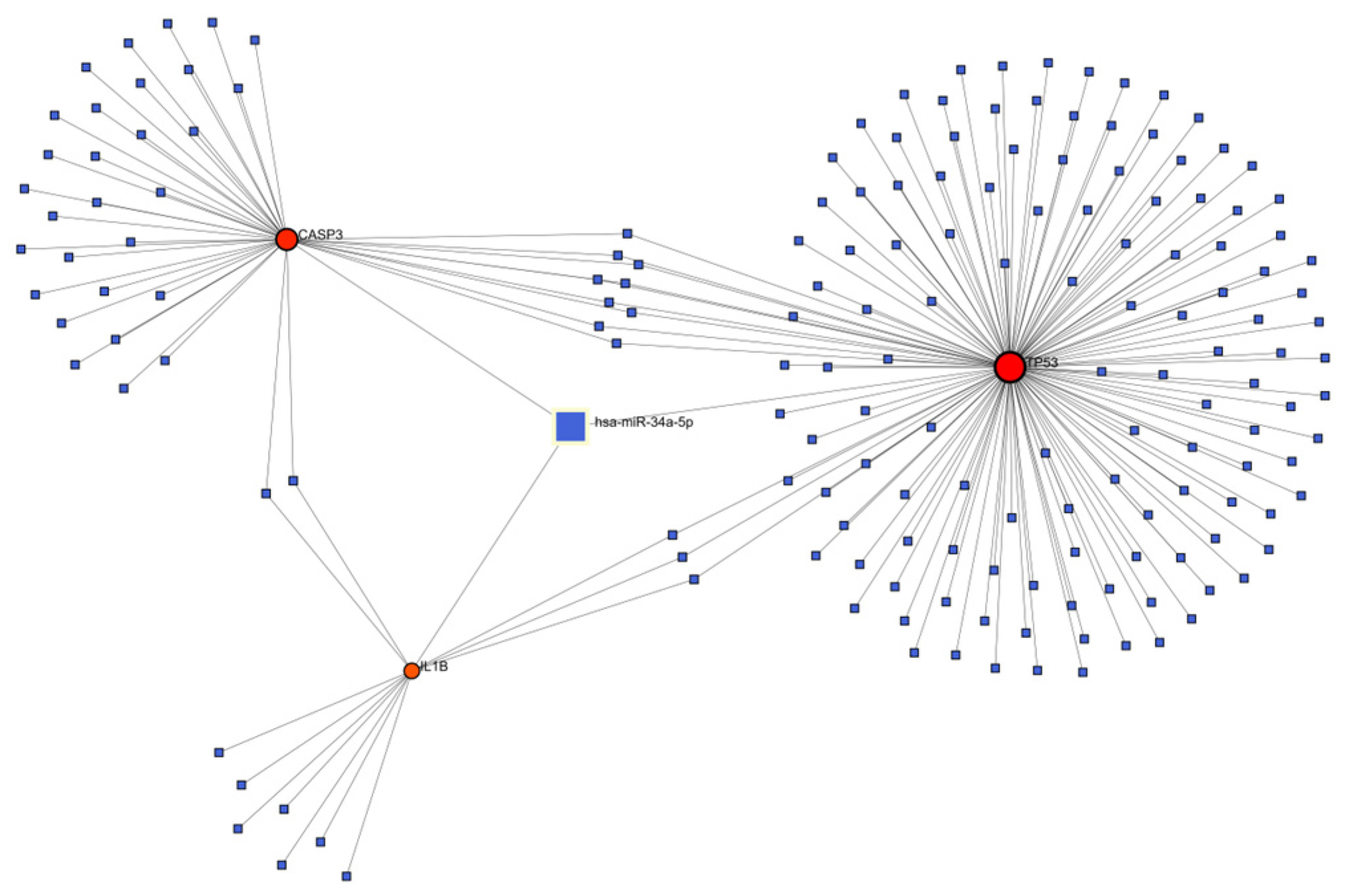
3.5. Construction of Genes-miRNA Network for Key Targets
3.6. The Results of Molecular Docking
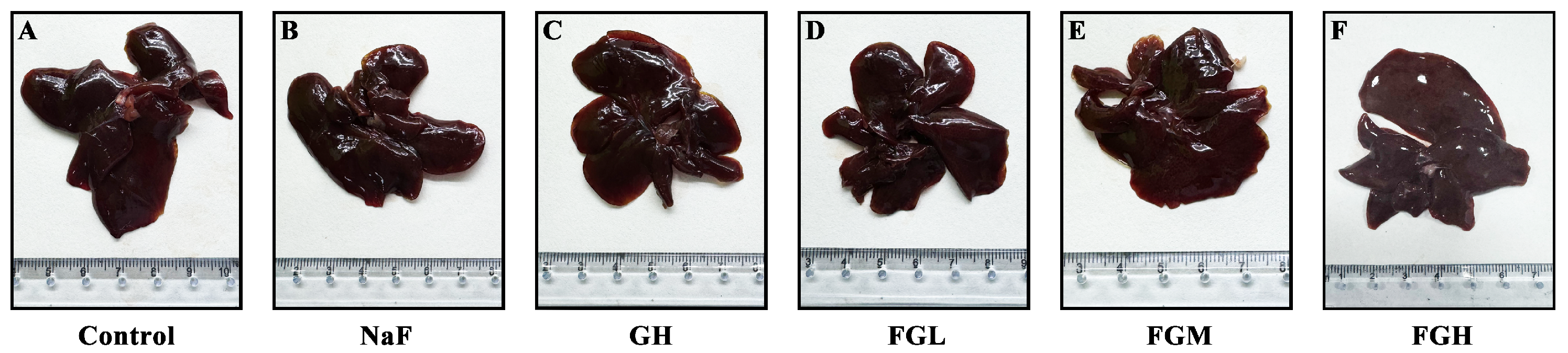
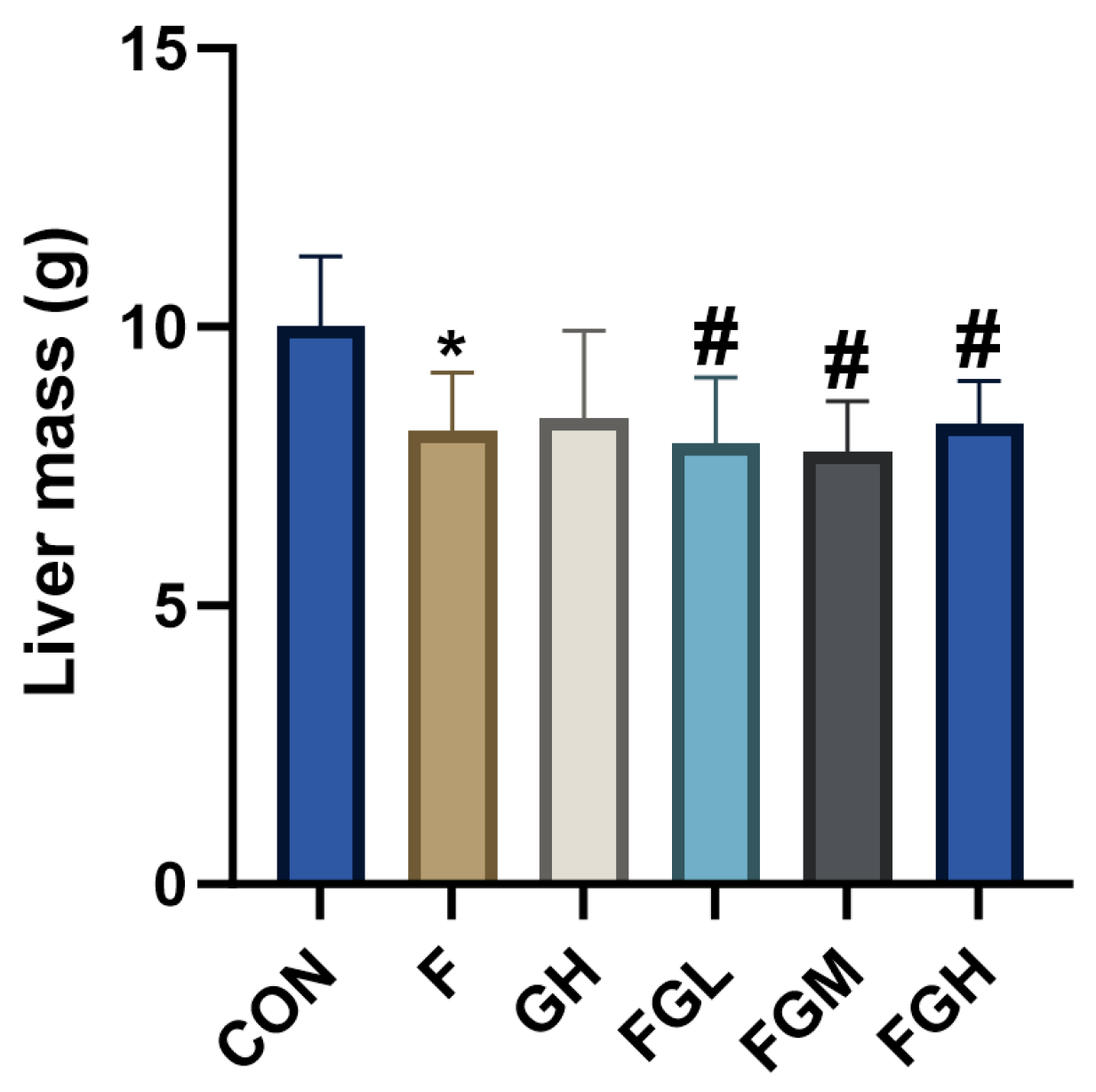
3.7. Histological Evidence of GSPE Alleviating Liver Injury in Rats Caused by Fluoride Poisoning
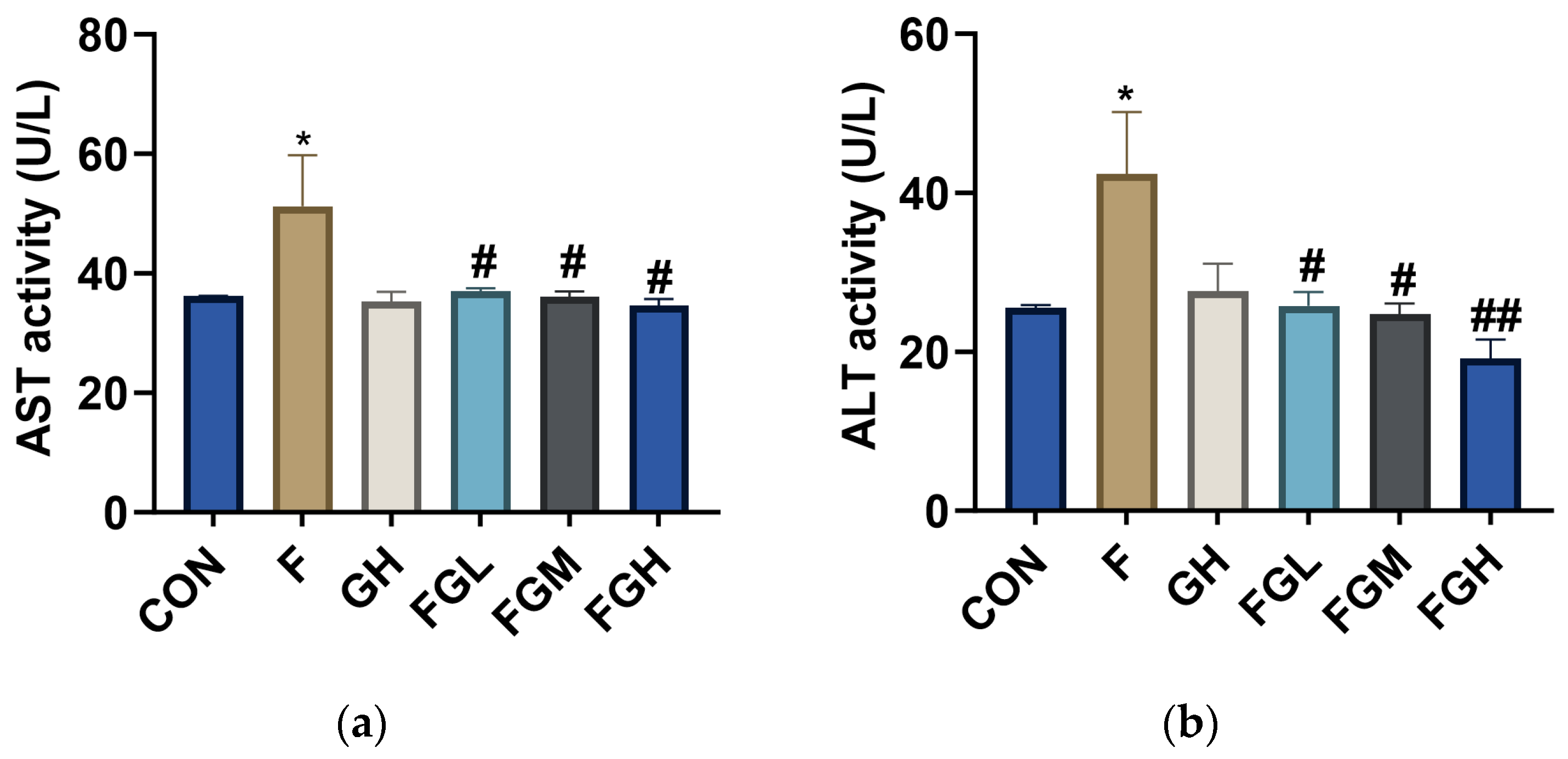
3.8. The Effects of NaF and GSPE on the Expression of ALT and AST in the Liver
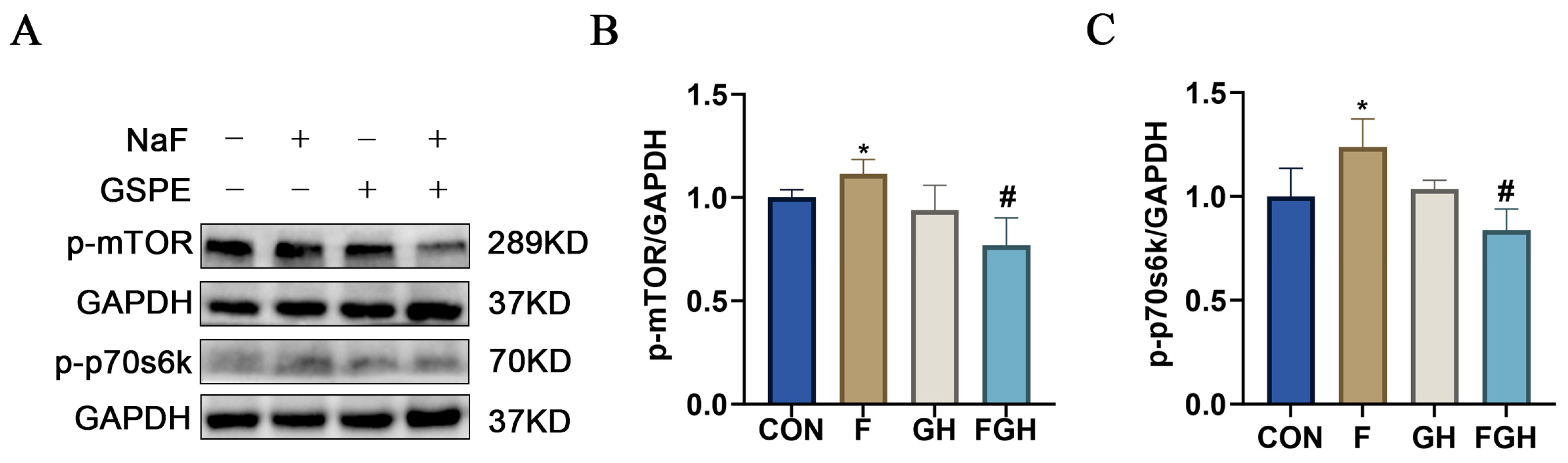
3.9. The Effects of NaF and GSPE on the Expression of the mTOR/P70s6k Signaling Pathway in the Liver
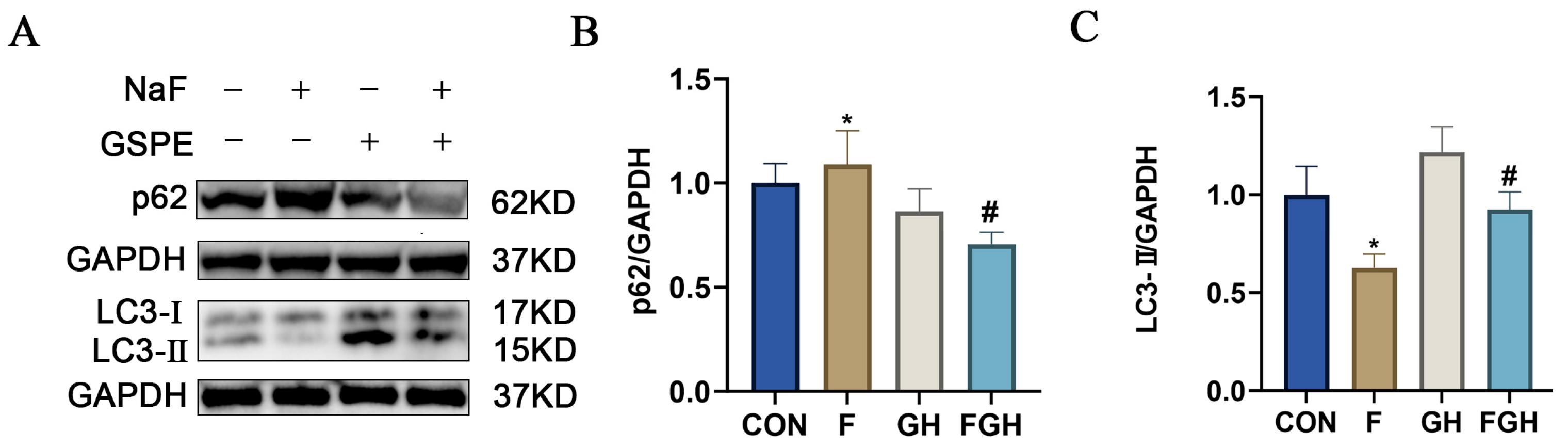
3.10. The Effects of NaF and GSPE on the Expression of Autophagy-Related Proteins in the Liver
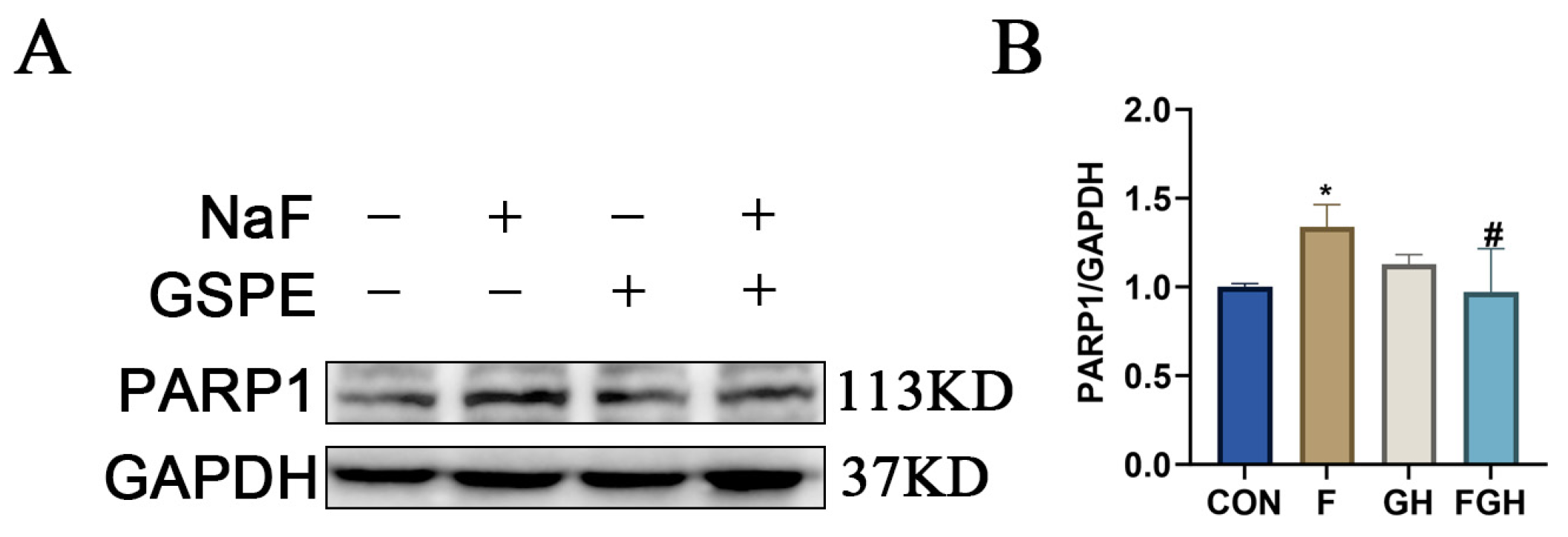
3.11. The Effects of NaF and GSPE on the Expression of Apoptosis-Related Proteins in the Liver
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NaF | Sodium fluoride |
| PC | Proanthocyanidin |
| GSPE | Grape seed proanthocyanidin extract |
| AST | Aspartate aminotransferase |
| ALT | Alanine aminotransferase |
| GO | Gene ontology |
| KEGG | Kyoto encyclopedia of genes and genome |
References
- Vasisth, D.; Mehra, P.; Yadav, L.; Kumari, V.; Bhatia, U.; Garg, R. Fluoride and its Implications on Oral Health: A Review. J. Pharm. Bioallied Sci. 2024, 16 (Suppl. S1), S49–S52. [Google Scholar] [CrossRef]
- Whelton, H.; Spencer, A.; Do, L.; Rugg-Gunn, A. Fluoride Revolution and Dental Caries: Evolution of Policies for Global Use. J. Dent. Res. 2019, 98, 837–846. [Google Scholar] [CrossRef] [PubMed]
- O’mullane, D.M.; Baez, R.J.; Jones, S.; Lennon, M.A.; Petersen, P.E.; Rugg-Gunn, A.J.; Whelton, H.; Whitford, G.M. Fluoride and Oral Health. Community Dent. Health 2016, 33, 69–99. [Google Scholar] [PubMed]
- Larsen, L.S.; Nyvad, B.; Baelum, V. Salivary fluoride levels after daily brushing with 5000 ppm fluoride toothpaste: A randomised, controlled clinical trial. Eur. J. Oral Sci. 2023, 131, e12934. [Google Scholar] [CrossRef] [PubMed]
- Adelakun, S.A.; Akintunde, O.W.; Ogunlade, B. Fluoride-induced testicular degeneration and sperm quality deteriorations: Salutary role of Cyperus esculentus tubers (tiger nut) extract in animal model. Rev. Int. Androl. 2021, 19, 201–212. [Google Scholar] [CrossRef]
- Yu, Y.-M.; Zhou, B.-H.; Yang, Y.-L.; Guo, C.-X.; Zhao, J.; Wang, H.-W. Estrogen Deficiency Aggravates Fluoride-Induced Liver Damage and Lipid Metabolism Disorder in Rats. Biol. Trace Elem. Res. 2022, 200, 2767–2776. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, X.; Zhan, X. Antagonistic effects of different selenium sources on growth inhibition, oxidative damage, and apoptosis induced by fluorine in broilers. Poult. Sci. 2018, 97, 3207–3217. [Google Scholar] [CrossRef]
- Tian, X.; Zhang, H.; Zhao, Y.; Mehmood, K.; Wu, X.; Chang, Z.; Luo, M.; Liu, X.; Ijaz, M.; Javed, M.T.; et al. Transcriptome analysis reveals the molecular mechanism of hepatic metabolism disorder caused by chromium poisoning in chickens. Environ. Sci. Pollut. Res. Int. 2018, 25, 15411–15421. [Google Scholar] [CrossRef]
- Fujioka, Y.; Noda, N.N. Biomolecular condensates in autophagy regulation. Curr. Opin. Cell Biol. 2021, 69, 23–29. [Google Scholar] [CrossRef]
- Xin, Y.; Jiang, F.; Yang, C.; Yan, Q.; Guo, W.; Huang, Q.; Zhang, L.; Jiang, G. Role of autophagy in regulating the radios ensitivity of tumor cells. J. Cancer Res. Clin. Oncol. 2017, 143, 2147–2157. [Google Scholar] [CrossRef]
- Ma, J.; Gao, S.-S.; Yang, H.-J.; Wang, M.; Cheng, B.-F.; Feng, Z.-W.; Wang, L. Neuroprotective Effects of Proanthocyanidins, Natural Flavonoids Derived From Plants, on Rotenone-Induced Oxidative Stress and Apoptotic Cell Death in Human Neuroblastoma SH-SY5Y Cells. Front. Neurosci. 2018, 12, 369. [Google Scholar] [CrossRef]
- Niu, Q.; Mu, L.; Li, S.; Xu, S.; Ma, R.; Guo, S. Proanthocyanidin Protects Human Embryo Hepatocytes from Fluoride-Induced Oxidative Stress by Regulating Iron Metabolism. Biol. Trace Elem. Res. 2016, 169, 174–179. [Google Scholar] [CrossRef]
- Rodríguez-Pérez, C.; García-Villanova, B.; Guerra-Hernández, E.; Verardo, V. Grape Seeds Proanthocyanidins: An Overview of In Vivo Bioactivity in Animal Models. Nutrients 2019, 11, 2435. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, T.; Liu, Z.; Zhu, X.; Wu, Q.; Meng, C.; Deng, Q. Air pollution and prostate cancer: Unraveling the connection through network toxicology and machine learning. Ecotoxicol. Environ. Saf. 2025, 292, 117966. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Y.; Sun, Y.; Wei, Z.; Wang, D.; Chen, S.; Yang, F.; Wang, J.; Kang, X. Assessing the toxicological impact of PET-MPs exposure on IVDD: Insights from network toxicology and molecular docking. J. Environ. Manag. 2025, 373, 123830. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; Guo, C.; Guo, J. Deciphering CSU pathogenesis: Network toxicologyand molecular dynamics of DOTP exposure. Ecotoxicol. Environ. Saf. 2025, 291, 117864. [Google Scholar] [CrossRef]
- Huang, S. Efficient analysis of toxicity and mechanisms of environmental pollutants with network toxicology and molecular docking strategy: Acetyl tributyl citrate as an example. Sci. Total Environ. 2023, 905, 167904. [Google Scholar] [CrossRef]
- Lv, Y.; Zhang, T.; Cai, J.; Huang, C.; Zhan, S.; Liu, J. Bioinformatics and systems biology approach to identify the pathogenetic link of Long COVID and Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Front. Immunol. 2022, 13, 952987. [Google Scholar] [CrossRef]
- Gao, K.; Hua, K.; Wang, S.; Chen, X.; Zhu, T. Exploring the reproductive exposure risks of phthalates and organophosphates in atmospheric particulate matter based on quantitative structure-activity relationships and network toxicology models. J. Hazard. Mater. 2025, 488, 137395. [Google Scholar] [CrossRef]
- Feng, J.; Qiu, S.; Zhou, S.; Tan, Y.; Bai, Y.; Cao, H.; Guo, J.; Su, Z. mTOR: A Potential New Target in Nonalcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2022, 23, 9196. [Google Scholar] [CrossRef]
- Kim, Y.C.; Guan, K.L. mTOR: A pharmacologic target for autophagy regulation. J. Clin. Investig. 2015, 125, 25–32. [Google Scholar] [CrossRef]
- Chao, X.; Wang, H.; Jaeschke, H.; Ding, W. Role and mechanisms of autophagy in acetaminophen-induced liver injury. Liver Int. 2018, 38, 1363–1374. [Google Scholar] [CrossRef]
- Opydo-Szymaczek, J.; Pawlaczyk-Kamieńska, T.; Borysewicz-Lewicka, M. Fluoride Intake and Salivary Fluoride Retention after Using High-Fluoride Toothpaste Followed by Post-Brushing Water Rinsing and Conventional (1400–1450 ppm) Fluoride Toothpastes Used without Rinsing. Int. J. Environ. Res. Public Health 2022, 19, 13235. [Google Scholar] [CrossRef]
- Srivastava, S.; Flora, S.J.S. Fluoride in Drinking Water and Skeletal Fluorosis: A Review of the Global Impact. Curr. Environ. Health Rep. 2020, 7, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Li, H.-H.; Zhang, C.-Y.; Song, X.-C. Effects of chronic fluorosis on the brain. Ecotoxicol. Environ. Saf. 2022, 244, 114021. [Google Scholar] [CrossRef]
- Nabavi, S.F.; Moghaddam, A.H.; Setzer, W.N.; Mirzaei, M. Effect of silymarin on sodium fluoride-induced toxicity and oxidative stress in rat cardiac tissues. An. Acad. Bras. Ciências 2012, 84, 1121–1126. [Google Scholar] [CrossRef]
- Yang, S.; Song, D.; Wang, R.; Liu, M.; Tan, T.; Wang, Y.; Xie, Q.; Wang, L. Sodium fluoride-induced autophagy of ameloblast-like cells via the p-ULk1/ATG13/LC3B pathway in vitro. Oral Dis. 2024, 30, 4518–4527. [Google Scholar] [CrossRef]
- Voskarides, K.; Giannopoulou, N. The Role of TP53 in Adaptation and Evolution. Cells 2023, 12, 512. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Ling, S.; Hong, J.; Zhang, L.; Zhou, W.; Yin, L.; Xu, S.; Que, Q.; Wu, Y.; Zhan, Q.; et al. TP53/mTORC1-mediated bidirectional regulation of PD-L1 modulates immune evasion in hepatocellular carcinoma. J. Immunother. Cancer 2023, 11, e007479. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Wang, X.; Tang, W. IL-1β-induced increase in NO production and decrease in mitochondrial membrane potential in rat hepatocytes. J. Southeast Univ. (Med. Sci. Ed.) 2009, 28, 253–256. [Google Scholar]
- Bliźniewska-Kowalska, K.; Gałecki, P.; Szemraj, J.; Su, K.-P.; Chang, J.P.-C.; Gałecka, M. CASP3 gene expression and the role of caspase 3 in the pathogenesis of depressive disorders. BMC Psychiatry 2023, 23, 656. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.-H.; Li, J.; Zhang, X.-Y.; Shi, S.; Wang, L.; Yuan, M.-L.; Liu, Y.-P.; Wang, Y.-D. Confusoside from Anneslea fragrans Alleviates Acetaminophen-Induced Liver Injury in HepG2 via PI3K-CASP3 Signaling Pathway. Molecules 2023, 28, 1932. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, G.; Yuan, J.; Li, N.; Yan, B.; Yan, J.; Sheng, Y. hsa-miR-34a-5p Ameliorates Hepatic Ischemia/Reperfusion Injury Via Targeting HNF4α. Turk. J. Gastroenterol. 2022, 33, 596–605. [Google Scholar] [CrossRef]
- Li, D.; Qian, J.; Li, J.; Wang, J.; Liu, W.; Li, Q.; Wu, D. Dexmedetomidine attenuates acute stress-induced liver injury in rats by regulating the miR-34a-5p/ROS/JNK/p38 signaling pathway. J. Toxicol. Sci. 2022, 47, 169–181. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, X. Research progress of mTOR inhibitors. Eur. J. Med. Chem. 2020, 208, 112820. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, Y.; Shi, Y.; Han, Y.; Liang, C.; Feng, Z.; Zheng, H.; Eng, M.; Wang, J. Fluoride-Induced Autophagy via the Regulation of Phosphorylation of Mammalian Targets of Rapamycin in Mice Leydig Cells. J. Agric. Food Chem. 2017, 65, 8966–8976. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.C.; Tian, Y.; Yuan, H.; Park, H.W.; Chang, Y.-Y.; Kim, J.; Kim, H.; Neufeld, T.P.; Dillin, A.; Guan, K.-L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013, 15, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Di, R.; Yang, Z.; Xu, P.; Xu, Y. Silencing PDK1 limits hypoxia-induced pulmonary arterial hypertension in mice via the Akt/p70S6K signaling pathway. Exp. Ther. Med. 2019, 18, 699–704. [Google Scholar] [CrossRef]
- Sangaunchom, P.; Dharmasaroja, P. Caffeine Potentiates Ethanol-Induced Neurotoxicity Through mTOR/p70S6K/4E-BP1 Inhibition in SH-SY5Y Cells. Int. J. Toxicol. 2020, 39, 131–140. [Google Scholar] [CrossRef]
- Alemasova, E.E.; Lavrik, O.I. Poly(ADP-ribosyl)ation by PARP1: Reaction mechanism and regulatory proteins. Nucleic Acids Res. 2019, 47, 3811–3827. [Google Scholar] [CrossRef]
- Xi, H.; Wang, S.; Wang, B.; Hong, X.; Liu, X.; Li, M.; Shen, R.; Dong, Q. The role of interaction between autophagy and apoptosis in tumorigenesis (Review). Oncol. Rep. 2022, 48, 208. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Y.; Yi, J.; Li, Y.; Yang, B.; Shang, P.; Mehmood, K.; Bilal, R.M.; Zhang, H.; Chang, Y.-F.; et al. The potential risks of chronic fluoride exposure on nephrotoxic via altering glucolipid metabolism and activating autophagy and apoptosis in ducks. Toxicology 2021, 461, 152906. [Google Scholar] [CrossRef] [PubMed]
- Angwa, L.M.; Nyadanu, S.D.; Kanyugo, A.M.; Adampah, T.; Pereira, G. Fluoride-induced apoptosis in non-skeletal tissues of experimental animals: A systematic review and meta-analysis. Heliyon 2023, 9, e18646. [Google Scholar] [CrossRef] [PubMed]




| Protein Receptor | Vina Score (kcal/mol) | Cavity Volume (Å3) |
|---|---|---|
| TP53 | −7.0 | 78 |
| IL1B | −8.2 | 185 |
| CASP3 | −8.7 | 1412 |
| IL6 | −9.0 | 134 |
| ALB | −9.5 | 1986 |
| BCL2 | −7.4 | 285 |
| PARP1 | −10.9 | 2969 |
| mTOR | −9.8 | 7094 |
| Group | AST (U/L) | ALT (U/L) |
|---|---|---|
| Control | 36.14 ± 0.08 | 25.52 ± 0.24 |
| NaF (100 mg/L) | 51.25 ± 6.00 * | 42.42 ± 5.47 * |
| GSPE (400 mg/L) | 35.26 ± 1.17 | 27.6 ± 2.47 |
| NaF (100 mg/L) + GSPE (100 mg/kg·bw) | 36.98 ± 0.36 # | 25.74 ± 1.25 # |
| NaF (100 mg/L) + GSPE (200 mg/kg·bw) | 36.04 ± 0.63 # | 24.71 ± 0.97 # |
| NaF (100 mg/L) + GSPE (400 mg/kg·bw) | 34.63 ± 0.78 # | 19.13 ± 1.71 ## |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Z.; Xiao, M.; Gong, Z.; Wang, B.; Zhao, W.; Guo, Y.; Yang, L. Identifying Key Genes of Proanthocyanidin Intervention in Fluoride-Induced Liver Injury: Integrated Molecular Docking and Experimental Validation. Genes 2025, 16, 1037. https://doi.org/10.3390/genes16091037
Wu Z, Xiao M, Gong Z, Wang B, Zhao W, Guo Y, Yang L. Identifying Key Genes of Proanthocyanidin Intervention in Fluoride-Induced Liver Injury: Integrated Molecular Docking and Experimental Validation. Genes. 2025; 16(9):1037. https://doi.org/10.3390/genes16091037
Chicago/Turabian StyleWu, Zhiyu, Menghuan Xiao, Zelin Gong, Benjie Wang, Wenxin Zhao, Yiyuan Guo, and Lu Yang. 2025. "Identifying Key Genes of Proanthocyanidin Intervention in Fluoride-Induced Liver Injury: Integrated Molecular Docking and Experimental Validation" Genes 16, no. 9: 1037. https://doi.org/10.3390/genes16091037
APA StyleWu, Z., Xiao, M., Gong, Z., Wang, B., Zhao, W., Guo, Y., & Yang, L. (2025). Identifying Key Genes of Proanthocyanidin Intervention in Fluoride-Induced Liver Injury: Integrated Molecular Docking and Experimental Validation. Genes, 16(9), 1037. https://doi.org/10.3390/genes16091037






