Half the Chromosome It Used to Be: Identifying Cancer Treatments Targeting Aneuploid Losses
Abstract
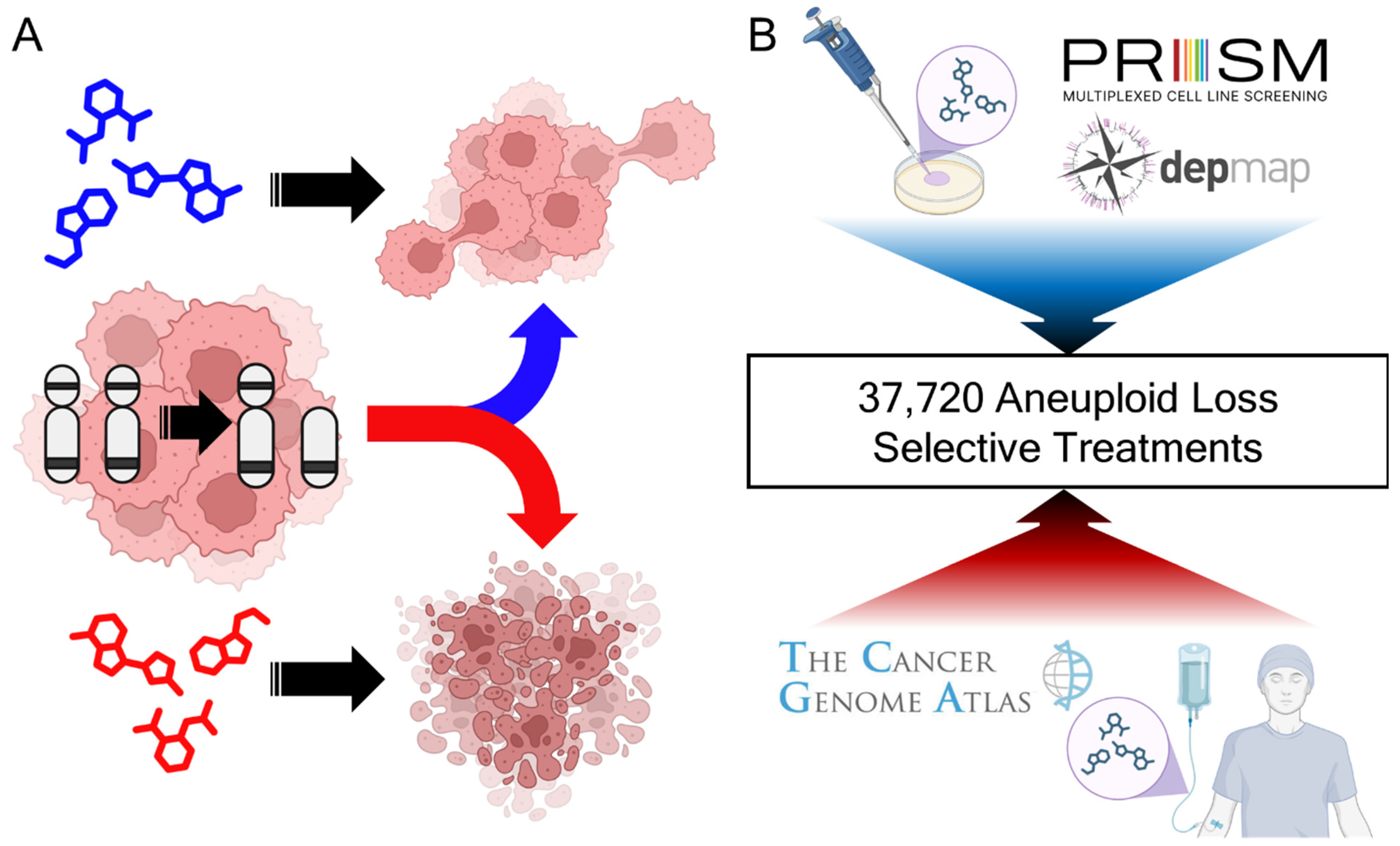
1. Introduction
2. Materials and Methods
2.1. Datasets
2.2. TCGA Aneuploidy Normalization
2.3. TCGA Survival Analyses
2.4. Analysis of Whole Genome Duplication Versus Aneuploid Loss on Prognosis
2.5. Aneuploidy and Breast Cancer Subtype Prognosis Impact Analysis
2.6. Broad PRISM ANOVA Analyses
2.7. TCGA MOA Mapping
2.8. Overlapping Findings Between Datasets
2.9. Other Plots
2.10. Software Tools and Packages
3. Results
3.1. Derived Insights from Aneuploidy Treatment Landscape
3.2. Aneuploid Losses Are Associated with Worse Outcomes on Current Chemotherapies
3.3. Positive Prognosis Associations from the TCGA
3.4. From Patient to Petri Dish: Cancer Cytotoxic Mechanisms of Action
3.5. Putative Novel or Repurposed Arm-Loss Selective Chemotherapies
3.6. Chemotherapies Selective for Multiple Concurrent Aneuploidies in Ovarian Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANOVA | Analysis of variance |
| BH | Benjamini–Hochberg |
| CIN | Chromosome instability |
| CNA | Copy number alteration |
| GR | Glucocorticoid receptor |
| LLM | Large language models |
| MDPI | Multidisciplinary Digital Publishing Institute |
| MOA | Mechanism of action |
| MT | Metallothionein |
| NCCN | National Comprehensive Cancer Network |
| NR | Not reached |
| NSCLC | Non-small cell lung cancer |
| PRISM | Profiling Relative Inhibition Simultaneously in Mixtures |
| TCGA | The Cancer Genome Atlas |
| WGD | Whole-genome duplication |
References
- Kasperski, A. Life Entrapped in a Network of Atavistic Attractors: How to Find a Rescue. Int. J. Mol. Sci. 2022, 23, 4017. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Amon, A. Context is everything: Aneuploidy in cancer. Nat. Rev. Genet. 2020, 21, 44–62. [Google Scholar] [CrossRef] [PubMed]
- Simonetti, G.; Bruno, S.; Padella, A.; Tenti, E.; Martinelli, G. Aneuploidy: Cancer strength or vulnerability? Int. J. Cancer 2019, 144, 8–25. [Google Scholar] [CrossRef]
- Weaver, B.A.A.; Cleveland, D.W. Does aneuploidy cause cancer? Curr. Opin. Cell Biol. 2006, 18, 658–667. [Google Scholar] [CrossRef] [PubMed]
- Sansregret, L.; Swanton, C. The role of aneuploidy in cancer evolution. Cold Spring Harb. Perspect. Med. 2017, 7, a028373. [Google Scholar] [CrossRef]
- Potapova, T.A.; Zhu, J.; Li, R. Aneuploidy and chromosomal instability: A vicious cycle driving cellular evolution and cancer genome chaos. Cancer Metastasis Rev. 2013, 32, 377–389. [Google Scholar] [CrossRef]
- Kawakami, M.; Liu, X.; Dmitrovsky, E. New cell cycle inhibitors target aneuploidy in cancer therapy. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 361–377. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, C.; Xu, Z.; Scuoppo, C.; Rillahan, C.D.; Gao, J.; Spitzer, B.; Bosbach, B.; Kastenhuber, E.R.; Baslan, T. Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature 2016, 531, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Zucker, M.; Perry, M.A.; Gould, S.I.; Elkrief, A.; Safonov, A.; Thummalapalli, R.; Mehine, M.; Chakravarty, D.; Brannon, A.R.; Ladanyi, M.; et al. Pan-cancer analysis of biallelic inactivation in tumor suppressor genes identifies KEAP1 zygosity as a predictive biomarker in lung cancer. Cell 2024, 188, 851–867.e17. [Google Scholar] [CrossRef]
- Davoli, T.; Xu, A.W.; Mengwasser, K.E.; Sack, L.M.; Yoon, J.C.; Park, P.J.; Elledge, S.J. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 2013, 155, 948–962. [Google Scholar] [CrossRef]
- Girish, V.; Lakhani, A.A.; Thompson, S.L.; Scaduto, C.M.; Brown, L.M.; Hagenson, R.A.; Sausville, E.L.; Mendelson, B.E.; Kandikuppa, P.K.; Lukow, D.A.; et al. Oncogene-like addiction to aneuploidy in human cancers. Science 2023, 381, eadg4521. [Google Scholar] [CrossRef] [PubMed]
- Replogle, J.M.; Zhou, W.; Amaro, A.E.; McFarland, J.M.; Villalobos-Ortiz, M.; Ryan, J.; Letai, A.; Yilmaz, O.; Sheltzer, J.; Lippard, S.J.; et al. Aneuploidy increases resistance to chemotherapeutics by antagonizing cell division. Proc. Natl. Acad. Sci. USA 2020, 117, 30566–30576. [Google Scholar] [CrossRef] [PubMed]
- Lukow, D.A.; Sausville, E.L.; Suri, P.; Chunduri, N.K.; Wieland, A.; Leu, J.; Smith, J.C.; Girish, V.; Kumar, A.A.; Kendall, J. Chromosomal instability accelerates the evolution of resistance to anti-cancer therapies. Dev. Cell 2021, 56, 2427–2439.e4. [Google Scholar] [CrossRef]
- Davoli, T.; Uno, H.; Wooten, E.C.; Elledge, S.J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017, 355, eaaf8399. [Google Scholar] [CrossRef]
- Holland, A.J.; Cleveland, D.W. Boveri revisited: Chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 2009, 10, 478–487. [Google Scholar] [CrossRef]
- Sunshine, A.B.; Payen, C.; Ong, G.T.; Liachko, I.; Tan, K.M.; Dunham, M.J. The Fitness Consequences of Aneuploidy Are Driven by Condition-Dependent Gene Effects. PLoS Biol. 2015, 13, e1002155. [Google Scholar] [CrossRef] [PubMed]
- Shih, J.; Sarmashghi, S.; Zhakula-Kostadinova, N.; Zhang, S.; Georgis, Y.; Hoyt, S.H.; Cuoco, M.S.; Gao, G.F.; Spurr, L.F.; Berger, A.C.; et al. Cancer aneuploidies are shaped primarily by effects on tumour fitness. Nature 2023, 619, 793–800. [Google Scholar] [CrossRef]
- Blagosklonny, M.V. Molecular theory of cancer. Cancer Biol. Ther. 2005, 4, 621–627. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014, 159, 676–690. [Google Scholar] [CrossRef]
- Kuang, X.; Li, J. Chromosome instability and aneuploidy as context-dependent activators or inhibitors of antitumor immunity. Front. Immunol. 2022, 13, 895961. [Google Scholar] [CrossRef]
- Sheltzer, J.M.; Blank, H.M.; Pfau, S.J.; Tange, Y.; George, B.M.; Humpton, T.J.; Brito, I.L.; Hiraoka, Y.; Niwa, O.; Amon, A. Aneuploidy drives genomic instability in yeast. Science 2011, 333, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Torres, E.M.; Williams, B.R.; Amon, A. Aneuploidy: Cells losing their balance. Genetics 2008, 179, 737–746. [Google Scholar] [CrossRef]
- Matzke, M.A.; Mette, M.F.; Kanno, T.; Matzke, A.J. Does the intrinsic instability of aneuploid genomes have a causal role in cancer? Trends Genet. 2003, 19, 253–256. [Google Scholar] [CrossRef]
- Shukla, A.; Nguyen, T.H.M.; Moka, S.B.; Ellis, J.J.; Grady, J.P.; Oey, H.; Cristino, A.S.; Khanna, K.K.; Kroese, D.P.; Krause, L. Chromosome arm aneuploidies shape tumour evolution and drug response. Nat. Commun. 2020, 11, 449. [Google Scholar] [CrossRef] [PubMed]
- Trédan, O.; Galmarini, C.M.; Patel, K.; Tannock, I.F. Drug resistance and the solid tumor microenvironment. J. Natl. Cancer Inst. 2007, 99, 1441–1454. [Google Scholar] [CrossRef]
- Stopsack, K.H.; Whittaker, C.A.; Gerke, T.A.; Loda, M.; Kantoff, P.W.; Mucci, L.A.; Amon, A. Aneuploidy drives lethal progression in prostate cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 11390–11395. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef]
- Bowers, R.R.; Jones, C.M.; Paz, E.A.; Barrows, J.K.; Armeson, K.E.; Long, D.T.; Delaney, J.R. SWAN pathway-network identification of common aneuploidy-based oncogenic drivers. Nucleic Acids Res. 2022, 50, 3673–3692. [Google Scholar] [CrossRef]
- Delaney, J.R.; Patel, C.B.; Willis, K.M.; Haghighiabyaneh, M.; Axelrod, J.; Tancioni, I.; Lu, D.; Bapat, J.; Young, S.; Cadassou, O.; et al. Haploinsufficiency networks identify targetable patterns of allelic deficiency in low mutation ovarian cancer. Nat. Commun. 2017, 8, 14423. [Google Scholar] [CrossRef]
- Delaney, J.R.; Patel, C.B.; Bapat, J.; Jones, C.M.; Ramos-Zapatero, M.; Ortell, K.K.; Tanios, R.; Haghighiabyaneh, M.; Axelrod, J.; DeStefano, J.W. Autophagy gene haploinsufficiency drives chromosome instability, increases migration, and promotes early ovarian tumors. PLoS Genet. 2020, 16, e1008558. [Google Scholar] [CrossRef]
- Wise, H.M.; Hermida, M.A.; Leslie, N.R. Prostate cancer, PI3K, PTEN and prognosis. Clin. Sci. 2017, 131, 197–210. [Google Scholar] [CrossRef]
- Vidotto, T.; Melo, C.; Lautert-Dutra, W.; Chaves, L.; Reis, R.; Squire, J. Pan-cancer genomic analysis shows hemizygous PTEN loss tumors are associated with immune evasion and poor outcome. Sci. Rep. 2023, 13, 5049. [Google Scholar] [CrossRef]
- Molinari, F.; Frattini, M. Functions and regulation of the PTEN gene in colorectal cancer. Front. Oncol. 2014, 3, 326. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, X.; Han, C.; Wang, L.; Zhang, X.; He, X.; Lu, X. Targeting tumor suppressor genes for cancer therapy. Bioessays 2015, 37, 1277–1286. [Google Scholar] [CrossRef]
- Perdrix, A.; Najem, A.; Saussez, S.; Awada, A.; Journe, F.; Ghanem, G.; Krayem, M. PRIMA-1 and PRIMA-1Met (APR-246): From mutant/wild type p53 reactivation to unexpected mechanisms underlying their potent anti-tumor effect in combinatorial therapies. Cancers 2017, 9, 172. [Google Scholar] [CrossRef]
- Deininger, M.; Buchdunger, E.; Druker, B.J. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood 2005, 105, 2640–2653. [Google Scholar] [CrossRef]
- Lee, H.-S.; Lee, N.C.; Kouprina, N.; Kim, J.-H.; Kagansky, A.; Bates, S.; Trepel, J.B.; Pommier, Y.; Sackett, D.; Larionov, V. Effects of anticancer drugs on chromosome instability and new clinical implications for tumor-suppressing therapies. Cancer Res. 2016, 76, 902–911. [Google Scholar] [CrossRef]
- Sarhadi, V.K.; Armengol, G. Molecular biomarkers in cancer. Biomolecules 2022, 12, 1021. [Google Scholar] [CrossRef]
- Mermel, C.H.; Schumacher, S.E.; Hill, B.; Meyerson, M.L.; Beroukhim, R.; Getz, G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011, 12, R41. [Google Scholar] [CrossRef]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Corsello, S.M.; Nagari, R.T.; Spangler, R.D.; Rossen, J.; Kocak, M.; Bryan, J.G.; Humeidi, R.; Peck, D.; Wu, X.; Tang, A.A.; et al. Discovering the anticancer potential of non-oncology drugs by systematic viability profiling. Nat. Cancer 2020, 1, 235–248. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. Addendum: The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 492, 290. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Broad DepMap. DepMap 24Q4 Public; Figshare: London, UK, 2024. [Google Scholar] [CrossRef]
- Carter, S.L.; Cibulskis, K.; Helman, E.; McKenna, A.; Shen, H.; Zack, T.; Laird, P.W.; Onofrio, R.C.; Winckler, W.; Weir, B.A.; et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012, 30, 413–421. [Google Scholar] [CrossRef]
- Tschaharganeh, D.F.; Bosbach, B.; Lowe, S.W. Coordinated tumor suppression by chromosome 8p. Cancer Cell 2016, 29, 617–619. [Google Scholar] [CrossRef][Green Version]
- Lin, Y.W.; Sheu, J.C.; Liu, L.Y.; Chen, C.H.; Lee, H.S.; Huang, G.T.; Wang, J.T.; Lee, P.H.; Lu, F.J. Loss of heterozygosity at chromosome 13q in hepatocellular carcinoma: Identification of three independent regions. Eur. J. Cancer 1999, 35, 1730–1734. [Google Scholar] [CrossRef]
- Gatto, F.; Nookaew, I.; Nielsen, J. Chromosome 3p loss of heterozygosity is associated with a unique metabolic network in clear cell renal carcinoma. Proc. Natl. Acad. Sci. USA 2014, 111, E866–E875. [Google Scholar] [CrossRef]
- Grundy, P.E.; Breslow, N.E.; Li, S.; Perlman, E.; Beckwith, J.B.; Ritchey, M.L.; Shamberger, R.C.; Haase, G.M.; D’Angio, G.J.; Donaldson, M.; et al. Loss of Heterozygosity for Chromosomes 1p and 16q Is an Adverse Prognostic Factor in Favorable-Histology Wilms Tumor: A Report from the National Wilms Tumor Study Group. J. Clin. Oncol. 2005, 23, 7312–7321. [Google Scholar] [CrossRef]
- Helke, K.L.; Gudi, R.R.; Vasu, C.; Delaney, J.R. Combination of Autophagy Selective Therapeutics with Doxil: An Assessment of Pathological Toxicity. Front. Toxicol. 2022, 4, 937150. [Google Scholar] [CrossRef]
- Meng, K.; Song, J.; Qi, F.; Li, J.; Fang, Z.; Song, L. MT1G promotes iron autophagy and inhibits the function of gastric cancer cell lines by intervening in GPX4/SQSTM1. Sci. Rep. 2024, 14, 28539. [Google Scholar] [CrossRef]
- Beach, R.R.; Ricci-Tam, C.; Brennan, C.M.; Moomau, C.A.; Hsu, P.-h.; Hua, B.; Silberman, R.E.; Springer, M.; Amon, A. Aneuploidy causes non-genetic individuality. Cell 2017, 169, 229–242.e21. [Google Scholar] [CrossRef]
- Befani, C.; Liakos, P. The role of hypoxia-inducible factor-2 α in angiogenesis. J. Cell. Physiol. 2018, 233, 9087–9098. [Google Scholar] [CrossRef]
- Hruban, R.H.; Gaida, M.M.; Thompson, E.; Hong, S.-M.; Noë, M.; Brosens, L.A.A.; Jongepier, M.; Offerhaus, G.J.A.; Wood, L.D. Why is pancreatic cancer so deadly? The pathologist’s view. J. Pathol. 2019, 248, 131–141. [Google Scholar] [CrossRef]
- Li, D.; Zhang, J.; Yin, L.; Jin, Z.; Chen, X.; Meng, X. Etomidate inhibits cell proliferation and induces apoptosis in A549 non-small cell lung cancer cells via downregulating WWP2. Exp. Ther. Med. 2021, 22, 1254. [Google Scholar] [CrossRef]
- Xu, H.; Jesson, M.I.; Seneviratne, U.I.; Lin, T.H.; Sharif, M.N.; Xue, L.; Nguyen, C.; Everley, R.A.; Trujillo, J.I.; Johnson, D.S.; et al. PF-06651600, a Dual JAK3/TEC Family Kinase Inhibitor. ACS Chem. Biol. 2019, 14, 1235–1242. [Google Scholar] [CrossRef]
- Nagayama, D.; Saiki, A.; Shirai, K. The Anti-Cancer Effect of Pitavastatin May Be a Drug-Specific Effect: Subgroup Analysis of the TOHO-LIP Study. Vasc. Health Risk Manag. 2021, 17, 169–173. [Google Scholar] [CrossRef]
- Maeda, H.; Khatami, M. Analyses of repeated failures in cancer therapy for solid tumors: Poor tumor-selective drug delivery, low therapeutic efficacy and unsustainable costs. Clin. Transl. Med. 2018, 7, 11. [Google Scholar] [CrossRef]
- Decoster, G.; Stein, G.; Holdener, E. Responses and toxic deaths in phase I clinical trials. Ann. Oncol. 1990, 1, 175–181. [Google Scholar] [CrossRef]
- Scribano, C.M.; Wan, J.; Esbona, K.; Tucker, J.B.; Lasek, A.; Zhou, A.S.; Zasadil, L.M.; Molini, R.; Fitzgerald, J.; Lager, A.M.; et al. Chromosomal instability sensitizes patient breast tumors to multipolar divisions induced by paclitaxel. Sci. Transl. Med. 2021, 13, eabd4811. [Google Scholar] [CrossRef]
- Ding, R.-B.; Chen, P.; Rajendran, B.K.; Lyu, X.; Wang, H.; Bao, J.; Zeng, J.; Hao, W.; Sun, H.; Wong, A.H.-H.; et al. Molecular landscape and subtype-specific therapeutic response of nasopharyngeal carcinoma revealed by integrative pharmacogenomics. Nat. Commun. 2021, 12, 3046. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [Google Scholar] [CrossRef]
- Ogden, A.; Rida, P.C.G.; Knudsen, B.S.; Kucuk, O.; Aneja, R. Docetaxel-induced polyploidization may underlie chemoresistance and disease relapse. Cancer Lett. 2015, 367, 89–92. [Google Scholar] [CrossRef]
- Eastmond, D.A.; Tucker, J.D. Identification of aneuploidy-inducing agents using cytokinesis-blocked human lymphocytes and an antikinetochore antibody. Environ. Mol. Mutagen. 1989, 13, 34–43. [Google Scholar] [CrossRef]
- Chowdhury, A.; Chowdhury, S.; Tsai, M.Y. A novel Aurora kinase A inhibitor MK-8745 predicts TPX2 as a therapeutic biomarker in non-Hodgkin lymphoma cell lines. Leuk. Lymphoma 2012, 53, 462–471. [Google Scholar] [CrossRef]
- Nair, J.S.; Ho, A.L.; Schwartz, G.K. The induction of polyploidy or apoptosis by the Aurora A kinase inhibitor MK8745 is p53-dependent. Cell Cycle 2012, 11, 807. [Google Scholar] [CrossRef]
- Simões-Sousa, S.; Littler, S.; Thompson, S.L.; Minshall, P.; Whalley, H.; Bakker, B.; Belkot, K.; Moralli, D.; Bronder, D.; Tighe, A.; et al. The p38α Stress Kinase Suppresses Aneuploidy Tolerance by Inhibiting Hif-1α. Cell Rep. 2018, 25, 749–7606. [Google Scholar] [CrossRef]
- Canovas, B.; Nebreda, A.R. Diversity and versatility of p38 kinase signalling in health and disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 346–366. [Google Scholar] [CrossRef]
- Cánovas, B.; Igea, A.; Sartori, A.A.; Gomis, R.R.; Paull, T.T.; Isoda, M.; Pérez-Montoyo, H.; Serra, V.; González-Suárez, E.; Stracker, T.H.; et al. Targeting p38α Increases DNA Damage, Chromosome Instability, and the Anti-tumoral Response to Taxanes in Breast Cancer Cells. Cancer Cell 2018, 33, 1094–11108. [Google Scholar] [CrossRef]
- Nairismägi, M.-L.; Gerritsen, M.E.; Li, Z.M.; Wijaya, G.C.; Chia, B.K.H.; Laurensia, Y.; Lim, J.Q.; Yeoh, K.W.; Yao, X.S.; Pang, W.L.; et al. Oncogenic activation of JAK3-STAT signaling confers clinical sensitivity to PRN371, a novel selective and potent JAK3 inhibitor, in natural killer/T-cell lymphoma. Leukemia 2018, 32, 1147. [Google Scholar] [CrossRef]
- Faris, M.; Bot, A. In this issue: Tec kinases in the crosshairs. Int. Rev. Immunol. 2012, 31, 85–86. [Google Scholar] [CrossRef]
- Lien, E.C.; Dibble, C.C.; Toker, A. PI3K signaling in cancer: Beyond AKT. Curr. Opin. Cell Biol. 2017, 45, 62–71. [Google Scholar] [CrossRef]
- Yeung, S.-F.; Zhou, Y.; Zou, W.; Chan, W.-L.; Ching, Y.P. TEC kinase stabilizes PLK4 to promote liver cancer metastasis. Cancer Lett. 2022, 524, 70–81. [Google Scholar] [CrossRef]
- McCoy, R.C.; Demko, Z.; Ryan, A.; Banjevic, M.; Hill, M.; Sigurjonsson, S.; Rabinowitz, M.; Fraser, H.B.; Petrov, D.A. Common variants spanning PLK4 are associated with mitotic-origin aneuploidy in human embryos. Science 2015, 348, 235. [Google Scholar] [CrossRef]
- Wang, K.; Liu, J.; Zhao, X.; Li, H.; Luo, G.; Yu, Y.; Guo, Y.; Zhang, L.; Zhu, J.; Wang, S.; et al. WWP2 regulates proliferation of gastric cancer cells in a PTEN-dependent manner. Biochem. Biophys. Res. Commun. 2020, 521, 652–659. [Google Scholar] [CrossRef]
- Qin, Y.; Xu, S.-q.; Pan, D.-b.; Ye, G.-x.; Wu, C.-j.; Wang, S.; Wang, C.-j.; Jiang, J.-y.; Fu, J. Silencing of WWP2 inhibits adhesion, invasion, and migration in liver cancer cells. Tumor Biol. 2016, 37, 6787–6799. [Google Scholar] [CrossRef]
- Xu, S.-Q.; Qin, Y.; Pan, D.-B.; Ye, G.-X.; Wu, C.-J.; Wang, S.; Jiang, J.-Y.; Fu, J.; Wang, C.-J. Inhibition of WWP2 suppresses proliferation, and induces G1 cell cycle arrest and apoptosis in liver cancer cells. Mol. Med. Rep. 2016, 13, 2261–2266. [Google Scholar] [CrossRef]
- Yang, R.; He, Y.; Chen, S.; Lu, X.; Huang, C.; Zhang, G. Elevated expression of WWP2 in human lung adenocarcinoma and its effect on migration and invasion. Biochem. Biophys. Res. Commun. 2016, 479, 146–151. [Google Scholar] [CrossRef]
- Jung, J.-G.; Stoeck, A.; Guan, B.; Wu, R.-C.; Zhu, H.; Blackshaw, S.; Shih, I.-M.; Wang, T.-L. Notch3 interactome analysis identified WWP2 as a negative regulator of Notch3 signaling in ovarian cancer. PLoS Genet. 2014, 10, e1004751. [Google Scholar] [CrossRef]
- Soond, S.M.; Chantry, A. Selective targeting of activating and inhibitory Smads by distinct WWP2 ubiquitin ligase isoforms differentially modulates TGFβ signalling and EMT. Oncogene 2011, 30, 2451. [Google Scholar] [CrossRef]
- Huminiecki, L.; Goldovsky, L.; Freilich, S.; Moustakas, A.; Ouzounis, C.; Heldin, C.-H. Emergence, development and diversification of the TGF-β signalling pathway within the animal kingdom. BMC Evol. Biol. 2009, 9, 28. [Google Scholar] [CrossRef]
- Comaills, V.; Kabeche, L.; Morris, R.; Buisson, R.; Yu, M.; Madden, M.W.; LiCausi, J.A.; Boukhali, M.; Tajima, K.; Pan, S.; et al. Genomic Instability Is Induced by Persistent Proliferation of Cells Undergoing Epithelial-to-Mesenchymal Transition. Cell Rep. 2016, 17, 2632. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J. TGFβ in Cancer. Cell 2008, 134, 215–230. [Google Scholar] [CrossRef]
- Khadse, A.; Haakensen, V.D.; Silwal-Pandit, L.; Hamfjord, J.; Micke, P.; Botling, J.; Brustugun, O.T.; Lingjærde, O.C.; Helland, Å.; Kure, E.H. Prognostic Significance of the Loss of Heterozygosity of KRAS in Early-Stage Lung Adenocarcinoma. Front. Oncol. 2022, 12, 873532. [Google Scholar] [CrossRef]
- Zhang, X.; Ge, Y.-L.; Zhang, S.-P.; Yan, P.; Tian, R.-H. Downregulation of KDR expression induces apoptosis in breast cancer cells. Cell. Mol. Biol. Lett. 2014, 19, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Kitadai, Y.; Bucana, C.D.; Cleary, K.R.; Ellis, L.M. Expression of Vascular Endothelial Growth Factor and Its Receptor, KDR, Correlates with Vascularity, Metastasis, and Proliferation of Human Colon Cancer. Cancer Res. 1995, 55, 3964–3968. [Google Scholar]
- Oladipupo, S.S.; Kabir, A.U.; Smith, C.; Choi, K.; Ornitz, D.M. Impaired tumor growth and angiogenesis in mice heterozygous for Vegfr2 (Flk1). Sci. Rep. 2018, 8, 14724. [Google Scholar] [CrossRef]
- Smith, H.L.; Southgate, H.; Tweddle, D.A.; Curtin, N.J. DNA damage checkpoint kinases in cancer. Expert. Rev. Mol. Med. 2020, 22, e2. [Google Scholar] [CrossRef] [PubMed]
- Iyevleva, A.G.; Aleksakhina, S.N.; Sokolenko, A.P.; Baskina, S.V.; Venina, A.R.; Anisimova, E.I.; Bizin, I.V.; Ivantsov, A.O.; Belysheva, Y.V.; Chernyakova, A.P.; et al. Somatic loss of the remaining allele occurs approximately in half of CHEK2-driven breast cancers and is accompanied by a border-line increase of chromosomal instability. Breast Cancer Res. Treat. 2022, 192, 283–291. [Google Scholar] [CrossRef]
- Peng, L.; He, K.; Cao, Z.; Bi, L.; Yu, D.; Wang, Q.; Wang, J. CARD10 promotes the progression of renal cell carcinoma by regulating the NF-κB signaling pathway. Mol. Med. Rep. 2020, 21, 329–337. [Google Scholar] [CrossRef]
- Wang, L.; Guo, Y.; Huang, W.-J.; Ke, X.; Poyet, J.-L.; Manji, G.A.; Merriam, S.; Glucksmann, M.A.; DiStefano, P.S.; Alnemri, E.S.; et al. CARD10 Is a Novel Caspase Recruitment Domain/Membrane-associated Guanylate Kinase Family Member That Interacts with BCL10 and Activates NF-κB. J. Biol. Chem. 2001, 276, 21405–21409. [Google Scholar] [CrossRef]
- Lambert, M.P.; Arulselvan, A.; Schott, A.; Markham, S.J.; Crowley, T.B.; Zackai, E.H.; McDonald-McGinn, D.M. The 22q11.2 deletion syndrome: Cancer predisposition, platelet abnormalities and cytopenias. Am. J. Med. Genet. A 2018, 176, 2121–2127. [Google Scholar] [CrossRef] [PubMed]
- Matthews, L.C.; Berry, A.A.; Morgan, D.J.; Poolman, T.M.; Bauer, K.; Kramer, F.; Spiller, D.G.; Richardson, R.V.; Chapman, K.E.; Farrow, S.N.; et al. Glucocorticoid receptor regulates accurate chromosome segregation and is associated with malignancy. Proc. Natl. Acad. Sci. USA 2015, 112, 5479. [Google Scholar] [CrossRef] [PubMed]
- Pufall, M.A. Glucocorticoids and Cancer. Adv. Exp. Med. Biol. 2015, 872, 315–333. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Wang, J.; Wang, H.; Zhou, J.; Qi, H.; Naji, Y.; Zhao, L.; Tang, Y.; Dai, Y. Knockdown of NR3C1 inhibits the proliferation and migration of clear cell renal cell carcinoma through activating endoplasmic reticulum stress–mitophagy. J. Transl. Med. 2023, 21, 701. [Google Scholar] [CrossRef]
- Pan, D.; Kocherginsky, M.; Conzen, S.D. Activation of the glucocorticoid receptor is associated with poor prognosis in estrogen receptor-negative breast cancer. Cancer Res. 2011, 71, 6360. [Google Scholar] [CrossRef]
- Smoak, K.A.; Cidlowski, J.A. Mechanisms of glucocorticoid receptor signaling during inflammation. Mech. Ageing Dev. 2004, 125, 697–706. [Google Scholar] [CrossRef]
- Escoter-Torres, L.; Greulich, F.; Quagliarini, F.; Wierer, M.; Uhlenhaut, N.H. Anti-inflammatory functions of the glucocorticoid receptor require DNA binding. Nucleic Acids Res. 2020, 48, 8393–8407. [Google Scholar] [CrossRef]
- Dewhurst, S.M.; McGranahan, N.; Burrell, R.A.; Rowan, A.J.; Gronroos, E.; Endesfelder, D.; Joshi, T.; Mouradov, D.; Gibbs, P.; Ward, R.L.; et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov. 2014, 4, 175–185. [Google Scholar] [CrossRef]
- Quinton, R.J.; DiDomizio, A.; Vittoria, M.A.; Kotynkova, K.; Ticas, C.J.; Patel, S.; Koga, Y.; Vakhshoorzadeh, J.; Hermance, N.; Kuroda, T.S.; et al. Whole-genome doubling confers unique genetic vulnerabilities on tumour cells. Nature 2021, 590, 492–497. [Google Scholar] [CrossRef]
- Rao, S.; Stadanlick, J.E.; Cai, K.Q.; Wiest, D.L. Abstract 5215: Loss of Rpl22 promotes tumor progression through regulation of angiogenesis. Cancer Res. 2015, 75, 5215. [Google Scholar] [CrossRef]
- Inoue, K.; Fry, E.A. Haploinsufficient tumor suppressor genes. Adv. Med. Biol. 2017, 118, 83. [Google Scholar]
- Chen, L.; Zhang, J.J.; Rafii, S.; Huang, X.-Y. Suppression of Tumor Angiogenesis by Gα13 Haploinsufficiency. J. Biol. Chem. 2009, 284, 27409. [Google Scholar] [CrossRef] [PubMed]
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Disharoon, A.O.; Delaney, J.R. Half the Chromosome It Used to Be: Identifying Cancer Treatments Targeting Aneuploid Losses. Genes 2025, 16, 708. https://doi.org/10.3390/genes16060708
Disharoon AO, Delaney JR. Half the Chromosome It Used to Be: Identifying Cancer Treatments Targeting Aneuploid Losses. Genes. 2025; 16(6):708. https://doi.org/10.3390/genes16060708
Chicago/Turabian StyleDisharoon, Andrew O., and Joe R. Delaney. 2025. "Half the Chromosome It Used to Be: Identifying Cancer Treatments Targeting Aneuploid Losses" Genes 16, no. 6: 708. https://doi.org/10.3390/genes16060708
APA StyleDisharoon, A. O., & Delaney, J. R. (2025). Half the Chromosome It Used to Be: Identifying Cancer Treatments Targeting Aneuploid Losses. Genes, 16(6), 708. https://doi.org/10.3390/genes16060708





