piRNA-Mediated Maintenance of Genome Stability in Gametogenesis and Cancer
Abstract
1. Introduction
2. Transposable Elements
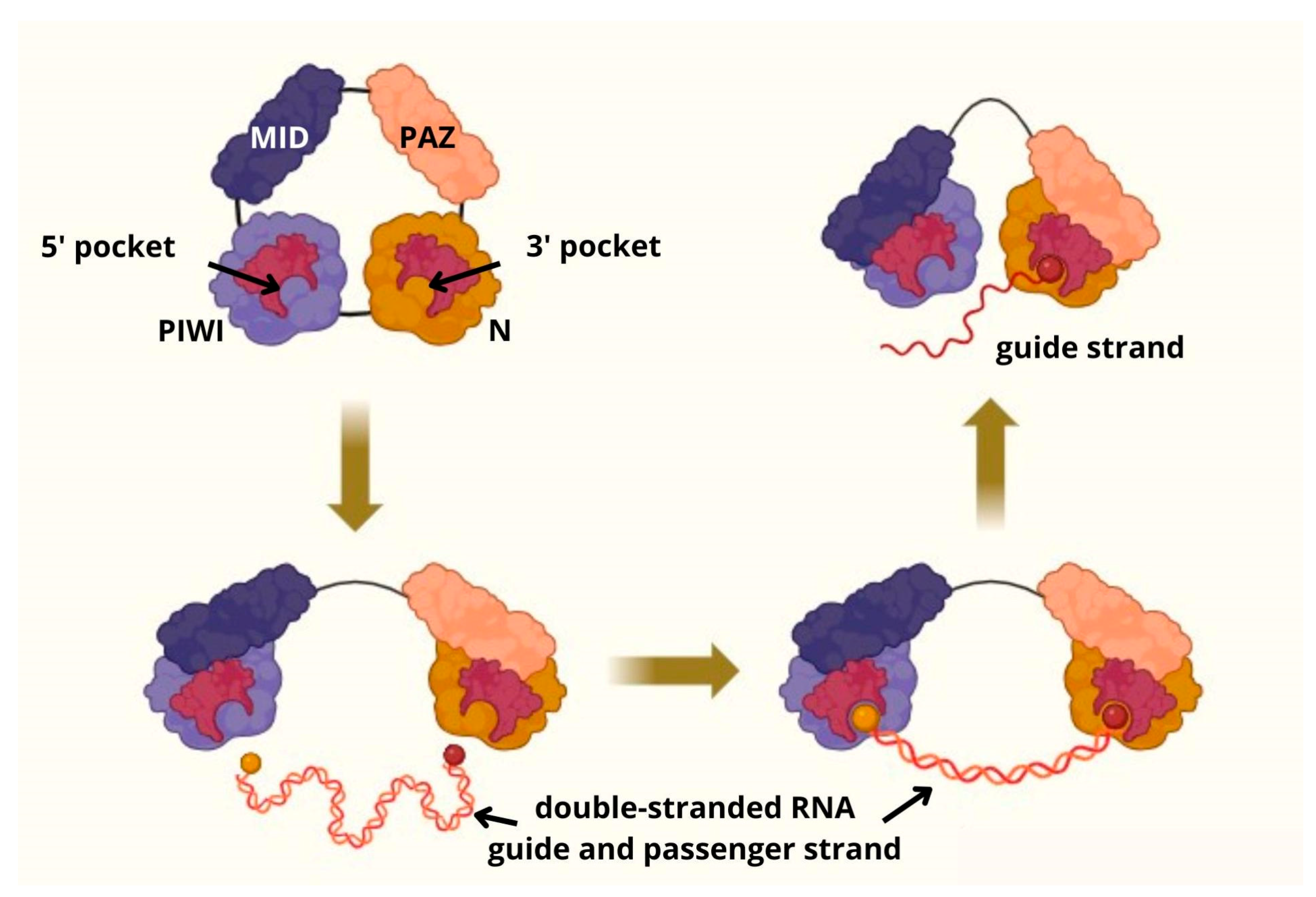
3. Origin and Structure of piRNA and PIWI Proteins Family
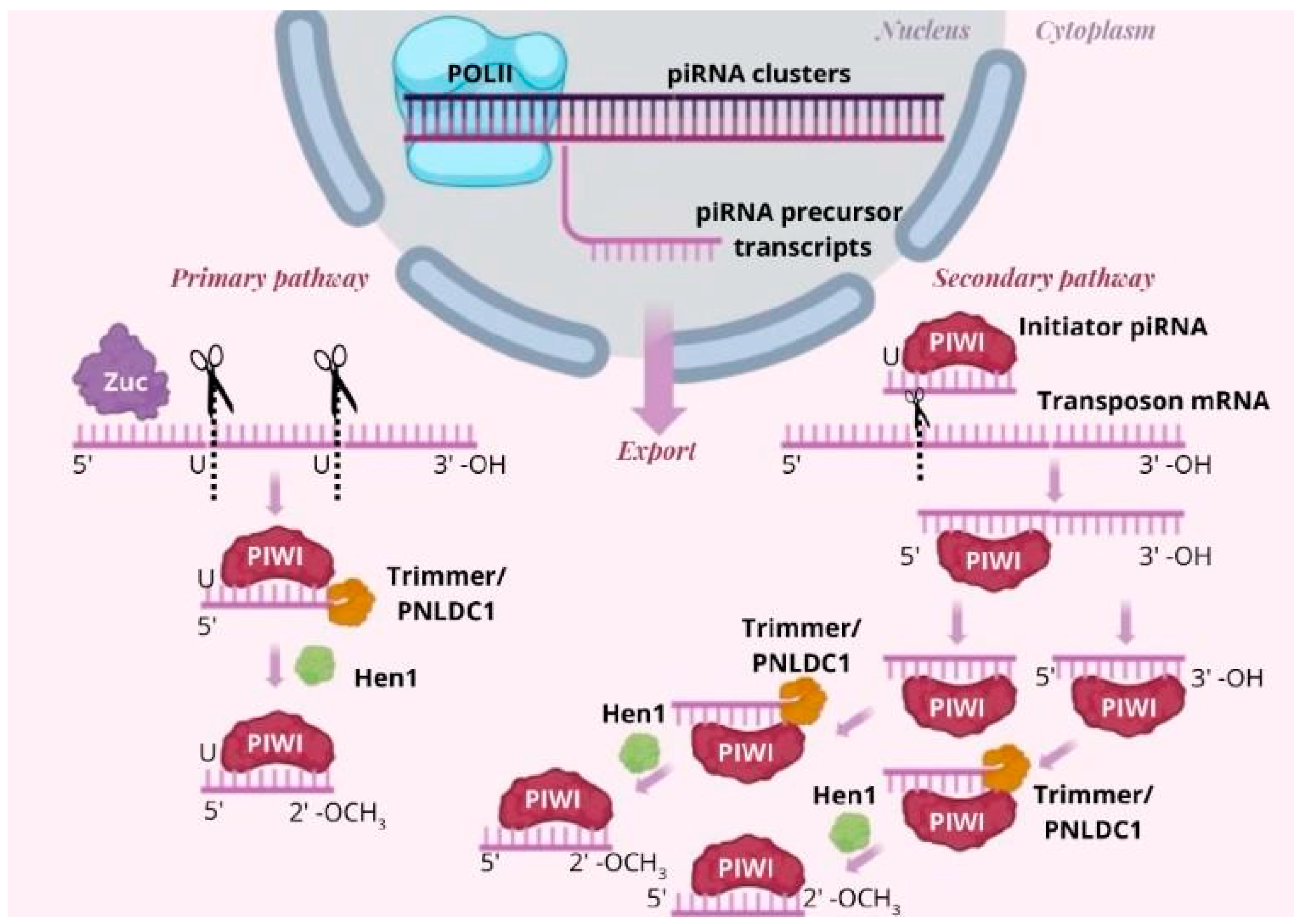
The piRNA Biogenesis Pathway
4. Functions of piRNAs
4.1. Transposon Silencing
4.2. Maintaining Genome Stability
4.3. Epigenetic Regulation of Germline Cell Development
5. Role in Physiology and Pathophysiology
5.1. Regulatory Functions of piRNAs and PIWI Proteins in Germline
5.2. Aberrant Expression of piRNAs and PIWI Proteins as Biomarkers in Cancer
5.3. Epigenetic Regulation by piRNAs: A Non-Complementary Mechanism of Gene Expression Control
| PIWI Protein | Role in Oogenesis [59,66] | Role in Spermatogenesis [61,63,64] | Role in Carcinogenesis [50,83,85,92,93] |
|---|---|---|---|
| PIWIL1 | Highly expressed. Involved in regulating transposon silencing and genomic stability. Crucial for proper oocyte development at the blastomere level. | Determines the proper sperm phenotype, ensuring proper spermatogenic development at the early stages. | Not described. |
| PIWIL2 | No apparent influence on female fertility. | Presence determines the occurrence of the spermatogenesis process. | Determines occurrence of the distant metastasis. |
| PIWIL3 | Highly expressed. Involved in regulating litter size and pregnancy rate. | Not found in spermatogenic cells. | Not described. |
| PIWIL4 | Not found in oocytes cells. | Involved in regulating male germ cell numbers and spermatogenesis arrest. | Crucial for the proliferation and apoptosis process of several cancers. |
6. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Podvalnaya, N.; Bronkhorst, A.W.; Lichtenberger, R.; Hellmann, S.; Nischwitz, E.; Falk, T.; Karaulanov, E.; Butter, F.; Falk, S.; Ketting, R.F. Author Correction: piRNA processing by a trimeric Schlafen-domain nuclease. Nature 2024, 632, E31. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Das, B.; Jain, N.; Mallick, B. piR-39980 promotes cell proliferation, migration and invasion, and inhibits apoptosis via repression of SERPINB1 in human osteosarcoma. Biol. Cell 2020, 112, 73–91. [Google Scholar] [CrossRef]
- Cornes, E.; Bourdon, L.; Singh, M.; Mueller, F.; Quarato, P.; Wernersson, E.; Bienko, M.; Li, B.; Cecere, G. piRNAs initiate transcriptional silencing of spermatogenic genes during C. elegans germline development. Dev. Cell 2022, 57, 180–196.e7. [Google Scholar] [CrossRef]
- Asif-Laidin, A.; Casier, K.; Ziriat, Z.; Boivin, A.; Viodé, E.; Delmarre, V.; Ronsseray, S.; Carré, C.; Teysset, L. Modeling early germline immunization after horizontal transfer of transposable elements reveals internal piRNA cluster heterogeneity. BMC Biol. 2023, 21, 117. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hayashi, R.; Schnabl, J.; Handler, D.; Mohn, F.; Ameres, S.L.; Brennecke, J. Genetic and mechanistic diversity of piRNA 3′-end formation. Nature 2016, 539, 588–592. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Aravin, A.A.; van der Heijden, G.W.; Castañeda, J.; Vagin, V.V.; Hannon, G.J.; Bortvin, A. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 2009, 5, e1000764. [Google Scholar] [CrossRef]
- Nakanishi, K.; Weinberg, D.E.; Bartel, D.P.; Patel, D.J. Structure of yeast Argonaute with guide RNA. Nature 2012, 486, 368–374. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Czech, B.; Munafò, M.; Ciabrelli, F.; Eastwood, E.L.; Fabry, M.H.; Kneuss, E.; Hannon, G.J. piRNA-Guided Genome Defense: From Biogenesis to Silencing. Annu. Rev. Genet. 2018, 52, 131–157. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Loubalova, Z.; Konstantinidou, P.; Haase, A.D. Themes and variations on piRNA-guided transposon control. Mob. DNA 2023, 14, 10. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Erber, R.; Meyer, J.; Taubert, H.; Fasching, P.A.; Wach, S.; Häberle, L.; Gaß, P.; Schulz-Wendtland, R.; Landgraf, L.; Olbricht, S.; et al. PIWI-Like 1 and PIWI-Like 2 Expression in Breast Cancer. Cancers 2020, 12, 2742. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cheng, J.; Deng, H.; Xiao, B.; Zhou, H.; Zhou, F.; Shen, Z.; Guo, J. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012, 315, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.C.; Lin, H. Beyond transposons: The epigenetic and somatic functions of the Piwi-piRNA mechanism. Curr. Opin. Cell Biol. 2013, 25, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Wu, F.; Li, B.; Ma, H.; Mai, L.; Peng, Y.; Feng, X.; Tan, X.; Fu, M.; Tan, Y.; et al. Cancer-associated fibroblasts-derived exosomal piR-35462 promotes the progression of oral squamous cell carcinoma via FTO/Twist1 pathway. BMC Oral Health 2025, 25, 840. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gupta, G.; Afzal, M.; Goyal, A.M.R.M.; Sharma, G.C.; Jayabalan, K.; Sahoo, S.; Devi, A.; Rana, M.; Rekha, A.; Goyal, K.; et al. piRNAs in leukemogenesis: Mechanisms, biomarkers, and therapeutic implications. Clin. Chim. Acta 2025, 571, 120220. [Google Scholar] [CrossRef]
- Stoyko, D.; Genzor, P.; Haase, A.D. Hierarchical length and sequence preferences establish a single major piRNA 3’-end. iScience 2022, 25, 104427. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ipsaro, J.J.; Haase, A.D.; Knott, S.R.; Joshua-Tor, L.; Hannon, G.J. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 2012, 491, 279–283. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, J.; Chen, S.; Liu, K. Structural insights into piRNA biogenesis. Biochim. Biophys. Acta Gene Regul Mech. 2022, 1865, 194799. [Google Scholar] [CrossRef] [PubMed]
- Loubalova, Z.; Fulka, H.; Horvat, F.; Pasulka, J.; Malik, R.; Hirose, M.; Ogura, A.; Svoboda, P. Formation of spermatogonia and fertile oocytes in golden hamsters requires piRNAs. Nat. Cell Biol. 2021, 23, 992–1001. [Google Scholar] [CrossRef]
- Saito, K.; Inagaki, S.; Mituyama, T.; Kawamura, Y.; Ono, Y.; Sakota, E.; Kotani, H.; Asai, K.; Siomi, H.; Siomi, M.C. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 2009, 461, 1296–1299. [Google Scholar] [CrossRef]
- Lee, S.K.; Shen, W.; Wen, W.; Joo, Y.; Xue, Y.; Park, A.; Qiang, A.; Su, S.; Zhang, T.; Zhang, M.; et al. Topoisomerase 3b facilitates piRNA biogenesis to promote transposon silencing and germ cell development. Cell Rep. 2025, 44, 115495. [Google Scholar] [CrossRef] [PubMed]
- Mcclintock, B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 1950, 36, 344–355. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mcclintock, B. Chromosome organization and genic expression. Cold Spring Harb. Symp. Quant. Biol. 1951, 16, 13–47. [Google Scholar] [CrossRef] [PubMed]
- Duret, L.; Marais, G.; Biémont, C. Transposons but not retrotransposons are located preferentially in regions of high recombination rate in Caenorhabditis elegans. Genetics 2000, 156, 1661–1669. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chalopin, D.; Naville, M.; Plard, F.; Galiana, D.; Volff, J.N. Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol. Evol. 2015, 7, 567–580. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Onishi, R.; Yamanaka, S.; Siomi, M.C. piRNA- and siRNA-mediated transcriptional repression in Drosophila, mice, and yeast: New insights and biodiversity. EMBO Rep. 2021, 22, e53062. [Google Scholar] [CrossRef]
- Mohn, F.; Sienski, G.; Handler, D.; Brennecke, J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 2014, 157, 1364–1379. [Google Scholar] [CrossRef] [PubMed]
- Vagin, V.V.; Sigova, A.; Li, C.; Seitz, H.; Gvozdev, V.; Zamore, P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 2006, 313, 320–324. [Google Scholar] [CrossRef]
- Houwing, S.; Kamminga, L.M.; Berezikov, E.; Cronembold, D.; Girard, A.; van den Elst, H.; Filippov, D.V.; Blaser, H.; Raz, E.; Moens, C.B.; et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 2007, 129, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y.; Liu, M. Knockout Gene-Based Evidence for PIWI-Interacting RNA Pathway in Mammals. Front. Cell Dev. Biol. 2021, 9, 681188. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, X.; Wong, G. An old weapon with a new function: PIWI-interacting RNAs in neurodegenerative diseases. Transl. Neurodegener. 2021, 10, 9. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wei, C.; Yan, X.; Mann, J.M.; Geng, R.; Wang, Q.; Xie, H.; Demireva, E.Y.; Sun, L.; Ding, D.; Chen, C. PNLDC1 catalysis and postnatal germline function are required for piRNA trimming, LINE1 silencing, and spermatogenesis in mice. PLoS Genet. 2024, 20, e1011429. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rayford, K.J.; Cooley, A.; Rumph, J.T.; Arun, A.; Rachakonda, G.; Villalta, F.; Lima, M.F.; Pratap, S.; Misra, S.; Nde, P.N. piRNAs as Modulators of Disease Pathogenesis. Int. J. Mol. Sci. 2021, 22, 2373. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, X.; Ramat, A.; Simonelig, M.; Liu, M.F. Emerging roles and functional mechanisms of PIWI-interacting RNAs. Nat. Rev. Mol. Cell Biol. 2023, 24, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Hutvagner, G.; Simard, M.J. Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008, 9, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Simon, B.; Kirkpatrick, J.P.; Eckhardt, S.; Reuter, M.; Rocha, E.A.; Andrade-Navarro, M.A.; Sehr, P.; Pillai, R.S.; Carlomagno, T. Recognition of 2’-O-methylated 3’-end of piRNA by the PAZ domain of a Piwi protein. Structure 2011, 19, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Sakaguchi, Y.; Suzuki, T.; Suzuki, T.; Siomi, H.; Siomi, M.C. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3’ ends. Genes Dev. 2007, 21, 1603–1608. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kirino, Y.; Mourelatos, Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat. Struct. Mol. Biol. 2007, 14, 347–348. [Google Scholar] [CrossRef]
- Mickute, M.; Nainyte, M.; Vasiliauskaite, L.; Plotnikova, A.; Masevicius, V.; Klimašauskas, S.; Vilkaitis, G. Animal Hen1 2’-O-methyltransferases as tools for 3′-terminal functionalization and labelling of single-stranded RNAs. Nucleic Acids Res. 2018, 46, e104. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Olotu, O.; Ahmedani, A.; Kotaja, N. Small Non-Coding RNAs in Male Reproduction. Semin. Reprod. Med. 2023, 41, 213–225. [Google Scholar] [CrossRef]
- Ho, S.; Theurkauf, W.; Rice, N. piRNA-Guided Transposon Silencing and Response to Stress in Drosophila Germline. Viruses 2024, 16, 714. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Castañeda, J.; Genzor, P.; Bortvin, A. piRNAs, transposon silencing, and germline genome integrity. Mutat. Res. 2011, 714, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Tóth, K.F.; Pezic, D.; Stuwe, E.; Webster, A. The piRNA Pathway Guards the Germline Genome Against Transposable Elements. Adv. Exp. Med. Biol. 2016, 886, 51–77. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Khurana, J.S.; Theurkauf, W. piRNAs, transposon silencing, and Drosophila germline development. J. Cell Biol. 2010, 191, 905–913. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ishizu, H.; Kinoshita, T.; Hirakata, S.; Komatsuzaki, C.; Siomi, M.C. Distinct and Collaborative Functions of Yb and Armitage in Transposon-Targeting piRNA Biogenesis. Cell Rep. 2019, 27, 1822–1835.e8. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Yu, X.; Zhang, S.; He, Y.; Guo, W. Novel roles of PIWI proteins and PIWI-interacting RNAs in human health and diseases. Cell Commun. Signal. 2023, 21, 343. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nishida, K.M.; Iwasaki, Y.W.; Murota, Y.; Nagao, A.; Mannen, T.; Kato, Y.; Siomi, H.; Siomi, M.C. Respective functions of two distinct Siwi complexes assembled during PIWI-interacting RNA biogenesis in Bombyx germ cells. Cell Rep. 2015, 10, 193–203. [Google Scholar] [CrossRef]
- Hyun, S. Small RNA Pathways That Protect the Somatic Genome. Int. J. Mol. Sci. 2017, 18, 912. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lewis, A.; Berkyurek, A.C.; Greiner, A.; Sawh, A.N.; Vashisht, A.; Merrett, S.; Flamand, M.N.; Wohlschlegel, J.; Sarov, M.; Miska, E.A.; et al. A Family of Argonaute-Interacting Proteins Gates Nuclear RNAi. Mol. Cell 2020, 78, 862–875.e8. [Google Scholar] [CrossRef]
- Sindik, N.; Pereza, N.; Dević Pavlić, S. Epigenetics of oogenesis. Arch. Gynecol Obstet. 2025, 311, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dou, M.; Song, X.; Dong, Y.; Liu, S.; Liu, H.; Tao, J.; Li, W.; Yin, X.; Xu, W. The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 2019, 18, 123. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Malone, C.D.; Brennecke, J.; Dus, M.; Stark, A.; McCombie, W.R.; Sachidanandam, R.; Hannon, G.J. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 2009, 137, 522–535. [Google Scholar] [CrossRef]
- Santos, D.; Feng, M.; Kolliopoulou, A.; Taning, C.N.T.; Sun, J.; Swevers, L. What Are the Functional Roles of Piwi Proteins and piRNAs in Insects? Insects 2023, 14, 187. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ramos-Lopez, O.; Milagro, F.I.; Riezu-Boj, J.I.; Martinez, J.A. Epigenetic signatures underlying inflammation: An interplay of nutrition, physical activity, metabolic diseases, and environmental factors for personalized nutrition. Inflamm. Res. 2021, 70, 29–49. [Google Scholar] [CrossRef]
- Ducreux, B.; Ferreux, L.; Patrat, C.; Fauque, P. Overview of Gene Expression Dynamics during Human Oogenesis/Folliculogenesis. Int. J. Mol. Sci. 2023, 25, 33. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Krajnik, K.; Mietkiewska, K.; Skowronska, A.; Kordowitzki, P.; Skowronski, M.T. Oogenesis in Women: From Molecular Regulatory Pathways and Maternal Age to Stem Cells. Int. J. Mol. Sci. 2023, 24, 6837. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Geijsen, N.; Horoschak, M.; Kim, K.; Gribnau, J.; Eggan, K.; Daley, G.Q. Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature 2004, 427, 148–154. [Google Scholar] [CrossRef]
- Toyooka, Y.; Tsunekawa, N.; Akasu, R.; Noce, T. Embryonic stem cells can form germ cells in vitro. Proc. Natl. Acad. Sci. USA 2003, 100, 11457–11462. [Google Scholar] [CrossRef]
- Romualdez-Tan, M.V. Modelling in vitro gametogenesis using induced pluripotent stem cells: A review. Cell Regen. 2023, 12, 33. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Eppig, J.J. Coordination of nuclear and cytoplasmic oocyte maturation in eutherian mammals. Reprod. Fertil. Dev. 1996, 8, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, K.I.; Matsuura, K.; Tani, N.; Takeda, N.; Usuki, S.; Yamane, M.; Sugimoto, M.; Fujimura, S.; Hosokawa, M.; Chuma, S.; et al. MEIOSIN Directs the Switch from Mitosis to Meiosis in Mammalian Germ Cells. Dev. Cell. 2020, 52, 429–445.e10. [Google Scholar] [CrossRef]
- Ishiguro, K.I. Mechanisms of meiosis initiation and meiotic prophase progression during spermatogenesis. Mol. Asp. Med. 2024, 97, 101282. [Google Scholar] [CrossRef] [PubMed]
- Kubíková, J.; Reinig, R.; Salgania, H.K.; Jeske, M. LOTUS-domain proteins—Developmental effectors from a molecular perspective. Biol. Chem. 2020, 402, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Gainetdinov, I.V.; Skvortsova, Y.V.; Stukacheva, E.A.; Bychenko, O.S.; Kondratieva, S.A.; Zinovieva, M.V.; Azhikina, T.L. Expression profiles of PIWIL2 short isoforms differ in testicular germ cell tumors of various differentiation subtypes. PLoS ONE 2014, 9, e112528. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ferreira, H.J.; Heyn, H.; Garcia del Muro, X.; Vidal, A.; Larriba, S.; Muñoz, C.; Villanueva, A.; Esteller, M. Epigenetic loss of the PIWI/piRNA machinery in human testicular tumorigenesis. Epigenetics 2014, 9, 113–118. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Swain, N.; Thakur, M.; Pathak, J.; Swain, B. SOX2, OCT4 and NANOG: The core embryonic stem cell pluripotency regulators in oral carcinogenesis. J. Oral Maxillofac. Pathol. 2020, 24, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Yan, K.; Zhou, Y.; Liang, H.; Liang, J.; Zhao, W.; Dong, Z.; Ling, B. Piwil2 is reactivated by HPV oncoproteins and initiates cell reprogramming via epigenetic regulation during cervical cancer tumorigenesis. Oncotarget 2016, 7, 64575–64588. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, N.; Tan, H.Y.; Lu, Y.; Chan, Y.T.; Wang, D.; Guo, W.; Xu, Y.; Zhang, C.; Chen, F.; Tang, G.; et al. PIWIL1 governs the crosstalk of cancer cell metabolism and immunosuppressive microenvironment in hepatocellular carcinoma. Signal Transduct. Target. Ther. 2021, 6, 86. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Feng, J.; Yang, M.; Wei, Q.; Song, F.; Zhang, Y.; Wang, X.; Liu, B.; Li, J. Novel evidence for oncogenic piRNA-823 as a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. J. Cell Mol. Med. 2020, 24, 9028–9040. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Bu, P. Non-coding RNA in cancer. Essays Biochem. 2021, 65, 625–639. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Giebler, M.; Greither, T.; Behre, H.M. Differential Regulation of PIWI-LIKE 2 Expression in Primordial Germ Cell Tumor Cell Lines by Promoter Methylation. Front. Genet. 2018, 9, 375. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Scott, E.C.; Gardner, E.J.; Masood, A.; Chuang, N.T.; Vertino, P.M.; Devine, S.E. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res. 2016, 26, 745–755. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cajuso, T.; Sulo, P.; Tanskanen, T.; Katainen, R.; Taira, A.; Hänninen, U.A.; Kondelin, J.; Forsström, L.; Välimäki, N.; Aavikko, M.; et al. Retrotransposon insertions can initiate colorectal cancer and are associated with poor survival. Nat. Commun. 2019, 10, 4022. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kazazian, H.H., Jr.; Wong, C.; Youssoufian, H.; Scott, A.F.; Phillips, D.G.; Antonarakis, S.E. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 1988, 332, 164–166. [Google Scholar] [CrossRef] [PubMed]
- Van de Water, N.; Williams, R.; Ockelford, P.; Browett, P. A 20.7 kb deletion within the factor VIII gene associated with LINE-1 element insertion. Thromb. Haemost. 1998, 79, 938–942. [Google Scholar] [PubMed]
- Yan, H.; Wu, Q.L.; Sun, C.Y.; Ai, L.S.; Deng, J.; Zhang, L.; Chen, L.; Chu, Z.B.; Tang, B.; Wang, K.; et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia 2015, 29, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Taghizadeh, M.; Jafari-Koshki, T.; Jafarlou, V.; Raeisi, M.; Alizadeh, L.; Roosta, Y.; Matin, S.; Jabari, R.; Sur, D.; Karimi, A. The role of piRNAs in predicting and prognosing in cancer: A focus on piRNA-823 (a systematic review and meta-analysis). BMC Cancer 2024, 24, 484. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yao, J.; Wang, Y.W.; Fang, B.B.; Zhang, S.J.; Cheng, B.L. piR-651 and its function in 95-D lung cancer cells. Biomed. Rep. 2016, 4, 546–550. [Google Scholar] [CrossRef]
- Zhang, S.J.; Yao, J.; Shen, B.Z.; Li, G.B.; Kong, S.S.; Bi, D.D.; Pan, S.H.; Cheng, B.L. Role of piwi-interacting RNA-651 in the carcinogenesis of non-small cell lung cancer. Oncol. Lett. 2018, 15, 940–946. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Luo, Y.; Gao, Y.; Yang, Y.; Wang, Y.; Xu, Y.; Tan, S.; Zhang, Y.; Duan, J.; Yang, Y. piR-651 promotes tumor formation in non-small cell lung carcinoma through the upregulation of cyclin D1 and CDK4. Int. J. Mol. Med. 2016, 38, 927–936. [Google Scholar] [CrossRef]
- Liu, T.; Wang, J.; Sun, L.; Li, M.; He, X.; Jiang, J.; Zhou, Q. Piwi-interacting RNA-651 promotes cell proliferation and migration and inhibits apoptosis in breast cancer by facilitating DNMT1-mediated PTEN promoter methylation. Cell Cycle 2021, 20, 1603–1616. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.; Bhattacharjee, S.; Mandal, D.P. PIWI-interacting RNA (piRNA): A narrative review of its biogenesis, function, and emerging role in lung cancer. Asian Biomed. 2022, 16, 3–14. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, X.; Sun, Y.; Guo, J.; Ma, H.; Li, J.; Dong, B.; Jin, G.; Zhang, J.; Wu, J.; Meng, L.; et al. Expression of hiwi gene in human gastric cancer was associated with proliferation of cancer cells. Int. J. Cancer 2006, 118, 1922–1929. [Google Scholar] [CrossRef] [PubMed]
- Ameli Mojarad, M.; Ameli Mojarad, M.; Shojaee, B.; Nazemalhosseini-Mojarad, E. piRNA: A promising biomarker in early detection of gastrointestinal cancer. Pathol. Res. Pract. 2022, 230, 153757. [Google Scholar] [CrossRef]
- Mohamed, F.S.; Jalal, D.; Fadel, Y.M.; El-Mashtoly, S.F.; Khaled, W.Z.; Sayed, A.A.; Ghazy, M.A. Characterization and comparative profiling of piRNAs in serum biopsies of pediatric Wilms tumor patients. Cancer Cell Int. 2025, 25, 163. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mentis, A.A.; Dardiotis, E.; Romas, N.A.; Papavassiliou, A.G. PIWI family proteins as prognostic markers in cancer: A systematic review and meta-analysis. Cell. Mol. Life Sci. 2020, 77, 2289–2314. [Google Scholar] [CrossRef]
- Vogiatzoglou, A.P.; Moretto, F.; Makkou, M.; Papamatheakis, J.; Kretsovali, A. Promyelocytic leukemia protein (PML) and stem cells: From cancer to pluripotency. Int. J. Dev. Biol. 2022, 66, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Varzideh, F.; Gambardella, J.; Kansakar, U.; Jankauskas, S.S.; Santulli, G. Molecular Mechanisms Underlying Pluripotency and Self-Renewal of Embryonic Stem Cells. Int. J. Mol. Sci. 2023, 24, 8386. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lee, J.H.; Schütte, D.; Wulf, G.; Füzesi, L.; Radzun, H.J.; Schweyer, S.; Engel, W.; Nayernia, K. Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum. Mol. Genet. 2006, 15, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhang, K.; Li, C.; Yao, Y.; Tao, D.; Liu, Y.; Zhang, S.; Ma, Y. Piwil2 suppresses p53 by inducing phosphorylation of signal transducer and activator of transcription 3 in tumor cells. PLoS ONE 2012, 7, e30999. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Ren, Z.J.; Wang, F.; Liu, M.; Li, X.; Tang, H. PIWIL4 regulates cervical cancer cell line growth and is involved in down-regulating the expression of p14ARF and p53. FEBS Lett. 2012, 586, 1356–1362. [Google Scholar] [CrossRef]
- Sadoughi, F.; Mirhashemi, S.M.; Asemi, Z. Epigenetic roles of PIWI proteins and piRNAs in colorectal cancer. Cancer Cell Int. 2021, 21, 328. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Qiao, D.; Zeeman, A.M.; Deng, W.; Looijenga, L.H.; Lin, H. Molecular characterization of hiwi, a human member of the piwi gene family whose overexpression is correlated to seminomas. Oncogene 2002, 21, 3988–3999. [Google Scholar] [CrossRef] [PubMed]
- Taubert, H.; Greither, T.; Kaushal, D.; Würl, P.; Bache, M.; Bartel, F.; Kehlen, A.; Lautenschläger, C.; Harris, L.; Kraemer, K.; et al. Expression of the stem cell self-renewal gene Hiwi and risk of tumour-related death in patients with soft-tissue sarcoma. Oncogene 2007, 26, 1098–1100. [Google Scholar] [CrossRef]
- Wang, D.W.; Wang, Z.H.; Wang, L.L.; Song, Y.; Zhang, G.Z. Overexpression of hiwi promotes growth of human breast cancer cells. Asian Pac. J. Cancer Prev. 2014, 15, 7553–7558. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Yu, X.; Han, X.; Hao, J.; Zhao, J.; Bebek, G.; Bao, S.; Prayson, R.A.; Khalil, A.M.; Jankowsky, E.; et al. Piwil1 Regulates Glioma Stem Cell Maintenance and Glioblastoma Progression. Cell Rep. 2021, 34, 108522. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Perera, B.P.U.; Morgan, R.K.; Polemi, K.M.; Sala-Hamrick, K.E.; Svoboda, L.K.; Dolinoy, D.C. PIWI-Interacting RNA (piRNA) and Epigenetic Editing in Environmental Health Sciences. Curr. Environ. Health Rep. 2022, 9, 650–660. [Google Scholar] [CrossRef]
- Zhao, H.; Liu, Y.; Zhu, L.; Cheng, J.; Li, Y. MAD2L1-mediated NANOG nuclear translocation: A critical factor in lung cancer chemoresistance. Cell. Signal. 2025, 132, 111811. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zawalska, M.; Tarnowski, M. piRNA-Mediated Maintenance of Genome Stability in Gametogenesis and Cancer. Genes 2025, 16, 722. https://doi.org/10.3390/genes16070722
Zawalska M, Tarnowski M. piRNA-Mediated Maintenance of Genome Stability in Gametogenesis and Cancer. Genes. 2025; 16(7):722. https://doi.org/10.3390/genes16070722
Chicago/Turabian StyleZawalska, Martyna, and Maciej Tarnowski. 2025. "piRNA-Mediated Maintenance of Genome Stability in Gametogenesis and Cancer" Genes 16, no. 7: 722. https://doi.org/10.3390/genes16070722
APA StyleZawalska, M., & Tarnowski, M. (2025). piRNA-Mediated Maintenance of Genome Stability in Gametogenesis and Cancer. Genes, 16(7), 722. https://doi.org/10.3390/genes16070722






