Differential Expression of Epstein–Barr Virus Sequences in Various Breast Cancer Subtypes
Abstract
1. Introduction
2. Materials and Methods
2.1. RNA-Seq Datasets Utilized
2.2. RNA-Seq Data Analysis
2.3. Identification of EBV Gene Expressed Sequence Hotspots
2.4. Identification of Novel EBV miRNAs Potentially Generated from the EBV Gene-Expressed Sequence Hotspot Regions
2.5. Statistical Analysis
3. Results
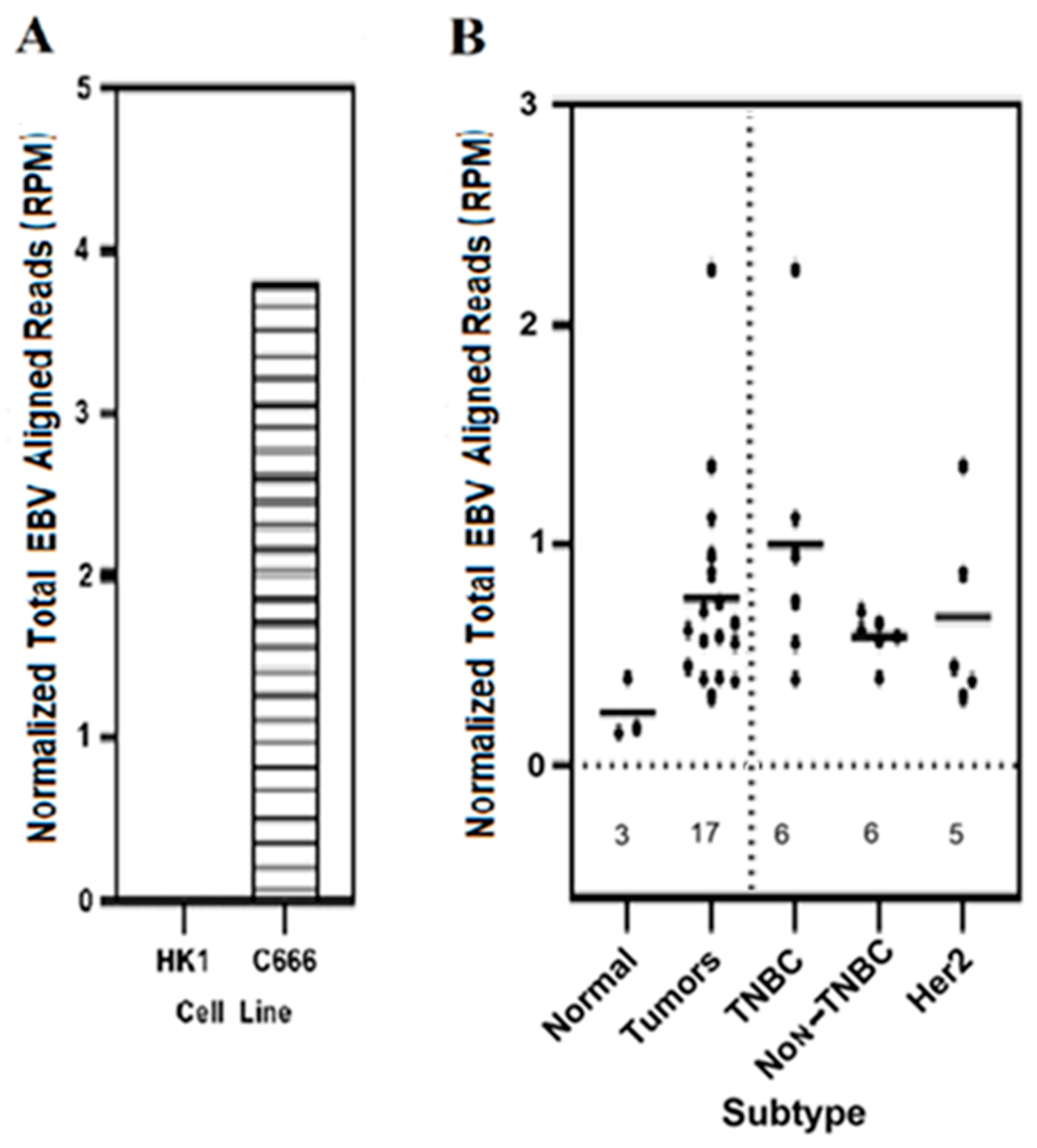
3.1. Expression of Total EBV Sequences Was Higher in Breast Cancer Tumors as Compared to the Results for Control Normal Breast Tissues
3.2. Expression of EBV Gene Transcript Sequences Were Elevated in Various Breast Cancer Tumor Subtypes as Compared to That in Control Normal Breast Tissues
3.3. Seven EBV Gene Transcript Sequences Were Downregulated in Breast Tumors Compared to in Control Normal Breast Tissue
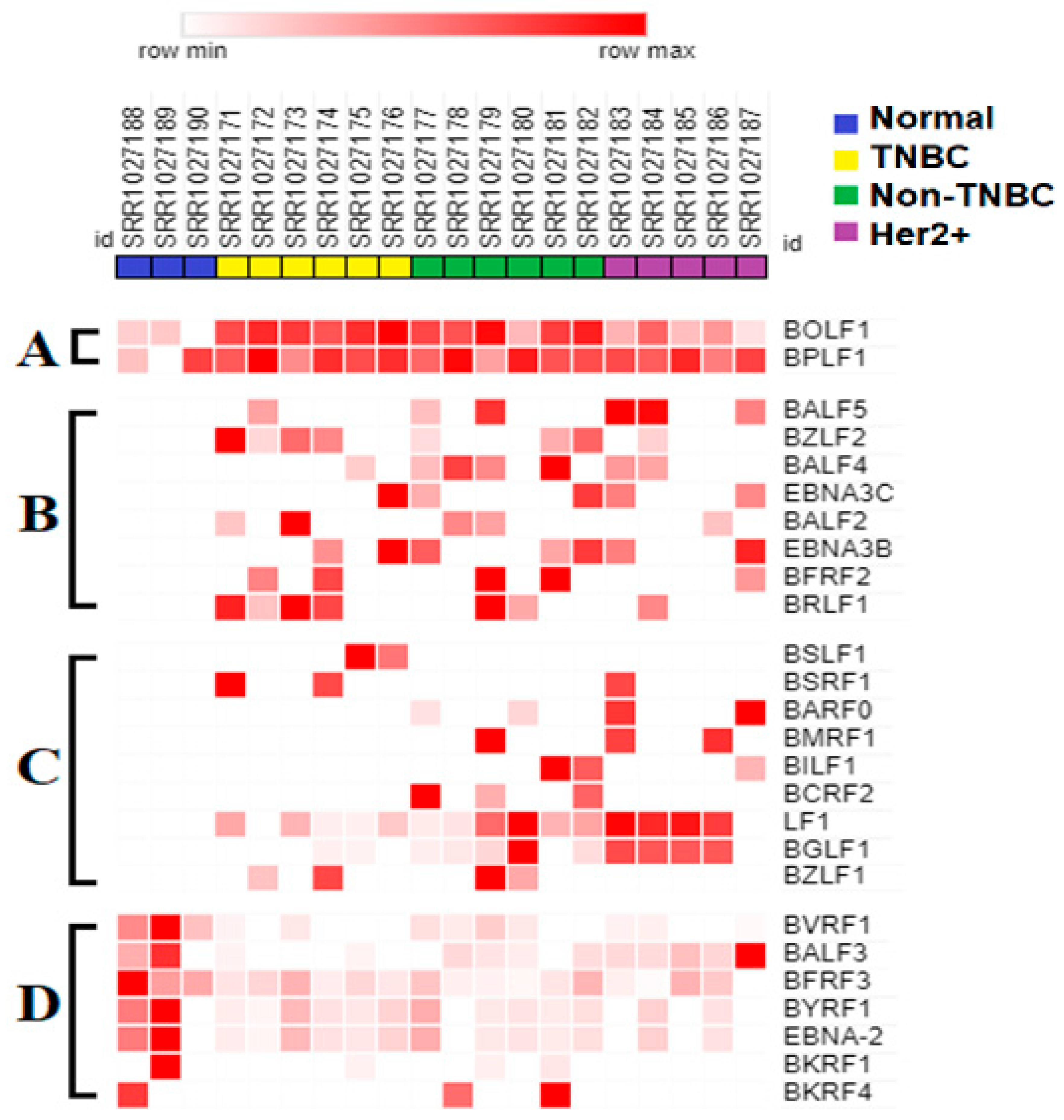
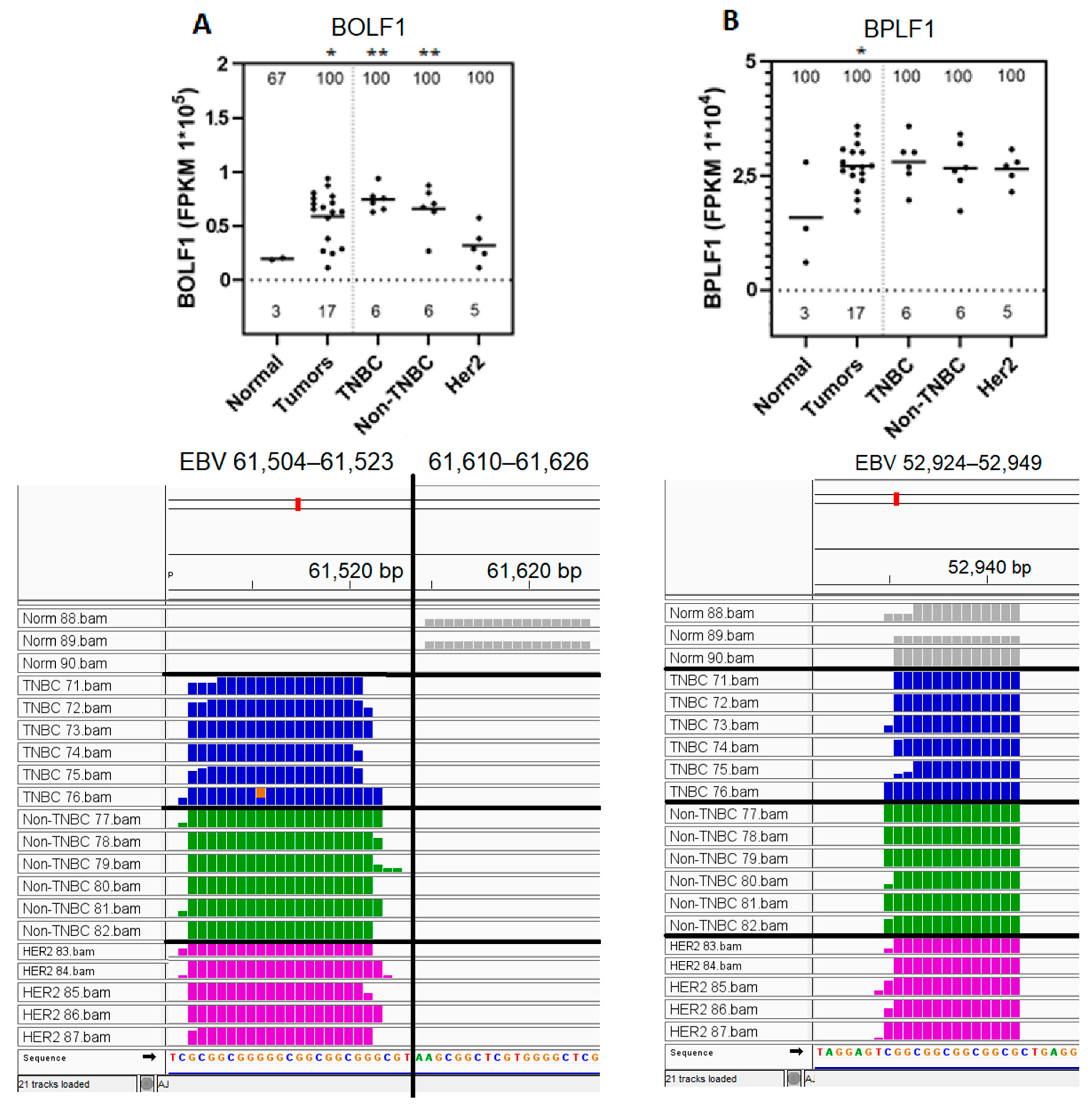
3.4. BOLF1 and BPLF1 EBV Gene Transcript Sequences Expressed in All Samples Are Upregulated in Breast Cancer Tumors Compared to in Normal Control Breast Tissue
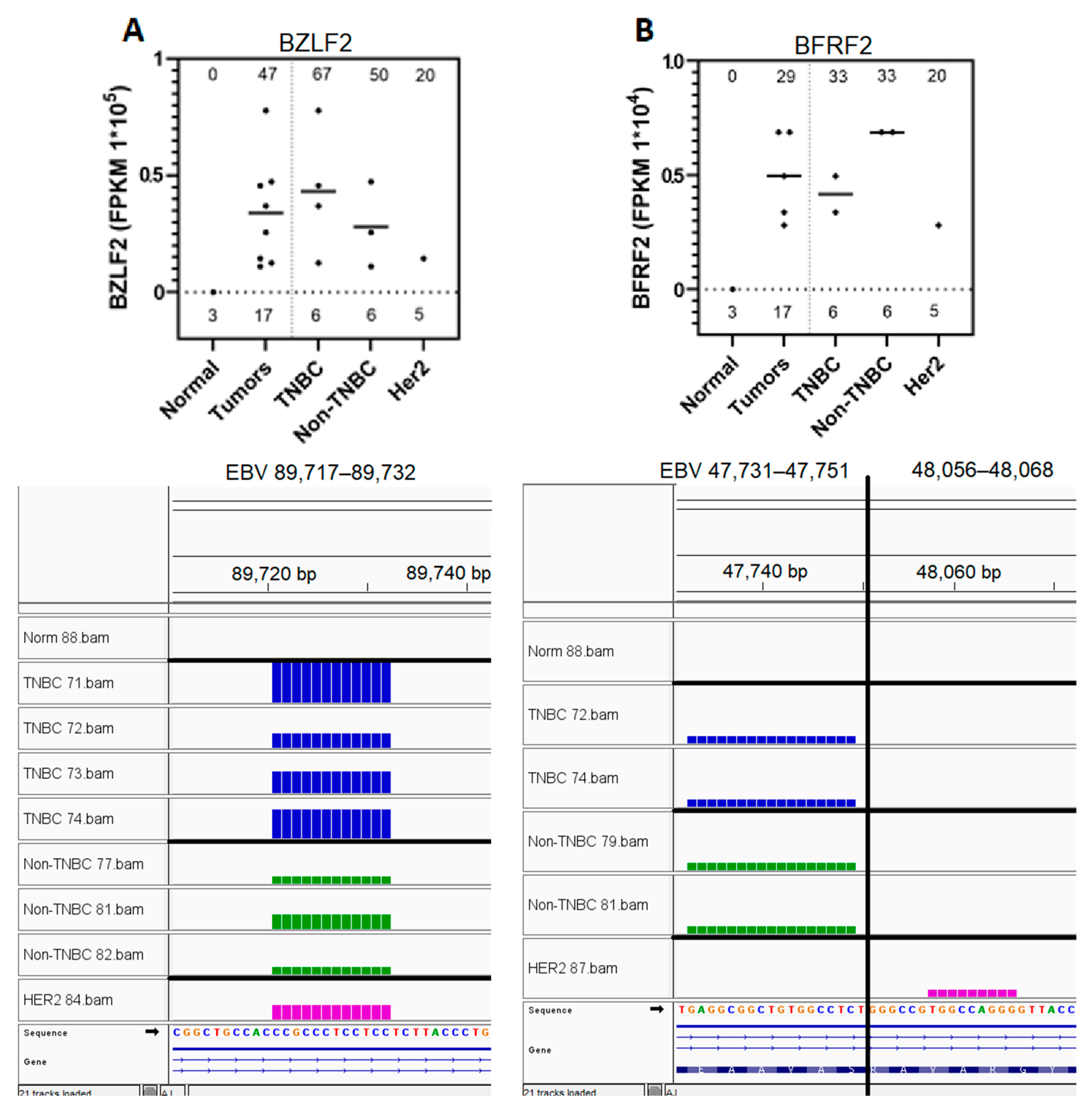
3.5. Eight EBV Gene Transcript Sequences Were Expressed in Breast Tumors and Not in Normal Breast Tissue Samples
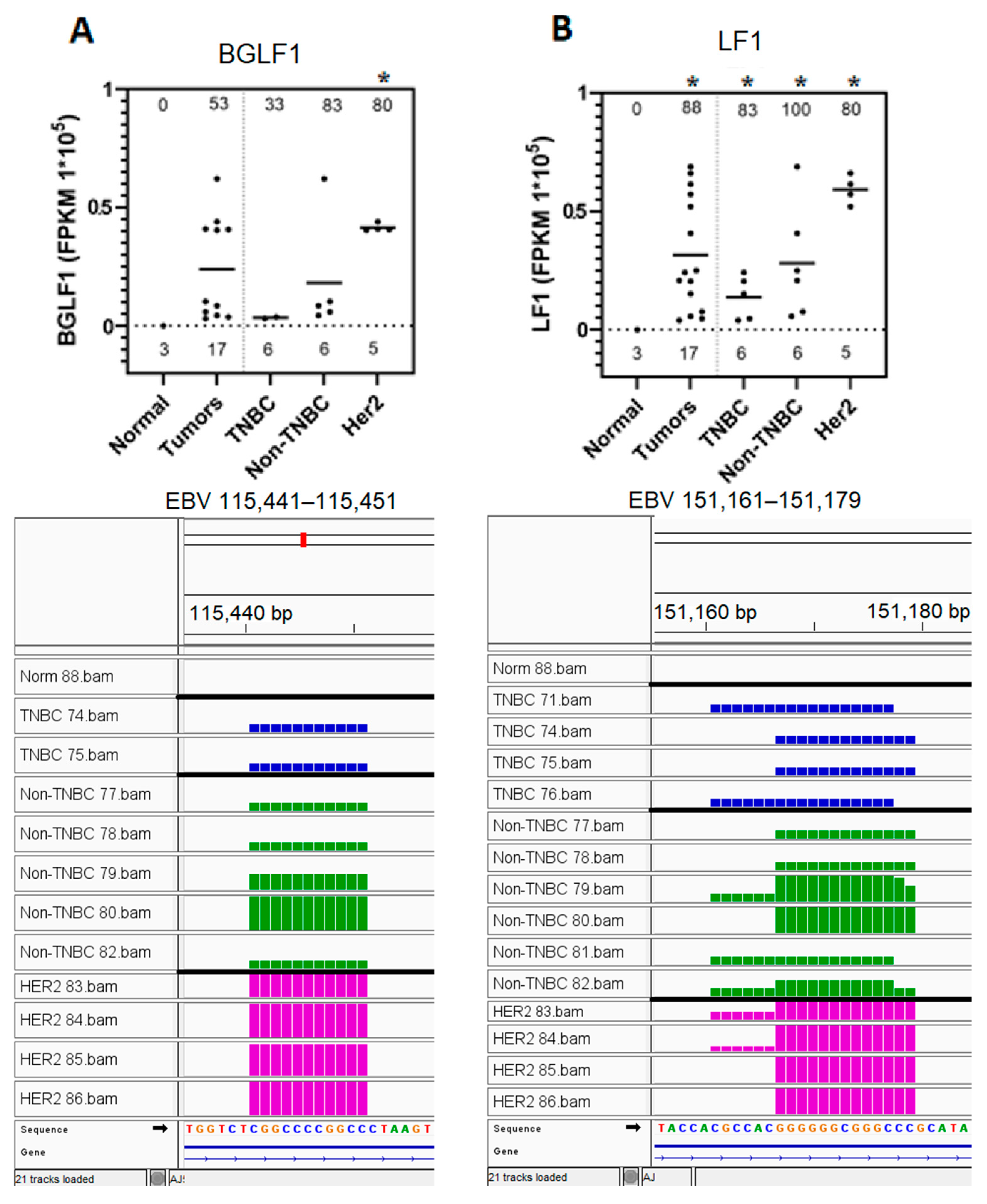
3.6. EBV BGLF1 and LF1 Gene Transcript Sequences Are Preferentially Expressed in HER2-Positive Tumors and Not in Normal Breast Tissue Controls
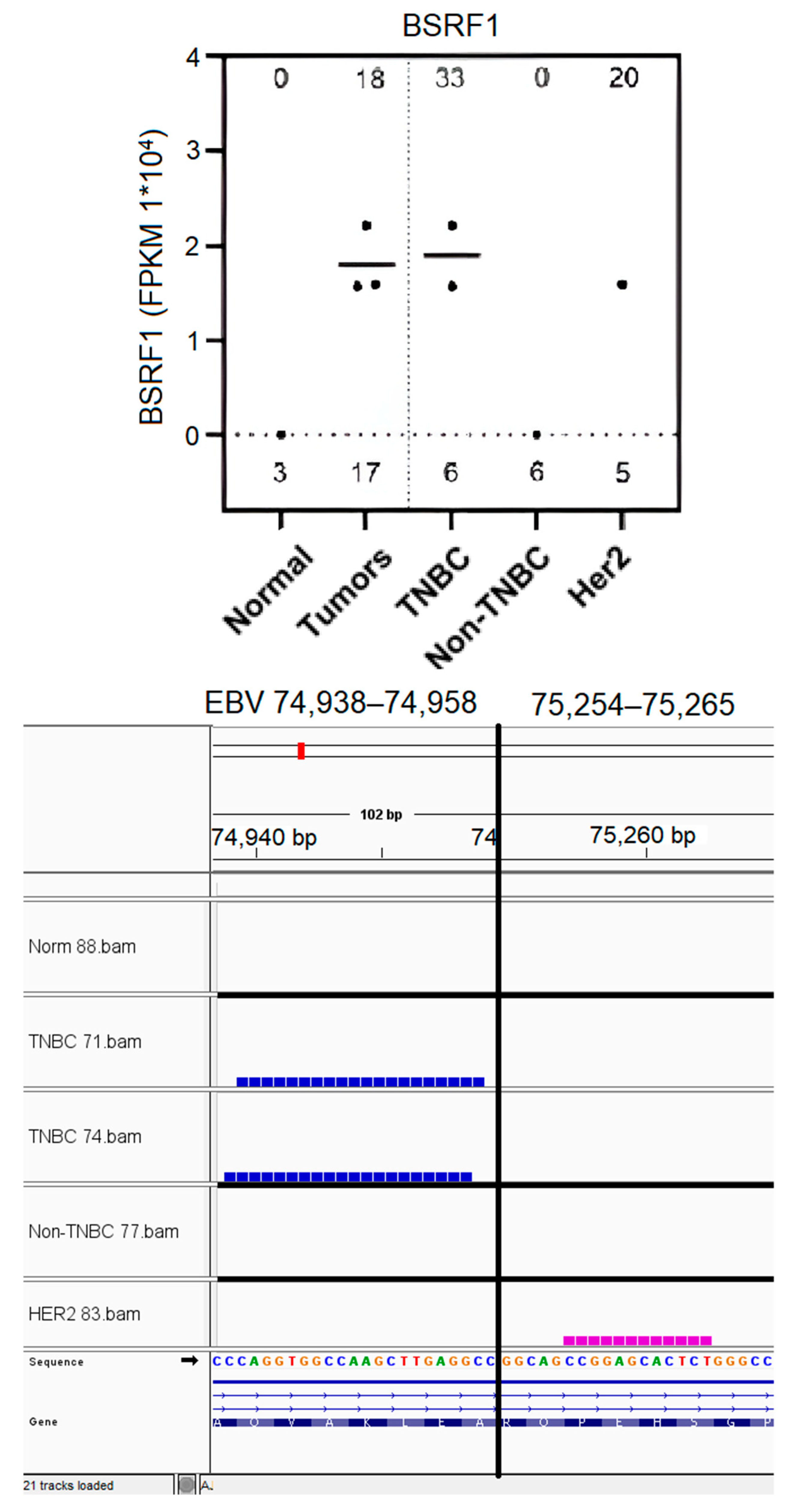
3.7. EBV BSRF1 Gene Transcript Sequences Were Preferentially Expressed in Triple-Negative Breast Cancer and HER2 Subtypes
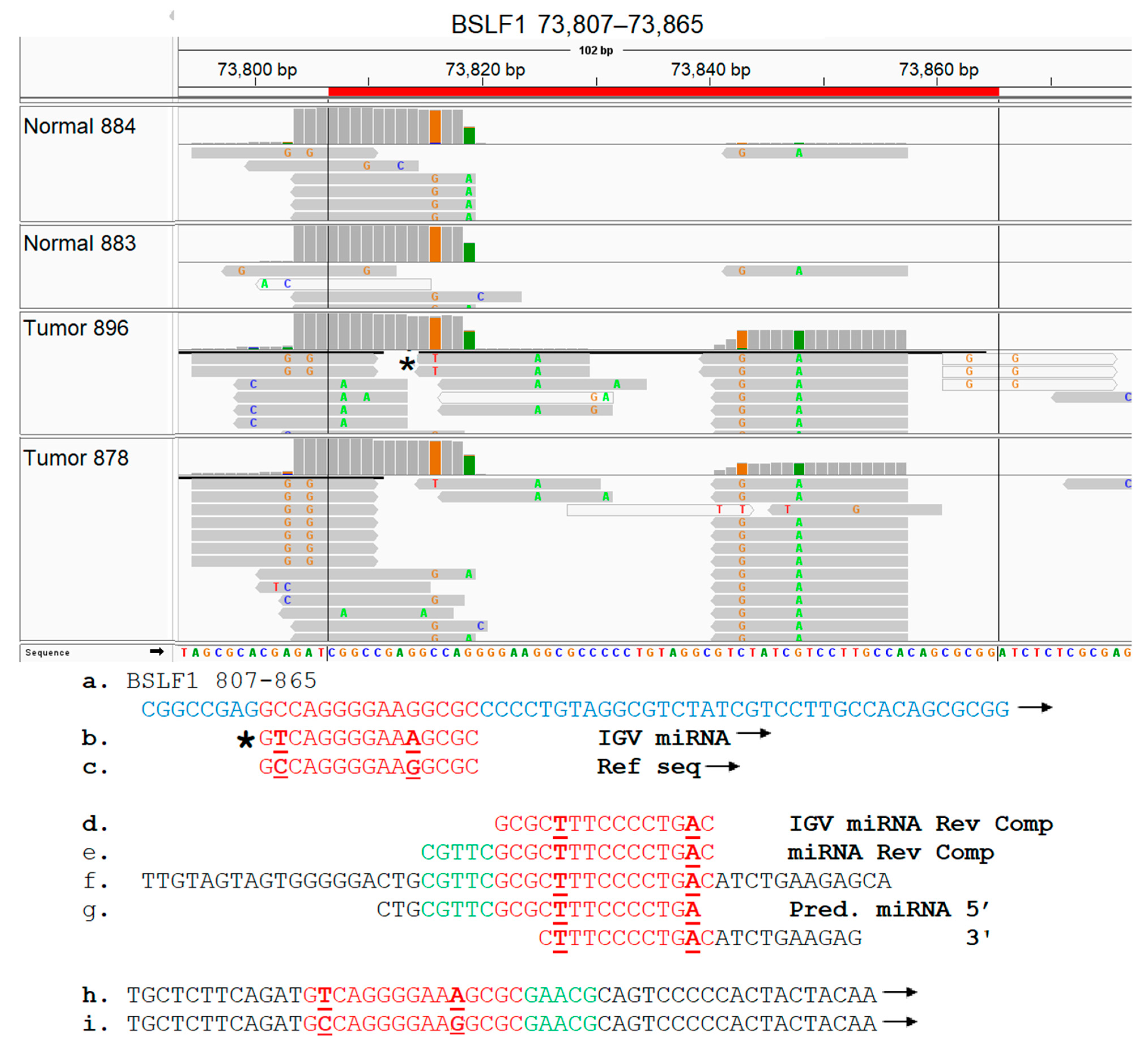
3.8. EBV BSLF1 Gene Transcript Sequences Are Expressed Specifically in Triple-Negative Breast Cancer Subtypes
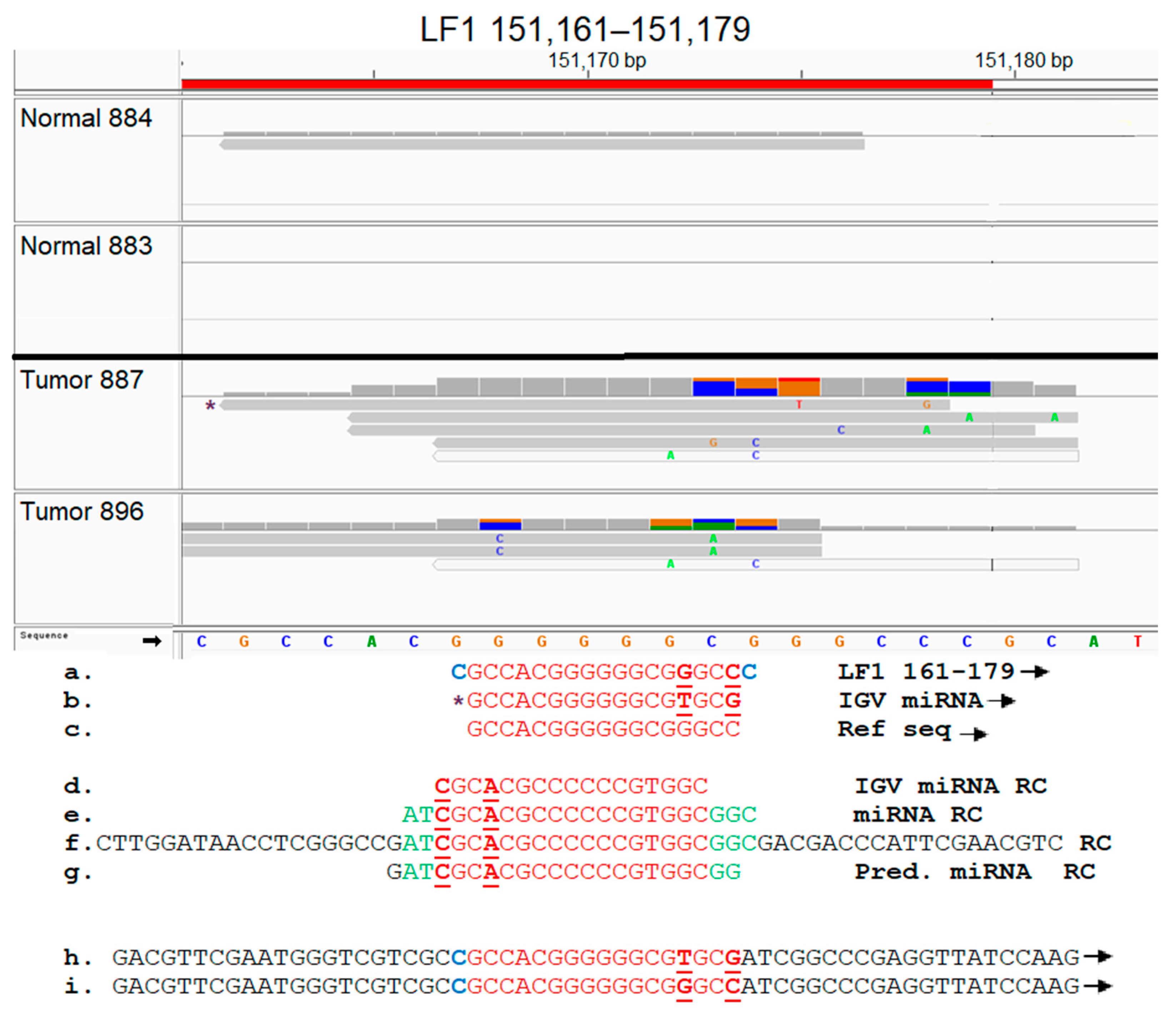
3.9. Functional Analysis of EBV Transcript Sequences Detects Putative Novel miRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.; Ji, Y.; Liu, S.; Li, J.; Wu, J.; Jin, Q.; Liu, X.; Duan, H.; Feng, Z.; Liu, Y.; et al. Global burden of female breast cancer: New estimates in 2022, temporal trend and future projections up to 2050 based on the latest release from GLOBOCAN. J. Natl. Cancer Cent. 2025, 5, 287–296. [Google Scholar] [CrossRef]
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. CA Cancer J. Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef] [PubMed]
- Swain, S.M.; Shastry, M.; Hamilton, E. Targeting HER2-positive breast cancer: Advances and future directions. Nat. Rev. Drug Discov. 2023, 22, 101–126. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Perou, C.M. Deconstructing the molecular portraits of breast cancer. Mol. Oncol. 2011, 5, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, E.; Canberk, S.; Schmitt, F.; Vale, N. Molecular Subtypes and Mechanisms of Breast Cancer: Precision Medicine Approaches for Targeted Therapies. Cancers 2025, 17, 1102. [Google Scholar] [CrossRef] [PubMed]
- Swarbrick, A.; Fernandez-Martinez, A.; Perou, C.M. Gene-expression profiling to decipher breast cancer inter-and intratumor heterogeneity. Cold Spring Harb. Perspect. Med. 2023, 14, a041320. [Google Scholar] [CrossRef] [PubMed]
- Turner, K.M.; Yeo, S.K.; Holm, T.M.; Shaughnessy, E.; Guan, J.-L. Heterogeneity within molecular subtypes of breast cancer. Am. J. Physiol.-Cell Physiol. 2021, 321, C343–C354. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Li, T.; Bai, Z.; Yang, Y.; Liu, X.; Zhan, J.; Shi, B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am. J. Cancer Res. 2015, 5, 2929–2943. [Google Scholar] [PubMed]
- Orrantia-Borunda, E.; Anchondo-Nuñez, P.; Acuña-Aguilar, L.E.; Gómez-Valles, F.O.; Ramírez-Valdespino, C.A. Subtypes of Breast Cancer. In Breast Cancer; Mayrovitz, H.N., Ed.; Exon Publications: Brisbane, Australia, 2022. [Google Scholar]
- Hou, Y.; Nitta, H.; Li, Z. HER2 Intratumoral Heterogeneity in Breast Cancer, an Evolving Concept. Cancers 2023, 15, 2664. [Google Scholar] [CrossRef] [PubMed]
- Manjunath, M.; Choudhary, B. Triple-negative breast cancer: A run-through of features, classification and current therapies. Oncol. Lett. 2021, 22, 512. [Google Scholar] [CrossRef] [PubMed]
- Marra, A.; Trapani, D.; Viale, G.; Criscitiello, C.; Curigliano, G. Practical classification of triple-negative breast cancer: Intratumoral heterogeneity, mechanisms of drug resistance, and novel therapies. NPJ Breast Cancer 2020, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Ma, D.; Xiao, Y.; Li, X.M.; Ma, J.L.; Zhang, H.; Xu, X.L.; Lv, H.; Jiang, W.H.; Yang, W.T.; et al. Molecular Subtyping of Triple-Negative Breast Cancers by Immunohistochemistry: Molecular Basis and Clinical Relevance. Oncologist 2020, 25, e1481–e1491. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Pietenpol, J.A.; Tan, A.R. Triple-negative breast cancer: Molecular subtypes and new targets for therapy. Am. Soc. Clin. Oncol. Educ. Book. 2015, 35, e31–e39. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Duan, J.-J.; Bian, X.-W.; Yu, S.-c. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Kong, D.; Liu, J.; Zhan, L.; Luo, L.; Zheng, W.; Zheng, Q.; Chen, C.; Sun, S. Breast cancer heterogeneity and its implication in personalized precision therapy. Exp. Hematol. Oncol. 2023, 12, 3. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhao, G.; Liu, X.L.; Zhang, G.; Zhao, S.Q.; Zhang, S.L.; Luo, L.H.; Yin, D.C.; Zhang, C.Y. Progress of non-coding RNAs in triple-negative breast cancer. Life Sci. 2021, 272, 119238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Lee, A.V.; Rosen, J.M. The Cellular Origin and Evolution of Breast Cancer. Cold Spring Harb. Perspect. Med. 2017, 7, a027128. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Dong, M.; Li, C.; Sheng, J. Unraveling Heterogeneity of Tumor Cells and Microenvironment and Its Clinical Implications for Triple Negative Breast Cancer. Front. Oncol. 2021, 11, 557477. [Google Scholar] [CrossRef] [PubMed]
- Esparza-López, J.; Escobar-Arriaga, E.; Soto-Germes, S.; Ibarra-Sánchez, M.J. Breast Cancer Intra-Tumor Heterogeneity: One Tumor, Different Entities. Rev. Investig. Clin. 2017, 69, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Shlyakhtina, Y.; Moran, K.L.; Portal, M.M. Genetic and Non-Genetic Mechanisms Underlying Cancer Evolution. Cancers 2021, 13, 1380. [Google Scholar] [CrossRef] [PubMed]
- Tsintarakis, A.; Papalouka, C.; Kontarini, C.; Zoumpourlis, P.; Karakostis, K.; Adamaki, M.; Zoumpourlis, V. The Intricate Interplay between Cancer Stem Cells and Oncogenic miRNAs in Breast Cancer Progression and Metastasis. Life 2023, 13, 1361. [Google Scholar] [CrossRef] [PubMed]
- Ciferri, M.C.; Tasso, R. Extracellular vesicle-mediated chemoresistance in breast cancer: Focus on miRNA cargo. Extracell. Vesicles Circ. Nucl. Acids 2025, 6, 112–127. [Google Scholar] [CrossRef] [PubMed]
- Ottaiano, A.; Ianniello, M.; Santorsola, M.; Ruggiero, R.; Sirica, R.; Sabbatino, F.; Perri, F.; Cascella, M.; Di Marzo, M.; Berretta, M.; et al. From Chaos to Opportunity: Decoding Cancer Heterogeneity for Enhanced Treatment Strategies. Biology 2023, 12, 1183. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Yajima, M.; Ikuta, K. Epstein-Barr virus strain variation and cancer. Cancer Sci. 2019, 110, 1132–1139. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Liu, Y.; Li, T.; Wang, C.; He, S.; Zhai, L.; Yang, Z.; Zhang, X.; Wu, Y.; Liu, Y. Viral oncogenesis in cancer: From mechanisms to therapeutics. Signal Transduct. Target. Ther. 2025, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.; Meehan, M.T.; Burrows, S.R.; Doolan, D.L.; Miles, J.J. Estimating the global burden of Epstein–Barr virus-related cancers. J. Cancer Res. Clin. Oncol. 2022, 148, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Rosemarie, Q.; Sugden, B. Epstein-Barr Virus: How Its Lytic Phase Contributes to Oncogenesis. Microorganisms 2020, 8, 1824. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lee, C.-Y.; Delecluse, H.-J. Epstein–Barr virus lytic replication and cancer. Curr. Opin. Virol. 2025, 70, 101438. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.d.M.; Alves, C.E.d.C.; Pontes, G.S. Epstein-Barr virus: The mastermind of immune chaos. Front. Immunol. 2024, 15, 1297994. [Google Scholar] [CrossRef] [PubMed]
- Salih, M.M.; Higgo, A.A.; Khalifa, A.S.; Eed, E.M. Incidence of Epstein-Barr Virus Among Women With Breast Cancer Using Monoclonal Antibodies for Latent Membrane Protein 1 (LMP1). Vivo 2022, 36, 1513–1518. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Liu, W.; Luo, B. Role of Oxidative Stress in the Epstein–Barr Virus Lifecycle and Tumorigenicity. Future Virol. 2023, 18, 465–477. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, W.L.; Zhu, Q.; Zhang, S.; Yao, Y.Y.; Xiang, T.; Feng, Q.S.; Zhang, Z.; Peng, R.J.; Jia, W.H.; et al. Genome-wide profiling of Epstein-Barr virus integration by targeted sequencing in Epstein-Barr virus associated malignancies. Theranostics 2019, 9, 1115–1124. [Google Scholar] [CrossRef] [PubMed]
- Friedenson, B. Evidence of lesions from Epstein-Barr virus infection in human breast cancer genomes. medRxiv 2024. [Google Scholar] [CrossRef]
- Torne, A.S.; Robertson, E.S. Epigenetic Mechanisms in Latent Epstein-Barr Virus Infection and Associated Cancers. Cancers 2024, 16, 991. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Tang, Y.; Wang, J.; Xiong, F.; Guo, C.; Wang, Y.; Xiang, B.; Zhou, M.; Li, X.; Wu, X.; et al. The emerging role of Epstein-Barr virus encoded microRNAs in nasopharyngeal carcinoma. J. Cancer 2018, 9, 2852–2864. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Xie, Y.; Wang, T.; Wang, L. New insights into Epstein-Barr virus-associated tumors: Exosomes (Review). Oncol. Rep. 2022, 47, 13. [Google Scholar] [CrossRef] [PubMed]
- Agolli, A.; Ishak, A.; Viswanathan, M.; Co, E.L.; Shivakumar, J.; Agolli, O. Epstein-Barr Viral Infection and the Risk for Breast Cancer: A Systematic Review. Int. J. Hematol. Oncol. Stem Cell Res. 2023, 17, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, M.; Guinebretiere, J.M.; Kremmer, E.; Grunewald, V.; Benhamou, E.; Contesso, G.; Joab, I. Detection of Epstein-Barr virus in invasive breast cancers. J. Natl. Cancer Inst. 1999, 91, 1376–1381. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Luo, M.L.; Desmedt, C.; Nabavi, S.; Yadegarynia, S.; Hong, A.; Konstantinopoulos, P.A.; Gabrielson, E.; Hines-Boykin, R.; Pihan, G.; et al. Epstein-Barr Virus Infection of Mammary Epithelial Cells Promotes Malignant Transformation. EBioMedicine 2016, 9, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Mazouni, C.; Fina, F.; Romain, S.; Ouafik, L.; Bonnier, P.; Brandone, J.M.; Martin, P.M. Epstein-Barr virus as a marker of biological aggressiveness in breast cancer. Br. J. Cancer 2011, 104, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.D.; Alghareeb, R.; Hussain, A.; Maheshwari, M.V.; Khalid, N. The Association of Epstein-Barr Virus With Cancer. Cureus 2022, 14, e26314. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; He, C.; Wu, J.; Yang, D.; Yi, W. Epstein barr virus encodes miRNAs to assist host immune escape. J. Cancer 2020, 11, 2091–2100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, B.; Zheng, L.; Fu, Z.; Fu, Y.; Huang, W.; Cheng, A. Advances in Research on microRNAs Related to the Invasion and Metastasis of Nasopharyngeal Carcinoma. Curr. Mol. Pharmacol. 2022, 15, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Torres, K.; Landeros, N.; Wichmann, I.A.; Polakovicova, I.; Aguayo, F.; Corvalan, A.H. EBV miR-BARTs and human lncRNAs: Shifting the balance in competing endogenous RNA networks in EBV-associated gastric cancer. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166049. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, W.; Luo, B. The roles of miRNAs and lncRNAs in Epstein-Barr virus associated epithelial cell tumors. Virus Res. 2021, 291, 198217. [Google Scholar] [CrossRef] [PubMed]
- Iizasa, H.; Kim, H.; Kartika, A.V.; Kanehiro, Y.; Yoshiyama, H. Role of Viral and Host microRNAs in Immune Regulation of Epstein-Barr Virus-Associated Diseases. Front. Immunol. 2020, 11, 367. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, T.; Zhou, Y.; Cheng, A.S.L.; Yu, J.; To, K.F.; Kang, W. The oncogenic role of Epstein-Barr virus-encoded microRNAs in Epstein-Barr virus-associated gastric carcinoma. J. Cell Mol. Med. 2018, 22, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ye, Y.; Fu, M.; Zheng, B.; Qiu, Q.; Huang, Z. Implication of viral microRNAs in the genesis and diagnosis of Epstein-Barr virus-associated tumors. Oncol. Lett. 2019, 18, 3433–3442. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, J.; Zhang, Z.; Tang, F. Epstein-Barr Virus-Encoded Products Promote Circulating Tumor Cell Generation: A Novel Mechanism of Nasopharyngeal Carcinoma Metastasis. Onco Targets Ther. 2019, 12, 11793–11804. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Ye, Y.; Jiang, Q.; Chen, Y.; Lyu, X.; Li, J.; Wang, S.; Liu, T.; Cai, H.; Yao, K.; et al. Epstein–Barr virus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma. Nat. Commun. 2015, 6, 7353. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Wang, J.; Wei, L.; Peng, Q.; Gao, Y.; Fu, Y.; Lu, Y.; Qin, Z.; Zhang, X.; Lu, J.; et al. Epstein-Barr Virus MicroRNA miR-BART5-3p Inhibits p53 Expression. J. Virol. 2018, 92, e01022-18. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.J.; Chen, G.H.; Chen, Y.H.; Liu, C.Y.; Chang, K.P.; Chang, Y.S.; Chen, H.C. Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS ONE 2010, 5, e12745. [Google Scholar] [CrossRef] [PubMed]
- Szeto, C.Y.; Lin, C.H.; Choi, S.C.; Yip, T.T.; Ngan, R.K.; Tsao, G.S.; Li Lung, M. Integrated mRNA and microRNA transcriptome sequencing characterizes sequence variants and mRNA-microRNA regulatory network in nasopharyngeal carcinoma model systems. FEBS Open Bio. 2014, 4, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Arias-Calvachi, C.; Blanco, R.; Calaf, G.M.; Aguayo, F. Epstein-Barr Virus Association with Breast Cancer: Evidence and Perspectives. Biology 2022, 11, 799. [Google Scholar] [CrossRef] [PubMed]
- Borozan, I.; Zapatka, M.; Frappier, L.; Ferretti, V. Analysis of Epstein-Barr virus genomes and expression profiles in gastric adenocarcinoma. J. Virol. 2018, 92, e01239-17. [Google Scholar] [CrossRef] [PubMed]
- Fimereli, D.; Gacquer, D.; Fumagalli, D.; Salgado, R.; Rothé, F.; Larsimont, D.; Sotiriou, C.; Detours, V. No significant viral transcription detected in whole breast cancer transcriptomes. BMC Cancer 2015, 15, 147. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Pananá, E.M.; Larios-Serrato, V.; Méndez-Tenorio, A.; Morales-Sánchez, A.; Arias, C.F.; Torres, J. Assessment of Epstein-Barr virus nucleic acids in gastric but not in breast cancer by next-generation sequencing of pooled Mexican samples. Mem. Inst. Oswaldo Cruz 2016, 111, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Khoury, J.D.; Tannir, N.M.; Williams, M.D.; Chen, Y.; Yao, H.; Zhang, J.; Thompson, E.J.; Meric-Bernstam, F.; Medeiros, L.J.; Weinstein, J.N.; et al. Landscape of DNA virus associations across human malignant cancers: Analysis of 3,775 cases using RNA-Seq. J. Virol. 2013, 87, 8916–8926. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, W.; Pan, Y.; Ji, J.; Lu, Z.; Ke, Y. Genome-wide analysis of Epstein-Barr virus (EBV) isolated from EBV-associated gastric carcinoma (EBVaGC). Oncotarget 2015, 7, 4903. [Google Scholar] [CrossRef] [PubMed]
- Rossi, C.; Inzani, F.S.; Cesari, S.; Rizzo, G.; Paulli, M.; Pedrazzoli, P.; Lasagna, A.; Lucioni, M. The Role of Oncogenic Viruses in the Pathogenesis of Sporadic Breast Cancer: A Comprehensive Review of the Current Literature. Pathogens 2024, 13, 451. [Google Scholar] [CrossRef] [PubMed]
- Song, K.-A.; Yang, S.-D.; Hwang, J.; Kim, J.-I.; Kang, M.-S. The full-length DNA sequence of Epstein Barr virus from a human gastric carcinoma cell line, SNU-719. Virus Genes. 2015, 51, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.-W.; Alaei-Mahabadi, B.; Samuelsson, T.; Lindh, M.; Larsson, E. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nat. Commun. 2013, 4, 2513. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.W.; Larsson, E. Tumour virology in the era of high-throughput genomics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160265. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.; Zeng, Z.; Qi, P.; Li, X.; Yu, Z.; Guo, C.; Xiong, F.; Xiang, B.; Zhou, M.; Gong, Z. Genome-wide analysis of 18 Epstein-Barr viruses isolated from primary nasopharyngeal carcinoma biopsy specimens. J. Virol. 2017, 91, e00301-17. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Yao, Y.; Chen, H.; Zhang, S.; Cao, S.-M.; Zhang, Z.; Luo, B.; Liu, Z.; Li, Z.; Xiang, T. Genome sequencing analysis identifies Epstein–Barr virus subtypes associated with high risk of nasopharyngeal carcinoma. Nat. Genet. 2019, 51, 1131–1136. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.-S.; Li, D.-J.; Liu, Q.-L.; Song, L.-B.; Li, M.-Z.; Zhang, R.-H.; Yu, X.-J.; Wang, H.-M.; Ernberg, I.; Zeng, Y.-X. Genomic sequence analysis of Epstein-Barr virus strain GD1 from a nasopharyngeal carcinoma patient. J. Virol. 2005, 79, 15323–15330. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Xu, M.; Liang, L.; Zhang, H.; Xu, R.; Feng, Q.; Feng, L.; Luo, B.; Zeng, Y.X. Genome-wide analysis of Epstein-Barr virus identifies variants and genes associated with gastric carcinoma and population structure. Tumour Biol. 2017, 39, 1010428317714195. [Google Scholar] [CrossRef] [PubMed]
- Glaser, S.L.; Canchola, A.J.; Keegan, T.H.; Clarke, C.A.; Longacre, T.A.; Gulley, M.L. Variation in risk and outcomes of Epstein-Barr virus-associated breast cancer by epidemiologic characteristics and virus detection strategies: An exploratory study. Cancer Causes Control. 2017, 28, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Mashaly, M.; Ghorab, D.; Hegazy, M.; Abdelkhalek, M.; Gaballah, K.; Elzehery, R. Association between Epstein-Barr Virus Gene Polymorphism and Breast Cancer Risk among Egyptian Females. Asian Pac. J. Cancer Prev. 2022, 23, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, A.J.; Moalwi, M.H.; Amoaten, T. Is EBV associated with breast cancer in specific geographic locations? Cancers 2021, 13, 819. [Google Scholar] [CrossRef] [PubMed]
- La Frazia, S.; Pauciullo, S.; Zulian, V.; Garbuglia, A.R. Viral Oncogenesis: Synergistic Role of Genome Integration and Persistence. Viruses 2024, 16, 1965. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, D.A.; Tayyar, Y.; Idris, A.; McMillan, N.A.J. A “hit-and-run” affair—A possible link for cancer progression in virally driven cancers. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188476. [Google Scholar] [CrossRef] [PubMed]
- Friedenson, B. Identifying Safeguards Disabled by Epstein-Barr Virus Infections in Genomes From Patients With Breast Cancer: Chromosomal Bioinformatics Analysis. JMIRx Med. 2025, 6, e50712. [Google Scholar] [CrossRef] [PubMed]
- Friedenson, B. Herpesvirus infections eliminate safeguards against breast cancer and its metastasis: Comparable to hereditary breast cancers. medRxiv 2023. [Google Scholar] [CrossRef]
- Hsu, Y.C.; Tsai, M.H.; Wu, G.; Liu, C.L.; Chang, Y.C.; Lam, H.B.; Su, P.Y.; Lung, C.F.; Yang, P.S. Role of Epstein-Barr Virus in Breast Cancer: Correlation with Clinical Outcome and Survival Analysis. J. Cancer 2024, 15, 2403–2411. [Google Scholar] [CrossRef] [PubMed]
- Mekrazi, S.; Kallel, I.; Jamai, D.; Yengui, M.; Khabir, A.; Gdoura, R. Epstein-Barr virus in breast carcinoma and in triple negative cases impact on clinical outcomes. Pathol. Res. Pr. Pract. 2023, 245, 154484. [Google Scholar] [CrossRef] [PubMed]
- Eswaran, J.; Cyanam, D.; Mudvari, P.; Reddy, S.D.N.; Pakala, S.B.; Nair, S.S.; Florea, L.; Fuqua, S.A.W.; Godbole, S.; Kumar, R. Transcriptomic landscape of breast cancers through mRNA sequencing. Sci. Rep. 2012, 2, 264. [Google Scholar] [CrossRef] [PubMed]
- Community, T.G. The Galaxy platform for accessible, reproducible, and collaborative data analyses: 2024 update. Nucleic Acids Res. 2024, 52, W83–W94. [Google Scholar] [CrossRef] [PubMed]
- Krueger, F. Trim Galore, v0.6.10; GitHub: San Francisco, CA, USA, 2021. [Google Scholar]
- Robinson, J.T.; Thorvaldsdottir, H.; Turner, D.; Mesirov, J.P. igv.js: An embeddable JavaScript implementation of the Integrative Genomics Viewer (IGV). Bioinformatics 2022, 39, btac830. [Google Scholar] [CrossRef] [PubMed]
- Gkirtzou, K.; Tsamardinos, I.; Tsakalides, P.; Poirazi, P. MatureBayes: A Probabilistic Algorithm for Identifying the Mature miRNA within Novel Precursors. PLoS ONE 2010, 5, e11843. [Google Scholar] [CrossRef] [PubMed]
- Hubalek, M.; Czech, T.; Müller, H. Biological Subtypes of Triple-Negative Breast Cancer. Breast Care 2017, 12, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Lawson, J.S.; Glenn, W.K. Catching viral breast cancer. Infect. Agents Cancer 2021, 16, 37. [Google Scholar] [CrossRef] [PubMed]
- Lawson, J.S.; Glenn, W.K. The viral origins of breast cancer. Infect. Agents Cancer 2024, 19, 39. [Google Scholar] [CrossRef] [PubMed]
- Al Hamad, M.; Matalka, I.; Al Zoubi, M.S.; Armogida, I.; Khasawneh, R.; Al-Husaini, M.; Sughayer, M.; Jaradat, S.; Al-Nasser, A.D.; Mazzanti, C.M. Human Mammary Tumor Virus, Human Papilloma Virus, and Epstein-Barr Virus Infection Are Associated With Sporadic Breast Cancer Metastasis. Breast Cancer 2020, 14, 1178223420976388. [Google Scholar] [CrossRef] [PubMed]
- Hachana, M.; Amara, K.; Ziadi, S.; Romdhane, E.; Gacem, R.B.; Trimeche, M. Investigation of Epstein–Barr virus in breast carcinomas in Tunisia. Pathol.-Res. Pract. 2011, 207, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Gupta, I.; Ulamec, M.; Peric-Balja, M.; Ramic, S.; Al Moustafa, A.E.; Vranic, S.; Al-Farsi, H.F. Presence of high-risk HPVs, EBV, and MMTV in human triple-negative breast cancer. Hum. Vaccin. Immunother. 2021, 17, 4457–4466. [Google Scholar] [CrossRef] [PubMed]
- Khasawneh, A.I.; Himsawi, N.; Sammour, A.; Al Shboul, S.; Alorjani, M.; Al-Momani, H.; Shahin, U.; Al-Momani, H.; Alotaibi, M.R.; Saleh, T. Association of Human Papilloma Virus, Cytomegalovirus, and Epstein-Barr Virus with Breast Cancer in Jordanian Women. Medicina 2024, 60, 699. [Google Scholar] [CrossRef] [PubMed]
- Golrokh Mofrad, M.; Kazeminezhad, B.; Faghihloo, E. Prevalence of Epstein–Barr virus (EBV) in Iranian Breast Carcinoma Patients. Asian Pac. J. Cancer Prev. 2020, 21, 133–137. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Joshi, D.; Quadri, M.; Gangane, N.; Joshi, R.; Gangane, N. Association of Epstein Barr virus infection (EBV) with breast cancer in rural Indian women. PLoS ONE 2009, 4, e8180. [Google Scholar] [CrossRef] [PubMed]
- Balfour, H.H., Jr.; Sifakis, F.; Sliman, J.A.; Knight, J.A.; Schmeling, D.O.; Thomas, W. Age-specific prevalence of Epstein-Barr virus infection among individuals aged 6-19 years in the United States and factors affecting its acquisition. J. Infect. Dis. 2013, 208, 1286–1293. [Google Scholar] [CrossRef] [PubMed]
- Joshi, D.; Buehring, G.C. Are viruses associated with human breast cancer? Scrutinizing the molecular evidence. Breast Cancer Res. Treat. 2012, 135, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Masud, H.; Watanabe, T.; Sato, Y.; Goshima, F.; Kimura, H.; Murata, T. The BOLF1 gene is necessary for effective Epstein-Barr viral infectivity. Virology 2019, 531, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Han, B.-W.; Liu, T.; Zhang, M.; Wu, Y.; Nie, J. Epstein-Barr virus deubiquitinating enzyme BPLF1 is involved in EBV carcinogenesis by affecting cellular genomic stability. Neoplasia 2024, 55, 101012. [Google Scholar] [CrossRef] [PubMed]
- Mund, R.; Whitehurst, C.B. Ubiquitin-Mediated Effects on Oncogenesis during EBV and KSHV Infection. Viruses 2024, 16, 1523. [Google Scholar] [CrossRef] [PubMed]
- Lui, W.Y.; Bharti, A.; Wong, N.M.; Jangra, S.; Botelho, M.G.; Yuen, K.S.; Jin, D.Y. Suppression of cGAS- and RIG-I-mediated innate immune signaling by Epstein-Barr virus deubiquitinase BPLF1. PLoS Pathog. 2023, 19, e1011186. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, A.; Yamashita, Y.; Kanda, T.; Murata, T.; Tsurumi, T. Epstein-Barr virus genome packaging factors accumulate in BMRF1-cores within viral replication compartments. PLoS ONE 2019, 14, e0222519. [Google Scholar] [CrossRef] [PubMed]
- Chiu, S.-H.; Wu, C.-C.; Fang, C.-Y.; Yu, S.-L.; Hsu, H.-Y.; Chow, Y.-H.; Chen, J.-Y. Epstein-Barr virus BALF3 mediates genomic instability and progressive malignancy in nasopharyngeal carcinoma. Oncotarget 2014, 5, 8583–8601. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Luo, B.; Zhao, P.; Huang, B.H. [Expression of Epstein-Barr virus genes in EBV-associated gastric carcinoma]. Ai Zheng 2004, 23, 782–787. [Google Scholar] [PubMed]
- Mrozek-Gorska, P.; Buschle, A.; Pich, D.; Schwarzmayr, T.; Fechtner, R.; Scialdone, A.; Hammerschmidt, W. Epstein-Barr virus reprograms human B lymphocytes immediately in the prelatent phase of infection. Proc. Natl. Acad. Sci. USA 2019, 116, 16046–16055. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Lee, J.H.; Seo, H.J.; Kim, K.H.; Kim, J.I.; An, C.H.; Park, W.C.; Song, B.J.; Oh, S.J.; Jung, S.S.; et al. Clinical Significance of Epstein-Barr Virus Expression in Breast Cancer. J. Korean Breast Cancer Soc. 2004, 7, 161. [Google Scholar] [CrossRef][Green Version]
- Greijer, A.E.; Ramayanti, O.; Verkuijlen, S.A.; Novalic, Z.; Juwana, H.; Middeldorp, J.M. Quantitative multi-target RNA profiling in Epstein-Barr virus infected tumor cells. J. Virol. Methods 2017, 241, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, Y.; Liu, Y.; Li, X.; Lao, S.; Long, Z.; Huang, C.; Huang, W.; Xu, C.; Chen, X.; et al. The Epstein-Barr virus small capsid protein BFRF3 disrupts the NF-small ka, CyrillicB signaling pathway by inhibiting p65 activity. FASEB J. 2024, 38, e23820. [Google Scholar] [CrossRef] [PubMed]
- Ragoczy, T.; Heston, L.; Miller, G. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 1998, 72, 7978–7984. [Google Scholar] [CrossRef] [PubMed]
- Rooney, C.M.; Rowe, D.T.; Ragot, T.; Farrell, P.J. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J. Virol. 1989, 63, 3109–3116. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Munz, C. Roles of Lytic Viral Replication and Co-Infections in the Oncogenesis and Immune Control of the Epstein-Barr Virus. Cancers 2021, 13, 2275. [Google Scholar] [CrossRef] [PubMed]
- Yap, L.F.; Wong, A.K.C.; Paterson, I.C.; Young, L.S. Functional Implications of Epstein-Barr Virus Lytic Genes in Carcinogenesis. Cancers 2022, 14, 5780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Strong, M.J.; Baddoo, M.; Lin, Z.; Wang, Y.-P.; Flemington, E.K.; Liu, Y.-Z. Interaction of Epstein-Barr virus genes with human gastric carcinoma transcriptome. Oncotarget 2017, 8, 38399. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, C.W.; Jheng, B.R.; Chen, B.S. Investigating genetic-and-epigenetic networks, and the cellular mechanisms occurring in Epstein-Barr virus-infected human B lymphocytes via big data mining and genome-wide two-sided NGS data identification. PLoS ONE 2018, 13, e0202537. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Divisconte, M.; Jiang, X.; Quink, C.; Wang, F. Epstein-Barr virus with the latent infection nuclear antigen 3B completely deleted is still competent for B-cell growth transformation in vitro. J. Virol. 2005, 79, 4506–4509. [Google Scholar] [CrossRef] [PubMed]
- Elkhalifa, A.M.E.; Nabi, S.U.; Shah, O.S.; Bashir, S.M.; Muzaffer, U.; Ali, S.I.; Wani, I.A.; Alzerwi, N.A.N.; Elderdery, A.Y.; Alanazi, A.; et al. Insight into Oncogenic Viral Pathways as Drivers of Viral Cancers: Implication for Effective Therapy. Curr. Oncol. 2023, 30, 1924–1944. [Google Scholar] [CrossRef] [PubMed]
- Tsurumi, T.; Kobayashi, A.; Tamai, K.; Yamada, H.; Daikoku, T.; Yamashita, Y.; Nishiyama, Y. Epstein-Barr virus single-stranded DNA-binding protein: Purification, characterization, and action on DNA synthesis by the viral DNA polymerase. Virology 1996, 222, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Chao, T.-Y.; Cheng, Y.-Y.; Wang, Z.-Y.; Fang, T.-F.; Chang, Y.-R.; Fuh, C.-S.; Su, M.-T.; Su, Y.-W.; Hsu, P.-H.; Su, Y.-C.; et al. Subcellular Distribution of BALF2 and the Role of Rab1 in the Formation of Epstein-Barr Virus Cytoplasmic Assembly Compartment and Virion Release. Microbiol. Spectr. 2023, 11, e0436922. [Google Scholar] [CrossRef] [PubMed]
- Kondo, S.; Okuno, Y.; Murata, T.; Dochi, H.; Wakisaka, N.; Mizokami, H.; Moriyama-Kita, M.; Kobayashi, E.; Kano, M.; Komori, T.; et al. EBV genome variations enhance clinicopathological features of nasopharyngeal carcinoma in a non-endemic region. Cancer Sci. 2022, 113, 2446–2456. [Google Scholar] [CrossRef] [PubMed]
- Munz, C. Latency and lytic replication in Epstein-Barr virus-associated oncogenesis. Nat. Rev. Microbiol. 2019, 17, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.H.; Tsai, C.H.; Chu, J.S.; Chen, J.Y.; Takada, K.; Shew, J.Y. Dysregulation of HER2/HER3 signaling axis in Epstein-Barr virus-infected breast carcinoma cells. J. Virol. 2007, 81, 5705–5713. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Xie, L.; Shi, F.; Tang, M.; Li, Y.; Hu, J.; Zhao, L.; Zhao, L.; Yu, X.; Luo, X.; et al. Targeting the signaling in Epstein-Barr virus-associated diseases: Mechanism, regulation, and clinical study. Signal Transduct. Target. Ther. 2021, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Robertson, E.S. The central role of the ubiquitin–proteasome system in EBV-mediated oncogenesis. Cancers 2022, 14, 611. [Google Scholar] [CrossRef] [PubMed]
- Schaeffner, M.; Mrozek-Gorska, P.; Buschle, A.; Woellmer, A.; Tagawa, T.; Cernilogar, F.M.; Schotta, G.; Krietenstein, N.; Lieleg, C.; Korber, P.; et al. BZLF1 interacts with chromatin remodelers promoting escape from latent infections with EBV. Life Sci. Alliance 2019, 2, e201800108. [Google Scholar] [CrossRef] [PubMed]
- Murata, T. Tegument proteins of Epstein-Barr virus: Diverse functions, complex networks, and oncogenesis. Tumour Virus Res. 2023, 15, 200260. [Google Scholar] [CrossRef] [PubMed]
- Friedenson, B. Chromosome breaks in breast cancers occur near herpes tumor virus sequences and explain why the cancer comes back. bioRxiv 2021. [Google Scholar] [CrossRef]
- Fülöp, Á.; Torma, G.; Moldován, N.; Szenthe, K.; Bánáti, F.; Almsarrhad, I.A.A.; Csabai, Z.; Tombácz, D.; Minárovits, J.; Boldogkői, Z. Integrative profiling of Epstein–Barr virus transcriptome using a multiplatform approach. Virol. J. 2022, 19, 7. [Google Scholar] [CrossRef] [PubMed]
- Mundo, L.; Ambrosio, M.R.; Picciolini, M.; Lo Bello, G.; Gazaneo, S.; Del Porro, L.; Lazzi, S.; Navari, M.; Onyango, N.; Granai, M.; et al. Unveiling Another Missing Piece in EBV-Driven Lymphomagenesis: EBV-Encoded MicroRNAs Expression in EBER-Negative Burkitt Lymphoma Cases. Front. Microbiol. 2017, 8, 229. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, P.; Kohli, A.S.; Dwivedi, M.; Hasan, S. Implications of EBV-Encoded and EBV-Related miRNAs in Tumors. Curr. Gene Ther. 2025, 25, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Afzal, S.; Fiaz, K.; Noor, A.; Sindhu, A.S.; Hanif, A.; Bibi, A.; Asad, M.; Nawaz, S.; Zafar, S.; Ayub, S.; et al. Interrelated Oncogenic Viruses and Breast Cancer. Front. Mol. Biosci. 2022, 9, 781111. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blanchard, A.; Elmalih, N.; Muganda, P. Differential Expression of Epstein–Barr Virus Sequences in Various Breast Cancer Subtypes. Genes 2025, 16, 756. https://doi.org/10.3390/genes16070756
Blanchard A, Elmalih N, Muganda P. Differential Expression of Epstein–Barr Virus Sequences in Various Breast Cancer Subtypes. Genes. 2025; 16(7):756. https://doi.org/10.3390/genes16070756
Chicago/Turabian StyleBlanchard, Alexander, Namarig Elmalih, and Perpetua Muganda. 2025. "Differential Expression of Epstein–Barr Virus Sequences in Various Breast Cancer Subtypes" Genes 16, no. 7: 756. https://doi.org/10.3390/genes16070756
APA StyleBlanchard, A., Elmalih, N., & Muganda, P. (2025). Differential Expression of Epstein–Barr Virus Sequences in Various Breast Cancer Subtypes. Genes, 16(7), 756. https://doi.org/10.3390/genes16070756





