Abnormal ERV Expression and Its Clinical Relevance in Colon Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Manually Compiled ERV Set
2.2. RNA-Seq Data
2.3. RNA-Seq Data Pre-Processing and Analysis
2.4. ERV-Defined Tumor Subtypes and Their Association with Clinical and Other Molecular Phenotypes
2.5. ERV Expression Association with Patient Survival
3. Results
3.1. Comparable ERV Expression Measurement in CRC Cell Lines Between Total RNA rRNA Depleted- and Poly a-Enriched RNA-Seq Data
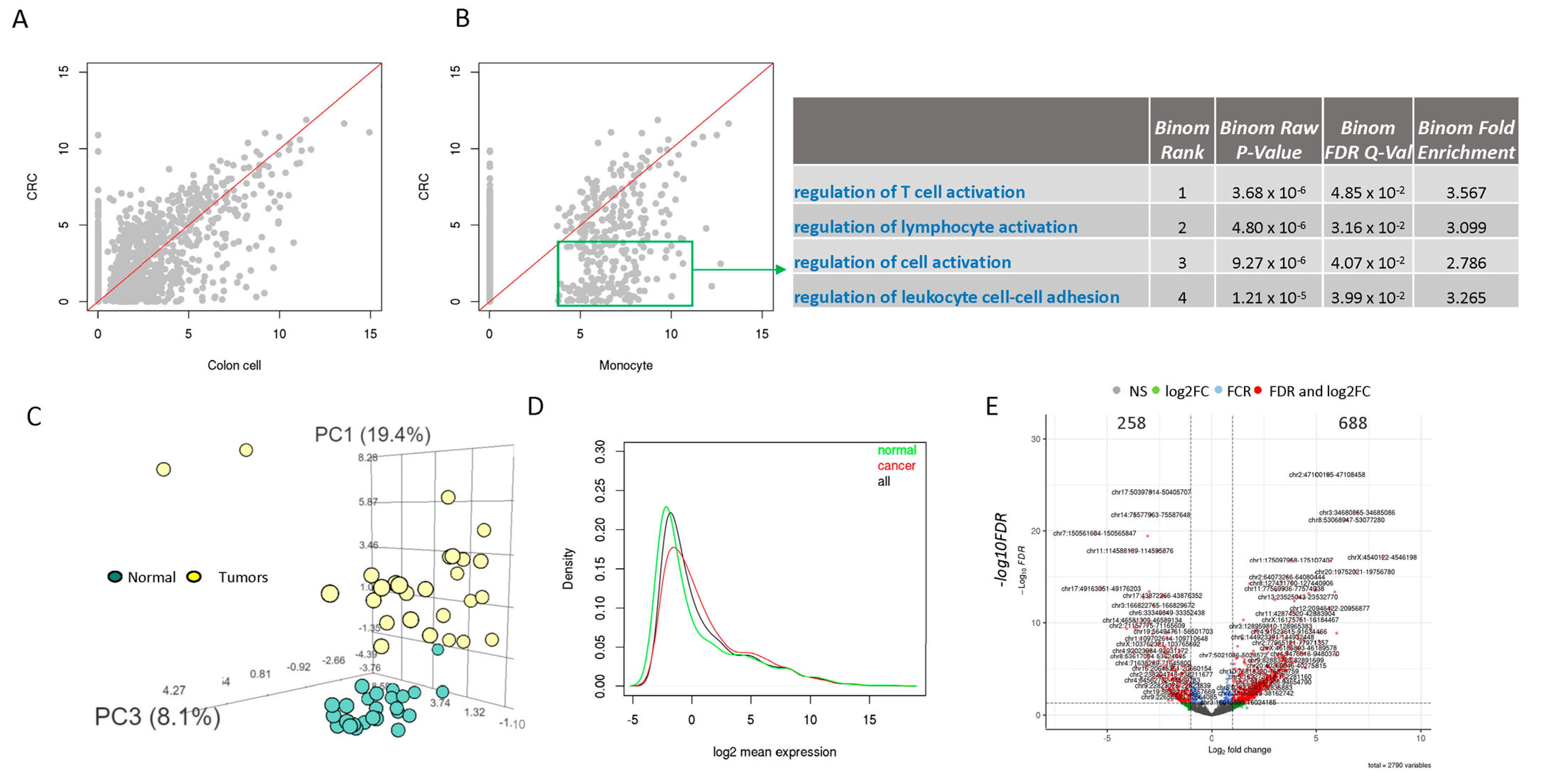
3.2. Low Number and Expression of ERVs in Normal Cells Compared to CRC Cells
3.3. Most ERVs Are Overexpressed in Primary Tumor Tissue Samples Compared to Their Paired Normal Colon Samples
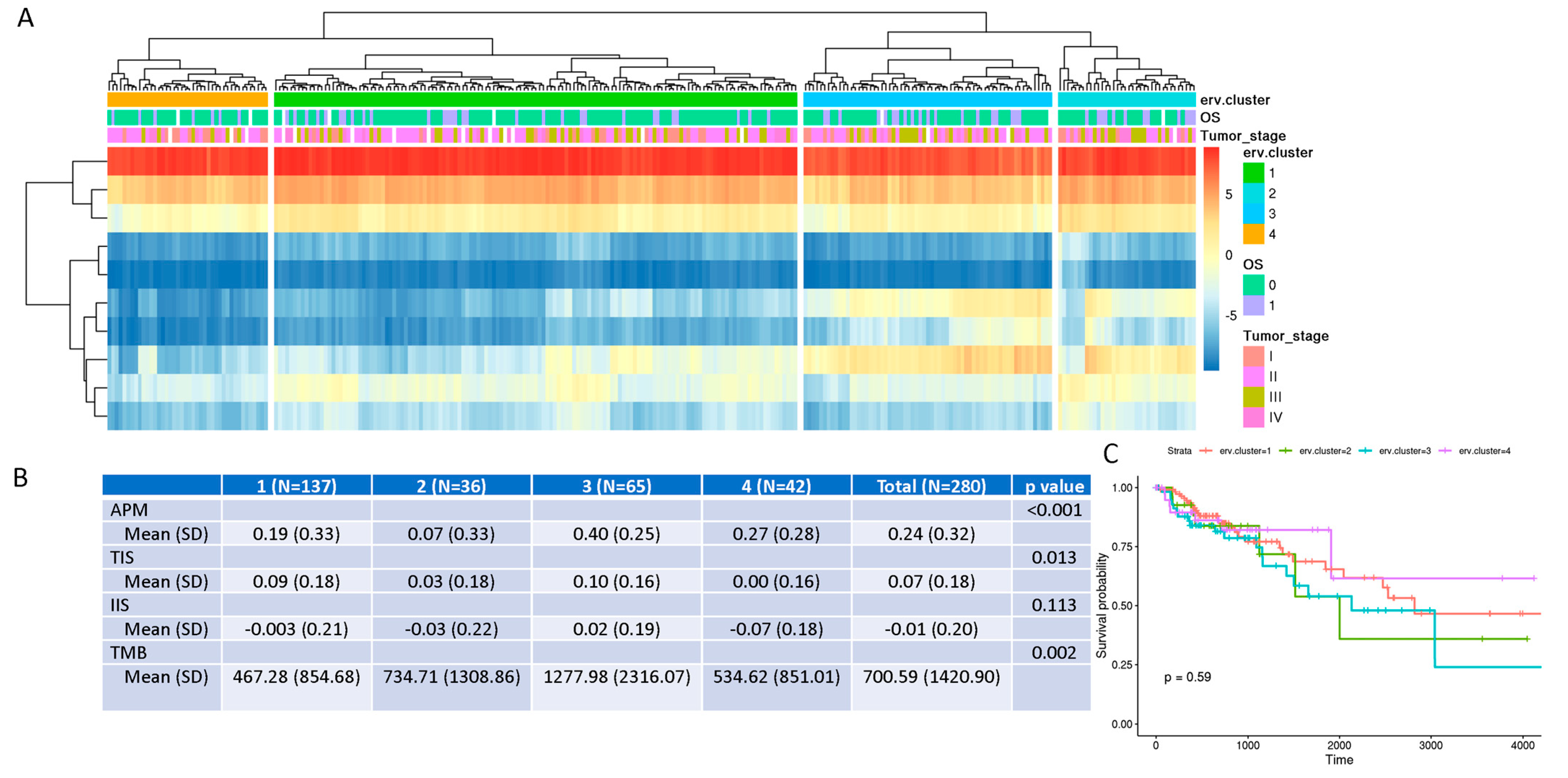
3.4. ERV Expression-Defined “Subtypes” of COAD Are Associated with Immunogenic Scores and Tumor Mutation Burden
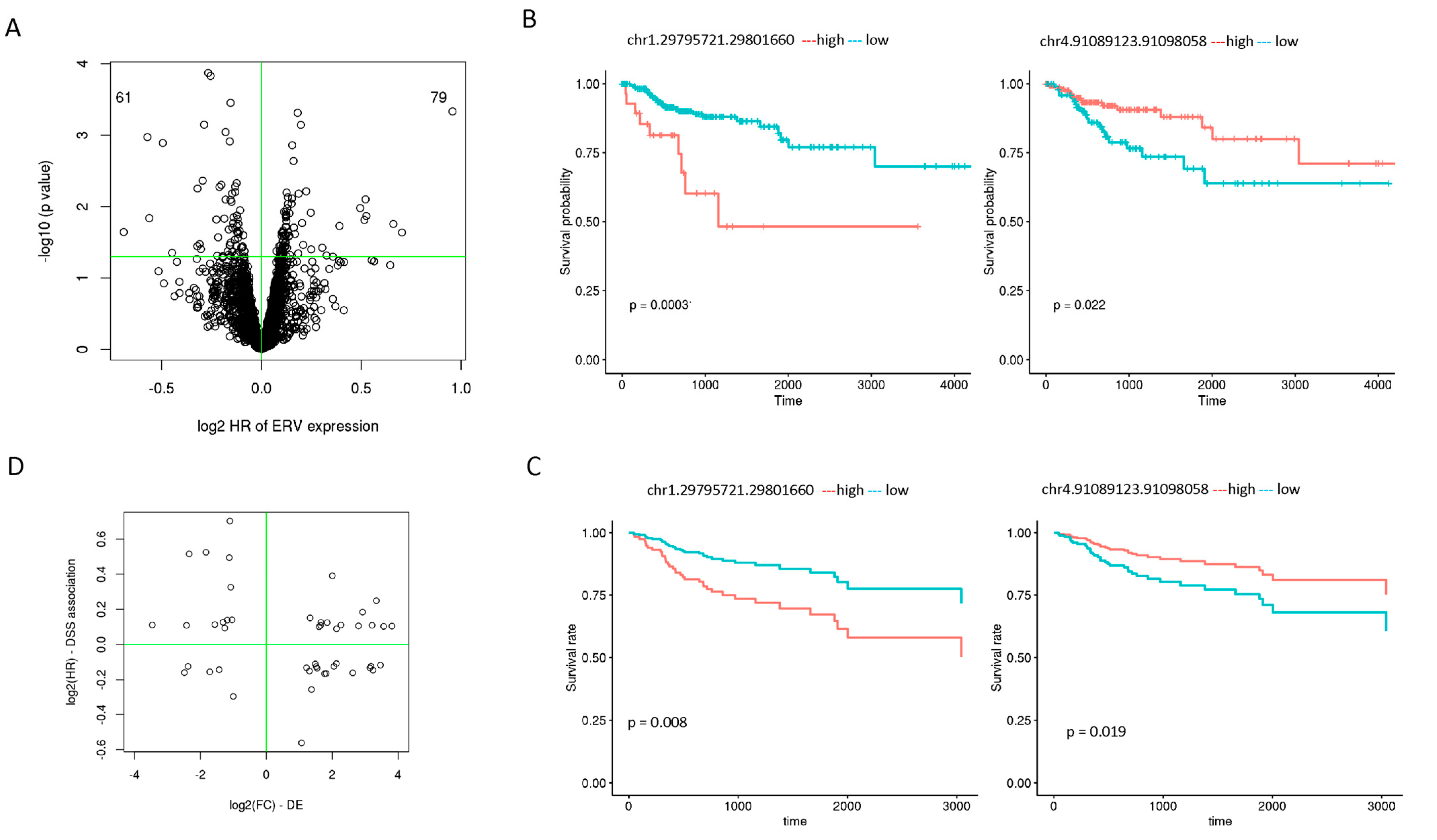
3.5. Individual ERVs Associated with Disease Specific Survival
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ERVs | Human endogenous retroviruses |
| COAD/CRC | Colon adenocarcinoma/colorectal carcinoma |
| TCGA | The Cancer Genome Atlas |
| DSS | Disease-specific survival |
| GEO | Gene expression omnibus |
| IIS | Immune infiltration score |
| APM | Antigen processing and presenting machinery |
| TMB | Tumor mutation burden |
| RPKM | Reads per kilobase of transcript per million mapped reads |
| GO | Gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
References
- Jansz, N.; Faulkner, G.J. Endogenous retroviruses in the origins and treatment of cancer. Genome Biol. 2021, 22, 147. [Google Scholar] [CrossRef] [PubMed]
- Leung, D.C.; Lorincz, M.C. Silencing of endogenous retroviruses: When and why do histone marks predominate? Trends Biochem. Sci. 2012, 37, 127–133. [Google Scholar] [CrossRef]
- Topham, J.T.; Titmuss, E.; Pleasance, E.D.; Williamson, L.M.; Karasinska, J.M.; Culibrk, L.; Lee, M.K.C.; Mendis, S.; Denroche, R.E.; Jang, G.H.; et al. Endogenous Retrovirus Transcript Levels Are Associated with Immunogenic Signatures in Multiple Metastatic Cancer Types. Mol. Cancer Ther. 2020, 19, 1889–1897. [Google Scholar] [CrossRef] [PubMed]
- Steiner, M.C.; Marston, J.L.; Iñiguez, L.P.; Bendall, M.L.; Chiappinelli, K.B.; Nixon, D.F.; Crandall, K.A. Locus-Specific Characterization of Human Endogenous Retrovirus Expression in Prostate, Breast, and Colon Cancers. Cancer Res. 2021, 81, 3449–3460. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.C.; Beckermann, K.E.; Bortone, D.S.; De Cubas, A.A.; Bixby, L.M.; Lee, S.J.; Panda, A.; Ganesan, S.; Bhanot, G.; Wallen, E.M.; et al. Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J. Clin. Investig. 2018, 128, 4804–4820. [Google Scholar] [CrossRef] [PubMed]
- Tokuyama, M.; Kong, Y.; Song, E.; Jayewickreme, T.; Kang, I.; Iwasaki, A. ERVmap analysis reveals genome-wide transcription of human endogenous retroviruses. Proc. Natl. Acad. Sci. USA 2018, 115, 12565–12572. [Google Scholar] [CrossRef] [PubMed]
- Petrizzo, A.; Ragone, C.; Cavalluzzo, B.; Mauriello, A.; Manolio, C.; Tagliamonte, M.; Buonaguro, L. Human Endogenous Retrovirus Reactivation: Implications for Cancer Immunotherapy. Cancers 2021, 13, 1999. [Google Scholar] [CrossRef] [PubMed]
- Panda, A.; de Cubas, A.A.; Stein, M.; Riedlinger, G.; Kra, J.; Mayer, T.; Smith, C.C.; Vincent, B.G.; Serody, J.S.; Beckermann, K.E.; et al. Endogenous retrovirus expression is associated with response to immune checkpoint blockade in clear cell renal cell carcinoma. JCI Insight 2018, 3, 121522. [Google Scholar] [CrossRef] [PubMed]
- Lorenzi, L.; Chiu, H.S.; Avila Cobos, F.; Gross, S.; Volders, P.J.; Cannoodt, R.; Nuytens, J.; Vanderheyden, K.; Anckaert, J.; Lefever, S.; et al. The RNA Atlas expands the catalog of human non-coding RNAs. Nat. Biotechnol. 2021, 39, 1453–1465. [Google Scholar] [CrossRef] [PubMed]
- Kalari, K.R.; Nair, A.A.; Bhavsar, J.D.; O’Brien, D.R.; Davila, J.I.; Bockol, M.A.; Nie, J.; Tang, X.; Baheti, S.; Doughty, J.B.; et al. MAP-RSeq: Mayo Analysis Pipeline for RNA sequencing. BMC Bioinform. 2014, 15, 224. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416 e411. [Google Scholar] [CrossRef]
- Wang, S.; He, Z.; Wang, X.; Li, H.; Liu, X.S. Antigen presentation and tumor immunogenicity in cancer immunotherapy response prediction. eLife 2019, 8, e49020. [Google Scholar] [CrossRef] [PubMed]
- La Ferlita, A.; Distefano, R.; Alaimo, S.; Beane, J.D.; Ferro, A.; Croce, C.M.; Tsichlis, P.N.; Pulvirenti, A.; Nigita, G. Transcriptome Analysis of Human Endogenous Retroviruses at Locus-Specific Resolution in Non-Small Cell Lung Cancer. Cancers 2022, 14, 4433. [Google Scholar] [CrossRef]
- Jang, H.J.; Shah, N.M.; Maeng, J.H.; Liang, Y.; Basri, N.L.; Ge, J.; Qu, X.; Mahlokozera, T.; Tzeng, S.C.; Williams, R.B.; et al. Epigenetic therapy potentiates transposable element transcription to create tumor-enriched antigens in glioblastoma cells. Nat. Genet. 2024, 56, 1903–1913. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.; Scrimieri, F.; Ghosh, S.; Zhong, J.; Kim, M.S.; Ren, Y.R.; Morgan, R.A.; Iacobuzio-Donahue, C.A.; Pandey, A.; Kern, S.E. FAM190A deficiency creates a cell division defect. Am. J. Pathol. 2013, 183, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Bendall, M.L.; de Mulder, M.; Iniguez, L.P.; Lecanda-Sanchez, A.; Perez-Losada, M.; Ostrowski, M.A.; Jones, R.B.; Mulder, L.C.F.; Reyes-Teran, G.; Crandall, K.A.; et al. Telescope: Characterization of the retrotranscriptome by accurate estimation of transposable element expression. PLoS Comput. Biol. 2019, 15, e1006453. [Google Scholar] [CrossRef] [PubMed]


| ERV | DSS HR | Surv Pval | DE log2FC | DE Pval | DE Padj | DE Direction |
|---|---|---|---|---|---|---|
| chr14:73702885–73716167 | 1.63 | 2.31 × 10−2 | −1.11 | 3.18 × 10−7 | 4.82 × 10−6 | Down |
| chr19:56494761–56501703 | 1.44 | 1.36 × 10−2 | −1.83 | 1.79 × 10−11 | 8.44 × 10−10 | Down |
| chr17:43872266–43876352 | 1.43 | 1.55 × 10−2 | −2.34 | 1.00 × 10−15 | 1.08 × 10−13 | Down |
| chr19:38935163–38941835 | 1.41 | 1.05 × 10−2 | −1.12 | 6.47 × 10−6 | 6.43 × 10−5 | Down |
| chr12:76291617–76300850 | 1.31 | 1.87 × 10−2 | 2.00 | 1.82 × 10−6 | 2.12 × 10−5 | Up |
| chr19:36323747–36332422 | 1.25 | 4.85 × 10−2 | −1.08 | 1.16 × 10−6 | 1.49 × 10−5 | Down |
| chr16:86739328–86747655 | 1.19 | 3.98 × 10−2 | 3.34 | 2.06 × 10−17 | 3.59 × 10−15 | Up |
| chr6:144923391–144932448 | 1.14 | 4.54 × 10−2 | 2.92 | 8.27 × 10−11 | 3.30 × 10−9 | Up |
| chr7:4590929–4600359 | 1.11 | 9.14 × 10−3 | 1.33 | 3.19 × 10−3 | 1.16 × 10−2 | Up |
| chr15:51357586–51367580 | 1.10 | 2.55 × 10−2 | −1.04 | 1.01 × 10−2 | 3.01 × 10−2 | Down |
| chr2:64886311–64899831 | 1.10 | 2.55 × 10−2 | −1.18 | 1.30 × 10−3 | 5.55 × 10−3 | Down |
| chr4:179720511–179728321 | 1.09 | 1.92 × 10−2 | 1.84 | 7.08 × 10−4 | 3.36 × 10−3 | Up |
| chr6:118574055–118578291 | 1.09 | 6.39 × 10−3 | −1.31 | 4.95 × 10−4 | 2.49 × 10−3 | Down |
| chr8:12037971–12041523 | 1.09 | 1.60 × 10−2 | 1.66 | 4.30 × 10−3 | 1.47 × 10−2 | Up |
| chr1:247898555–247905892 | 1.08 | 2.17 × 10−2 | −2.42 | 6.05 × 10−6 | 6.12 × 10−5 | Down |
| chr11:23882492–23892875 | 1.08 | 3.05 × 10−2 | 2.80 | 7.56 × 10−6 | 7.30 × 10−5 | Up |
| chr12:38123075–38131103 | 1.08 | 3.44 × 10−2 | 3.21 | 1.14 × 10−6 | 1.47 × 10−5 | Up |
| chr19:54894290–54900597 | 1.08 | 2.55 × 10−2 | 2.26 | 3.24 × 10−4 | 1.74 × 10−3 | Up |
| chr21:38224030–38229830 | 1.08 | 4.06 × 10−2 | 4.42 | 3.08 × 10−10 | 1.06 × 10−8 | Up |
| chr4:4130683–4133908 | 1.08 | 1.19 × 10−2 | −1.56 | 3.73 × 10−3 | 1.32 × 10−2 | Down |
| chr6:123556843–123562657 | 1.08 | 2.29 × 10−2 | −3.46 | 4.42 × 10−12 | 2.46 × 10−10 | Down |
| chr6:123582329–123588017 | 1.08 | 3.02 × 10−2 | 1.65 | 7.54 × 10−3 | 2.36 × 10−2 | Up |
| chr4:11653551–11659272 | 1.07 | 3.66 × 10−2 | 3.81 | 1.21 × 10−7 | 2.05 × 10−6 | Up |
| chr8:11931841–11936717 | 1.07 | 2.34 × 10−2 | 1.60 | 1.30 × 10−3 | 5.55 × 10−3 | Up |
| chrX:77077094–77086082 | 1.07 | 4.51 × 10−2 | −1.26 | 5.94 × 10−3 | 1.95 × 10−2 | Down |
| chrY:8994259–9004253 | 1.07 | 4.17 × 10−2 | 3.56 | 1.29 × 10−5 | 1.14 × 10−4 | Up |
| chr11:89381303–89387559 | 1.06 | 3.61 × 10−2 | 2.13 | 4.48 × 10−4 | 2.26 × 10−3 | Up |
| chr12:4018995–4025863 | 0.93 | 1.13 × 10−2 | 2.13 | 3.79 × 10−5 | 2.87 × 10−4 | Up |
| chr8:60493460–60499925 | 0.93 | 2.21 × 10−2 | 1.48 | 1.84 × 10−3 | 7.30 × 10−3 | Up |
| chr18:51112855–51117176 | 0.92 | 2.42 × 10−2 | 3.46 | 5.00 × 10−8 | 9.42 × 10−7 | Up |
| chr4:23722376–23727827 | 0.92 | 2.25 × 10−2 | −2.38 | 1.00 × 10−9 | 3.07 × 10−8 | Down |
| chr5:34461058–34468430 | 0.92 | 1.89 × 10−2 | 2.05 | 1.34 × 10−6 | 1.65 × 10−5 | Up |
| chrX:151550923–151562615 | 0.92 | 1.34 × 10−2 | 3.17 | 1.32 × 10−8 | 2.91 × 10−7 | Up |
| chr8:7402289–7408615 | 0.92 | 3.54 × 10−2 | 1.51 | 1.36 × 10−2 | 3.84 × 10−2 | Up |
| chr20:25394210–25403221 | 0.91 | 6.26 × 10−3 | 1.22 | 8.64 × 10−3 | 2.66 × 10−2 | Up |
| chr6:137717275–137724946 | 0.91 | 5.27 × 10−3 | 3.14 | 1.20 × 10−10 | 4.65 × 10−9 | Up |
| chr14:103234947–103242530 | 0.91 | 2.92 × 10−2 | 1.54 | 6.25 × 10−5 | 4.41 × 10−4 | Up |
| chrX:97841727–97849389 | 0.91 | 1.71 × 10−2 | −1.43 | 1.78 × 10−3 | 7.10 × 10−3 | Down |
| chr9:64870612–64878934 | 0.90 | 1.98 × 10−2 | 3.24 | 1.45 × 10−9 | 4.26 × 10−8 | Up |
| chr11:58996950–59006462 | 0.90 | 8.61 × 10−3 | 1.31 | 5.78 × 10−4 | 2.82 × 10−3 | Up |
| chr4:115950630–115956482 | 0.90 | 3.20 × 10−2 | −1.72 | 2.70 × 10−3 | 1.01 × 10−2 | Down |
| chr6:63321317–63335844 | 0.90 | 1.23 × 10−3 | −2.48 | 5.14 × 10−10 | 1.67 × 10−8 | Down |
| chr4:77630582–77637015 | 0.90 | 3.47 × 10−2 | 2.62 | 1.47 × 10−9 | 4.28 × 10−8 | Up |
| chr4:58458021–58466147 | 0.89 | 2.10 × 10−2 | 1.81 | 3.38 × 10−3 | 1.22 × 10−2 | Up |
| chr9:29775512–29780684 | 0.89 | 4.75 × 10−2 | 1.77 | 2.88 × 10−3 | 1.06 × 10−2 | Up |
| chr22:30676787–30679926 | 0.84 | 1.49 × 10−4 | 1.37 | 4.00 × 10−6 | 4.26 × 10−5 | Up |
| chr3:11247257–11254838 | 0.82 | 4.36 × 10−3 | −1.00 | 4.07 × 10−5 | 3.03 × 10−4 | Down |
| chr11:17370179–17379293 | 0.68 | 1.46 × 10−2 | 1.07 | 7.14 × 10−6 | 6.97 × 10−5 | Up |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhagwate, A.; Taylor, W.; Kisiel, J.; Sun, Z. Abnormal ERV Expression and Its Clinical Relevance in Colon Cancer. Genes 2025, 16, 988. https://doi.org/10.3390/genes16080988
Bhagwate A, Taylor W, Kisiel J, Sun Z. Abnormal ERV Expression and Its Clinical Relevance in Colon Cancer. Genes. 2025; 16(8):988. https://doi.org/10.3390/genes16080988
Chicago/Turabian StyleBhagwate, Aditya, William Taylor, John Kisiel, and Zhifu Sun. 2025. "Abnormal ERV Expression and Its Clinical Relevance in Colon Cancer" Genes 16, no. 8: 988. https://doi.org/10.3390/genes16080988
APA StyleBhagwate, A., Taylor, W., Kisiel, J., & Sun, Z. (2025). Abnormal ERV Expression and Its Clinical Relevance in Colon Cancer. Genes, 16(8), 988. https://doi.org/10.3390/genes16080988






