Association Between UGT1A1 mRNA Expression and Cis-Acting Genetic Variants and Trans-Acting Transcriptional Regulators in Human Liver Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. Liver Samples
2.2. DNA and RNA Preparation
2.3. Gene Expression
2.4. Genotyping
2.5. Multiple Linear Regression
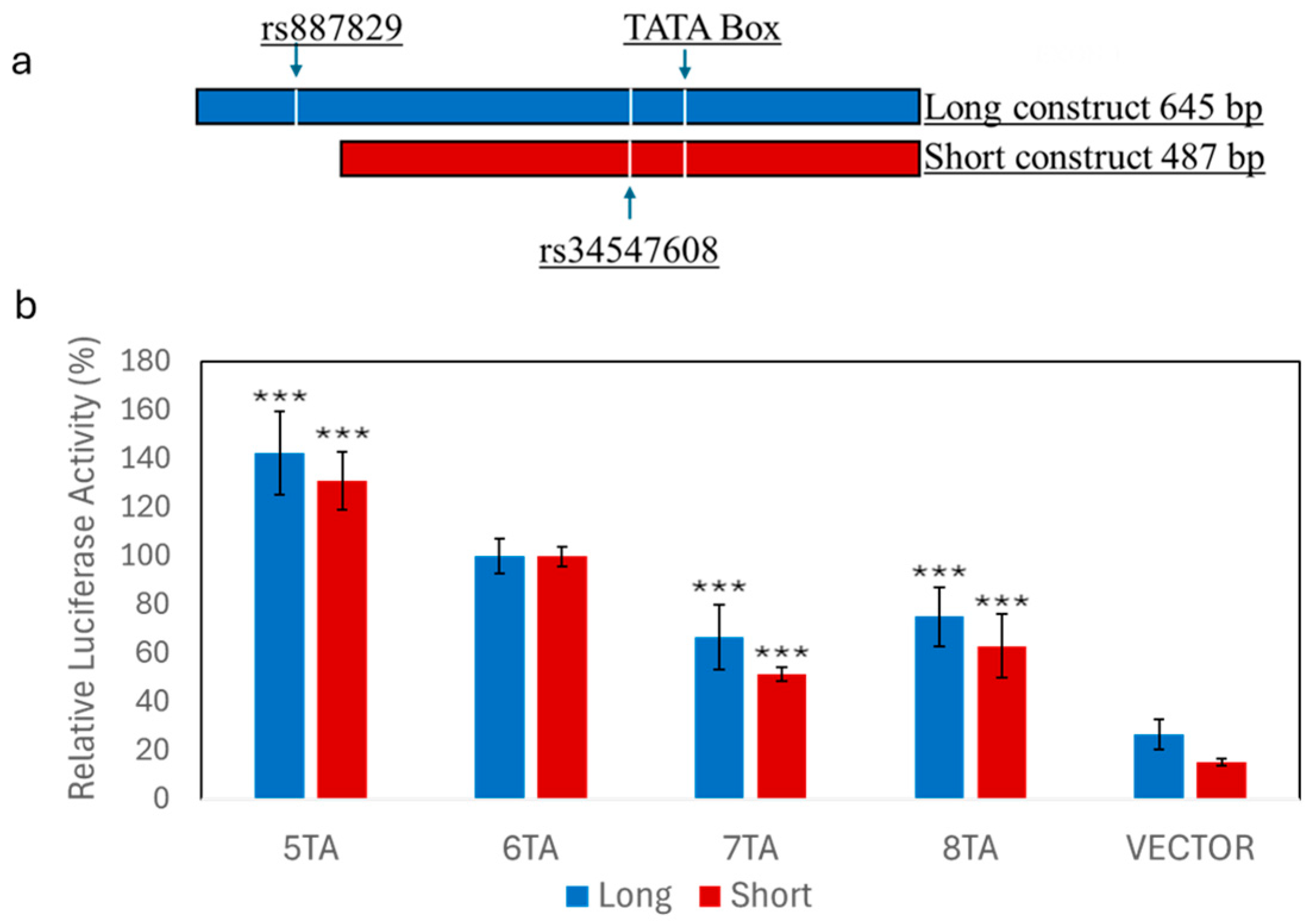
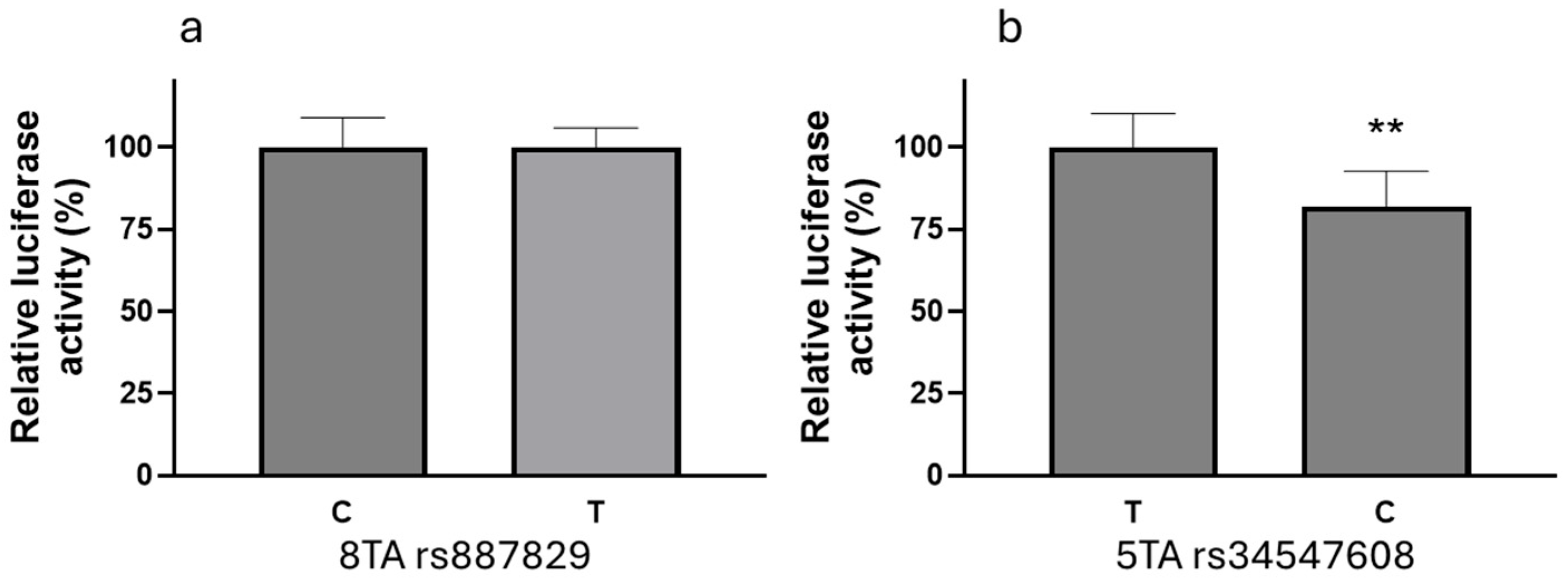
2.6. Reporter Gene Constructs
2.7. Site-Directed Mutagenesis
2.8. Cell Culture, Transfection, and Reporter Gene Assays
2.9. Statistical Analysis
3. Results
3.1. The Allele Frequencies of UGT1A1 *28, *37, *36, and rs887829 in Liver Samples
3.2. The Linkage Disequilibrium Between UGT1A1*28, *37, *36, and rs887829
3.3. Testing the Association Between UGT1A1 Expression and the Promoter TA Repeat Polymorphism or rs887829 in the Liver Samples
3.4. The Inclusion of Trans-Acting TFs Improves the Association Between Genotypes and UGT1A1 Expression in Liver Samples
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rowland, A.; Miners, J.O.; Mackenzie, P.I. The UDP-glucuronosyltransferases: Their role in drug metabolism and detoxification. Int. J. Biochem. Cell Biol. 2013, 45, 1121–1132. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Ramírez, J.; Gamazon, E.R.; Mirkov, S.; Chen, P.; Wu, K.; Sun, C.; Cox, N.J.; Cook, E.; Das, S.; et al. Genetic factors affecting gene transcription and catalytic activity of UDP-glucuronosyltransferases in human liver. Hum. Mol. Genet. 2014, 23, 5558–5569. [Google Scholar] [CrossRef]
- Mathijssen, R.H.; Van Alphen, R.J.; Verweij, J.; Loos, W.J.; Nooter, K.; Stoter, G.; Sparreboom, A. Clinical pharmacokinetics and metabolism of irinotecan (CPT-11). Clin. Cancer Res. 2001, 7, 2182–2194. [Google Scholar]
- Cottrell, M.L.; Hadzic, T.; Kashuba, A.D. Clinical pharmacokinetic, pharmacodynamic and drug-interaction profile of the integrase inhibitor dolutegravir. Clin. Pharmacokinet. 2013, 52, 981–994. [Google Scholar] [CrossRef]
- Goey, A.K.L.; Sissung, T.M.; Peer, C.J.; Trepel, J.B.; Lee, M.; Tomita, Y.; Ehrlich, S.; Bryla, C.; Balasubramaniam, S.; Piekarz, R.; et al. Effects of UGT1A1 genotype on the pharmacokinetics, pharmacodynamics, and toxicities of belinostat administered by 48-hour continuous infusion in patients with cancer. J. Clin. Pharmacol. 2016, 56, 461–473. [Google Scholar] [CrossRef]
- Marcuello, E.; Altés, A.; Menoyo, A.; Del Rio, E.; Gómez-Pardo, M.; Baiget, M. UGT1A1 gene variations and irinotecan treatment in patients with metastatic colorectal cancer. Br. J. Cancer 2004, 91, 678–682. (In English) [Google Scholar] [CrossRef]
- Wang, L.-Z.; Ramírez, J.; Yeo, W.; Chan, M.-Y.M.; Thuya, W.-L.; Lau, J.-Y.A.; Wan, S.-C.; Wong, A.L.-A.; Zee, Y.-K.; Lim, R.; et al. Glucuronidation by UGT1A1 is the dominant pathway of the metabolic disposition of belinostat in liver cancer patients. PLoS ONE 2013, 8, e54522. [Google Scholar] [CrossRef]
- Bosma, P.J.; Chowdhury, J.R.; Bakker, C.; Gantla, S.; De Boer, A.; Oostra, B.A.; Lindhout, D.; Tytgat, G.N.J.; Jansen, P.L.M.; Oude Elferink, R.P.; et al. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert’s syndrome. N. Engl. J. Med. 1995, 333, 1171–1175. [Google Scholar] [CrossRef]
- Lankisch, T.O.; Schulz, C.; Zwingers, T.; Erichsen, T.J.; Manns, M.P.; Heinemann, V.; Strassburg, C.P. Gilbert’s Syndrome and irinotecan toxicity: Combination with UDP-glucuronosyltransferase 1A7 variants increases risk. Cancer Epidemiol. Biomark. Prev. 2008, 17, 695–701. [Google Scholar] [CrossRef]
- Ribaudo, H.J.; Daar, E.S.; Tierney, C.; Morse, G.D.; Mollan, K.; Sax, P.E.; Fischl, M.A.; Collier, A.C.; Haas, D.W.; the AIDS Clinical Trials Group. Impact of UGT1A1 Gilbert variant on discontinuation of ritonavir-boosted atazanavir in AIDS Clinical Trials Group Study A5202. J. Infect. Dis. 2013, 207, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Gammal, R.S.; Court, M.H.; Haidar, C.E.; Iwuchukwu, O.F.; Gaur, A.H.; Alvarellos, M.; Guillemette, C.; Lennox, J.L.; Whirl-Carrillo, M.; Brummel, S.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for UGT1A1 and Atazanavir Prescribing. Clin. Pharmacol. Ther. 2016, 99, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Hulshof, E.C.; Deenen, M.J.; Nijenhuis, M.; Soree, B.; de Boer-Veger, N.J.; Buunk, A.-M.; Houwink, E.J.F.; Risselada, A.; Rongen, G.A.P.J.M.; van Schaik, R.H.N.; et al. Dutch pharmacogenetics working group (DPWG) guideline for the gene-drug interaction between UGT1A1 and irinotecan. Eur. J. Hum. Genet. 2023, 31, 982–987. [Google Scholar] [CrossRef]
- Bravo-Gómez, A.; Salvador-Martín, S.; Zapata-Cobo, P.; Sanjurjo-Sáez, M.; López-Fernández, L.A. Genotyping of UGT1A1*80 as an Alternative to UGT1A1*28 Genotyping in Spain. Pharmaceutics 2022, 14, 2082. [Google Scholar] [CrossRef]
- Leger, P.; Chirwa, S.; Nwogu, J.N.; Turner, M.; Richardson, D.M.; Baker, P.; Leonard, M.; Erdem, H.; Olson, L.; Haas, D.W. Race/ethnicity difference in the pharmacogenetics of bilirubin-related atazanavir discontinuation. Pharmacogenet. Genomics 2018, 28, 1–6. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, M.; Hu, M.; Cui, Y.; Zhong, Q.; Liang, L.; Huang, F. UGT1A1*6 and UGT1A1*28 polymorphisms are correlated with irinotecan-induced toxicity: A meta-analysis. Asia Pac. J. Clin. Oncol. 2018, 14, e479–e489. [Google Scholar] [CrossRef]
- Atasilp, C.; Biswas, M.; Jinda, P.; Nuntharadthanaphong, N.; Rachanakul, J.; Hongkaew, Y.; Vanwong, N.; Saokaew, S.; Sukasem, C. Association of UGT1A1*6, UGT1A1*28, or ABCC2 c.3972C>T genetic polymorphisms with irinotecan-induced toxicity in Asian cancer patients: Meta-analysis. Clin. Transl. Sci. 2022, 15, 1613–1633. [Google Scholar] [CrossRef]
- Hu, Z.Y.; Yu, Q.; Zhao, Y.S. Dose-dependent association between UGT1A1*28 polymorphism and irinotecan-induced diarrhoea: A meta-analysis. Eur. J. Cancer 2010, 46, 1856–1865. [Google Scholar] [CrossRef]
- Yagura, H.; Watanabe, D.; Kushida, H.; Tomishima, K.; Togami, H.; Hirano, A.; Takahashi, M.; Hirota, K.; Ikuma, M.; Kasai, D.; et al. Impact of UGT1A1 gene polymorphisms on plasma dolutegravir trough concentrations and neuropsychiatric adverse events in Japanese individuals infected with HIV-1. BMC Infect. Dis. 2017, 17, 622. [Google Scholar] [CrossRef]
- Griesel, R.; Sinxadi, P.; Kawuma, A.; Joska, J.; Sokhela, S.; Akpomiemie, G.; Venter, F.; Denti, P.; Haas, D.W.; Maartens, G. Pharmacokinetic and pharmacogenetic associations with dolutegravir neuropsychiatric adverse events in an African population. J. Antimicrob. Chemother. 2022, 77, 3110–3117. [Google Scholar] [CrossRef] [PubMed]
- Cindi, Z.; Kawuma, A.N.; Maartens, G.; Bradford, Y.; Venter, F.; Sokhela, S.; Chandiwana, N.; E Wasmann, R.; Denti, P.; Wiesner, L.; et al. Pharmacogenetics of Dolutegravir Plasma Exposure Among Southern Africans with Human Immunodeficiency Virus. J. Infect. Dis. 2022, 226, 1616–1625. [Google Scholar] [CrossRef] [PubMed]
- Mpofu, R.; Kawuma, A.N.; Wasmann, R.E.; Akpomiemie, G.; Chandiwana, N.; Sokhela, S.M.; Moorhouse, M.; Venter, W.D.F.; Denti, P.; Wiesner, L.; et al. Determinants of early change in serum creatinine after initiation of dolutegravir-based antiretroviral therapy in South Africa. Br. J. Clin. Pharmacol. 2024, 90, 1247–1257. [Google Scholar] [CrossRef]
- Feng, L.; Wang, Y.; Qin, J.; Fu, Y.; Guo, Z.; Zhang, J.; He, G.; Jiang, Z.; Xu, X.; Zhou, C.; et al. UGT1A Gene Family Members Serve as Potential Targets and Prognostic Biomarkers for Pancreatic Cancer. BioMed. Res. Int. 2021, 2021, 6673125. [Google Scholar] [CrossRef]
- Matsui, K.; Maruo, Y.; Sato, H.; Takeuchi, Y. Combined effect of regulatory polymorphisms on transcription of UGT1A1 as a cause of Gilbert syndrome. BMC Gastroenterol. 2010, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Ramos, E.; Adeyemo, A.; Shriner, D.; Zhou, J.; Doumatey, A.P.; Huang, H.; Erdos, M.R.; Gerry, N.P.; Herbert, A.; et al. UGT1A1 is a major locus influencing bilirubin levels in African Americans. Eur. J. Hum. Genet. 2012, 20, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Adeyemo, A.; Zhou, J.; Doumatey, A.P.; Bentley, A.R.; Ekoru, K.; Shriner, D.; Rotimi, C.N. A UGT1A1 variant is associated with serum total bilirubin levels, which are causal for hypertension in African-ancestry individuals. NPJ Genomic Med. 2021, 6, 44. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.A.; Burgos, A.E.; Flaherman, V.; Chung, E.K.; Simpson, E.A.; Goyal, N.K.; Von Kohorn, I.; Dhepyasuwan, N.; Better Outcomes through Research for Newborns Network. Discrepancies between transcutaneous and serum bilirubin measurements. Pediatrics 2015, 135, 224–231. [Google Scholar] [CrossRef]
- Collins, J.M.; Wang, D. Co-expression of drug metabolizing cytochrome P450 enzymes and estrogen receptor alpha (ESR1) in human liver: Racial differences and the regulatory role of ESR1. Drug Metab. Pers. Ther. 2021, 36, 205–214. [Google Scholar] [CrossRef]
- Zhou, L.; Montalvo, A.D.; Collins, J.M.; Wang, D. Quantitative analysis of the UDP-glucuronosyltransferase transcriptome in human tissues. Pharmacol. Res. Perspect. 2023, 11, e01154. [Google Scholar] [CrossRef]
- Montalvo, A.D.; Gong, Y.; Collins, J.M.; Wang, D. The Association Between Promoter Tandem Repeat Polymorphism (pVNTR) and CYP2C9 Gene Expression in Human Liver Samples. Genes 2025, 16, 213. [Google Scholar] [CrossRef]
- Beutler, E.; Gelbart, T.; Demina, A. Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: A balanced polymorphism for regulation of bilirubin metabolism? Proc. Natl. Acad. Sci. USA 1998, 95, 8170–8174. [Google Scholar] [CrossRef]
- Smith, A.; Cropp, C.D.; Vidal, G.; Pritchard, E.; Cordero, J.; Simpson, C.; Starlard-Davenport, A. Prevalence of the UGT1A1*28 promoter polymorphism and breast cancer risk among African American women in Memphis, TN. Cancer Health Disparities 2019, 3, e1–e12. [Google Scholar]
- Guillemette, C.; Millikan, R.C.; Newman, B.; Housman, D.E. Genetic polymorphisms in uridine diphospho-glucuronosyltransferase 1A1 and association with breast cancer among African Americans. Cancer Res. 2000, 60, 950–956. [Google Scholar]
- Sugatani, J.; Nishitani, S.; Yamakawa, K.; Yoshinari, K.; Sueyoshi, T.; Negishi, M.; Miwa, M. Transcriptional regulation of human UGT1A1 gene expression: Activated glucocorticoid receptor enhances constitutive androstane receptor/pregnane X receptor-mediated UDP-glucuronosyltransferase 1A1 regulation with glucocorticoid receptor-interacting protein 1. Mol. Pharmacol. 2005, 67, 845–855. [Google Scholar] [CrossRef]
- Yueh, M.F.; Huang, Y.H.; Hiller, A.; Chen, S.; Nguyen, N.; Tukey, R.H. Involvement of the xenobiotic response element (XRE) in Ah receptor-mediated induction of human UDP-glucuronosyltransferase 1A1. J. Biol. Chem. 2003, 278, 15001–15006. [Google Scholar] [CrossRef]
- Yueh, M.F.; Tukey, R.H. Nrf2-Keap1 signaling pathway regulates human UGT1A1 expression in vitro and in transgenic UGT1 mice. J. Biol. Chem. 2007, 282, 8749–8758. [Google Scholar] [CrossRef] [PubMed]
- Gallucci, G.M.; Hayes, C.M.; Boyer, J.L.; Barbier, O.; Assis, D.N.; Ghonem, N.S. PPAR-Mediated Bile Acid Glucuronidation: Therapeutic Targets for the Treatment of Cholestatic Liver Diseases. Cells 2024, 13, 1296. [Google Scholar] [CrossRef]
- Chiang, J.Y. Hepatocyte nuclear factor 4alpha regulation of bile acid and drug metabolism. Expert Opin. Drug Metab. Toxicol. 2009, 5, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Pavek, P. Pregnane X Receptor (PXR)-Mediated Gene Repression and Cross-Talk of PXR with Other Nuclear Receptors via Coactivator Interactions. Front. Pharmacol. 2016, 7, 456. [Google Scholar] [CrossRef] [PubMed]
- Bochkis, I.M.; Schug, J.; Rubins, N.E.; Chopra, A.R.; O’Malley, B.W.; Kaestner, K.H. Foxa2-dependent hepatic gene regulatory networks depend on physiological state. Physiol. Genomics 2009, 38, 186–195. [Google Scholar] [CrossRef]
- Khatri, R.; Fallon, J.K.; Sykes, C.; Kulick, N.; Rementer, R.J.B.; Miner, T.A.; Schauer, A.P.; Kashuba, A.D.M.; Boggess, K.A.; Brouwer, K.L.R.; et al. Pregnancy-Related Hormones Increase UGT1A1-Mediated Labetalol Metabolism in Human Hepatocytes. Front. Pharmacol. 2021, 12, 655320. [Google Scholar] [CrossRef]
- Pedroza, D.A.; Rajamanickam, V.; Subramani, R.; Bencomo, A.; Galvez, A.; Lakshmanaswamy, R. Progesterone receptor membrane component 1 promotes the growth of breast cancers by altering the phosphoproteome and augmenting EGFR/PI3K/AKT signalling. Br. J. Cancer 2020, 123, 1326–1335. [Google Scholar] [CrossRef]
- Thieffry, C.; Van Wynendaele, M.; Aynaci, A.; Maja, M.; Dupuis, C.; Loriot, A.; Marbaix, E.; Henriet, P. AG-205 Upregulates Enzymes Involved in Cholesterol Biosynthesis and Steroidogenesis in Human Endometrial Cells Independently of PGRMC1 and Related MAPR Proteins. Biomolecules 2021, 11, 1472. [Google Scholar] [CrossRef] [PubMed]
- Kimura, I.; Nakayama, Y.; Konishi, M.; Terasawa, K.; Ohta, M.; Itoh, N.; Fujimoto, M. Functions of MAPR (membrane-associated progesterone receptor) family members as heme/steroid-binding proteins. Curr. Protein Pept. Sci. 2012, 13, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Lu, R.; Rempala, G.; Sadee, W. Ligand-Free Estrogen Receptor α (ESR1) as Master Regulator for the Expression of CYP3A4 and Other Cytochrome P450 Enzymes in the Human Liver. Mol. Pharmacol. 2019, 96, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Bosma, P.J.; Seppen, J.; Goldhoorn, B.; Bakker, C.; Oude Elferink, R.P.; Chowdhury, J.R.; Chowdhury, N.R.; Jansen, P.L. Bilirubin UDP-glucuronosyltransferase 1 is the only relevant bilirubin glucuronidating isoform in man. J. Biol. Chem. 1994, 269, 17960–17964. [Google Scholar] [CrossRef]
- Piel, R.B., 3rd; Shiferaw, M.T.; Vashisht, A.A.; Marcero, J.R.; Praissman, J.L.; Phillips, J.D.; Wohlschlegel, J.A.; Medlock, A.E. A Novel Role for Progesterone Receptor Membrane Component 1 (PGRMC1): A Partner and Regulator of Ferrochelatase. Biochemistry 2016, 55, 5204–5217. [Google Scholar] [CrossRef]
- Kabe, Y.; Nakane, T.; Koike, I.; Yamamoto, T.; Sugiura, Y.; Harada, E.; Sugase, K.; Shimamura, T.; Ohmura, M.; Muraoka, K.; et al. Haem-dependent dimerization of PGRMC1/Sigma-2 receptor facilitates cancer proliferation and chemoresistance. Nat. Commun. 2016, 7, 11030. [Google Scholar] [CrossRef]
| AA + EA | AA | EA | |
|---|---|---|---|
| Variants | n = 257 | n = 138 | n = 119 |
| UGT1A1*28 | 0.35 | 0.37 | 0.32 |
| UGT1A1*37 | 0.03 | 0.05 | 0.008 |
| UGT1A1*36 | 0.05 | 0.07 | 0.013 |
| rs887829 (*80) | 0.39 | 0.45 | 0.33 |
| TA Repeats | AA | EA | ||
|---|---|---|---|---|
| D’ | r2 | D’ | r2 | |
| UGT1A1*28 | 0.99 | 0.77 | 0.98 | 0.93 |
| UGT1A1*37 | 0.99 | 0.08 | 0.99 | 0.02 |
| UGG1A1*36 | 0.68 | 0.03 | 0.71 | 0.03 |
| UGT1A1*28/*37 | 0.99 | 0.94 | 0.99 | 0.98 |
| Genetic Variants | AA + EA | AA | EA | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Beta Estimate | p-Value | r2 | Beta Estimate | p-Value | r2 | Beta Estimate | p-Value | r2 | |
| UGT1A1*28/*37 a | −0.33 | 1.65 × 10−8 | 0.13 | −0.336 | 4.18 × 10−5 | 0.12 | −0.343 | 2.62 × 10−5 | 0.16 |
| rs887829 a | −0.325 | 2.16 × 10−8 | 0.12 | −0.343 | 2.93 × 10−5 | 0.13 | −0.33 | 4.12 × 10−5 | 0.15 |
| UGT1A1*36 a,b | ND | ND | ND | 0.145 | 3.802 × 10−1 | 0.04 | ND | ND | ND |
| Race | Independent Variables | UGT1A1*28/*37 Genotype | rs887829 Genotype | ||||
|---|---|---|---|---|---|---|---|
| Beta Estimate | p Value | Partial R2 | Beta Estimate | p Value | Partial R2 | ||
| AA | *28/*37 | −0.3 | 1.30 × 10−5 | 0.134 | −0.312 | 6.16 × 10−6 | 0.142 |
| Age | 0.002 | 0.376 | 0.006 | 0.002 | 0.32 | 0.017 | |
| Sex | 0.065 | 0.468 | 0.004 | 0.08 | 0.371 | 0.014 | |
| PGRMC1 | 0.937 | 2.57 × 10−12 | 0.309 | 0.94 | 1.74 × 10−12 | 0.316 | |
| Total adjusted R2 = 39% | Total adjusted R2 = 39% | ||||||
| EA | *28/*37 | −0.341 | 9.29 × 10−8 | 0.233 | −0.334 | 1.16 × 10−7 | 0.23 |
| age | −0.002 | 0.626 | 0.002 | −0.001 | 0.664 | 0.002 | |
| sex | −0.125 | 0.129 | 0.021 | −0.13 | 0.115 | 0.023 | |
| ARNT | −1.517 | 3.34 × 10−6 | 0.182 | −1.554 | 2.08 × 10−6 | 0.189 | |
| AHR | 1.045 | 0.0001 | 0.13 | 1.065 | 8.66 × 10−5 | 0.134 | |
| PPARA | 0.677 | 0.0002 | 0.119 | 0.667 | 0.0002 | 0.116 | |
| ESR1 | 0.345 | 0.001 | 0.096 | 0.353 | 0.001 | 0.1 | |
| PGRMC1 | 0.632 | 0.004 | 0.076 | 0.633 | 0.004 | 0.076 | |
| NFE2L2 | −0.54 | 0.02 | 0.049 | −0.562 | 0.016 | 0.053 | |
| NR1I2 | −0.424 | 0.0456 | 0.036 | −0.401 | 0.059 | 0.033 | |
| Total adjusted R2 = 52% | Total adjusted R2 = 53% | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taylor, M.J.; Collins, J.M.; Montalvo, A.D.; Wang, D. Association Between UGT1A1 mRNA Expression and Cis-Acting Genetic Variants and Trans-Acting Transcriptional Regulators in Human Liver Samples. Genes 2025, 16, 971. https://doi.org/10.3390/genes16080971
Taylor MJ, Collins JM, Montalvo AD, Wang D. Association Between UGT1A1 mRNA Expression and Cis-Acting Genetic Variants and Trans-Acting Transcriptional Regulators in Human Liver Samples. Genes. 2025; 16(8):971. https://doi.org/10.3390/genes16080971
Chicago/Turabian StyleTaylor, Matthew J., Joseph M. Collins, Abelardo D. Montalvo, and Danxin Wang. 2025. "Association Between UGT1A1 mRNA Expression and Cis-Acting Genetic Variants and Trans-Acting Transcriptional Regulators in Human Liver Samples" Genes 16, no. 8: 971. https://doi.org/10.3390/genes16080971
APA StyleTaylor, M. J., Collins, J. M., Montalvo, A. D., & Wang, D. (2025). Association Between UGT1A1 mRNA Expression and Cis-Acting Genetic Variants and Trans-Acting Transcriptional Regulators in Human Liver Samples. Genes, 16(8), 971. https://doi.org/10.3390/genes16080971






