Phenotypic Characterization and Marker–Trait Association Analysis Using SCoT Markers in Chrysanthemum (Chrysanthemum morifolium Ramat.) Germplasm
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Phenotypic Trait Measurement and Analysis
2.3. DNA Extraction
2.4. SCoT-PCR Amplification
2.5. Genetic Diversity and Population Structure Analysis
2.6. Association Analysis
3. Results
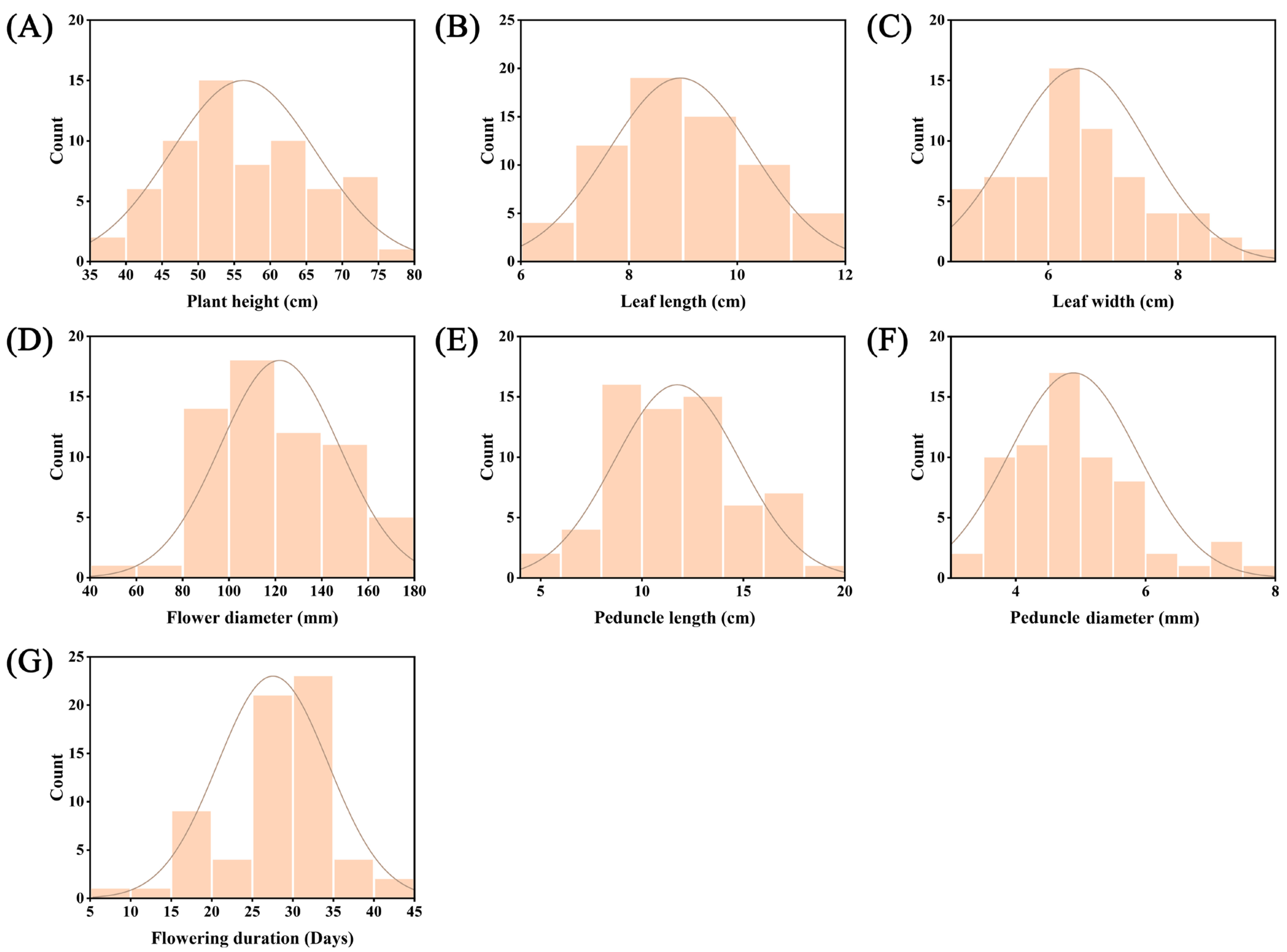
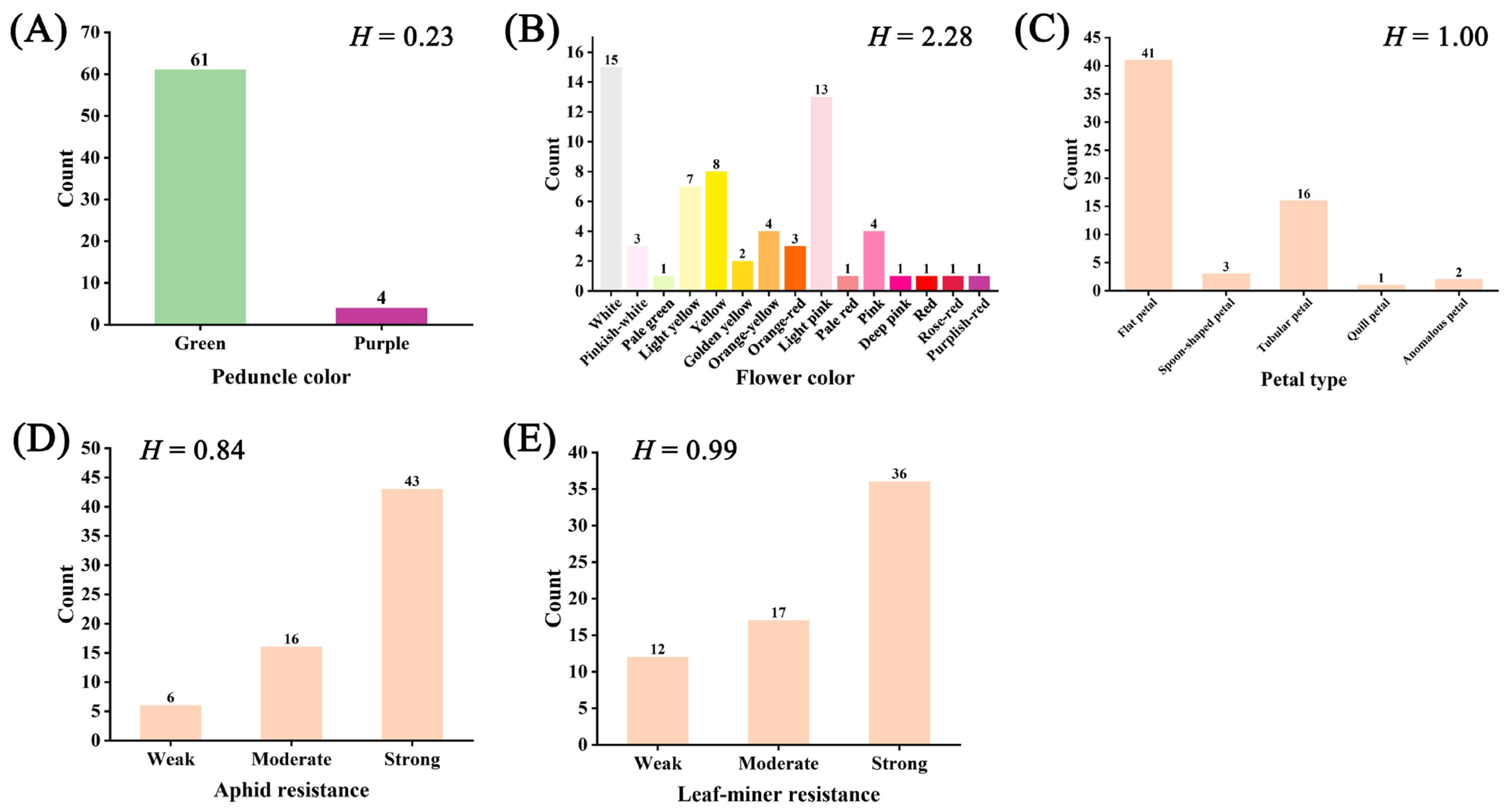
3.1. Phenotypic Diversity Analysis
3.1.1. Variation in Quantitative Traits
3.1.2. Diversity of Qualitative Traits
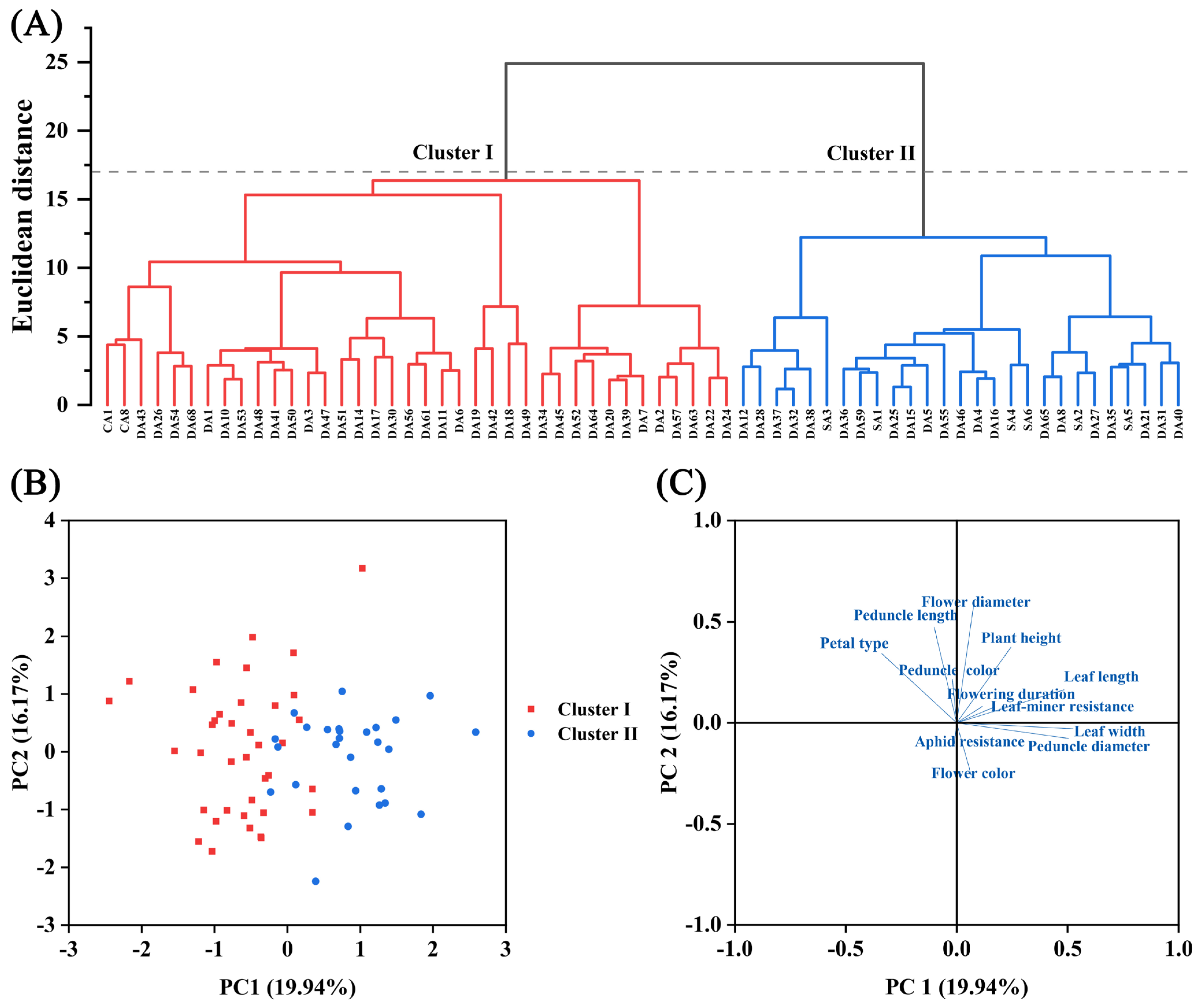
3.1.3. Multivariate Analysis
3.2. Genetic Diversity Analysis Based on SCoT Markers
3.2.1. Genetic Diversity Parameters
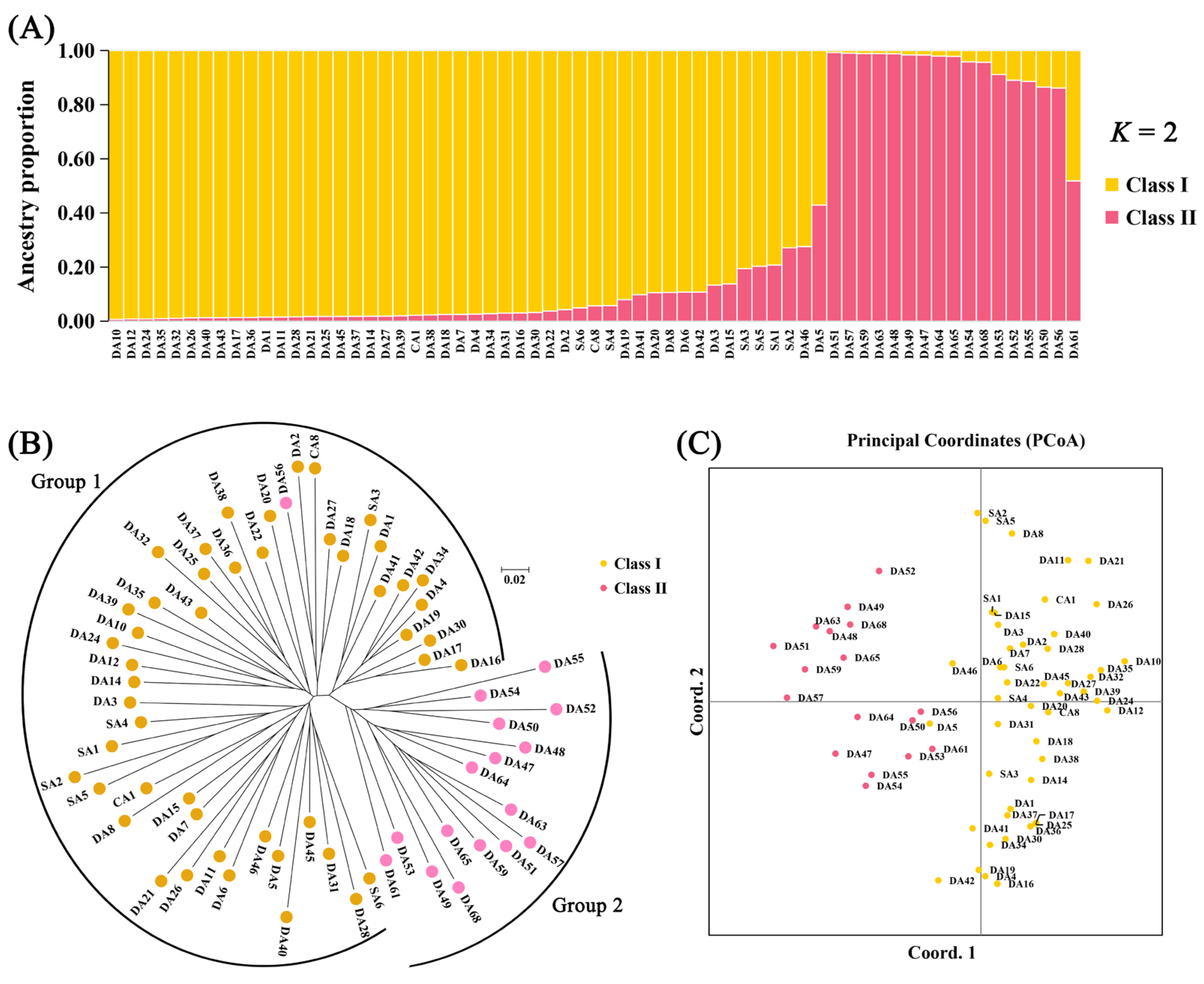
3.2.2. Population Structure, PCoA, and Cluster Analysis
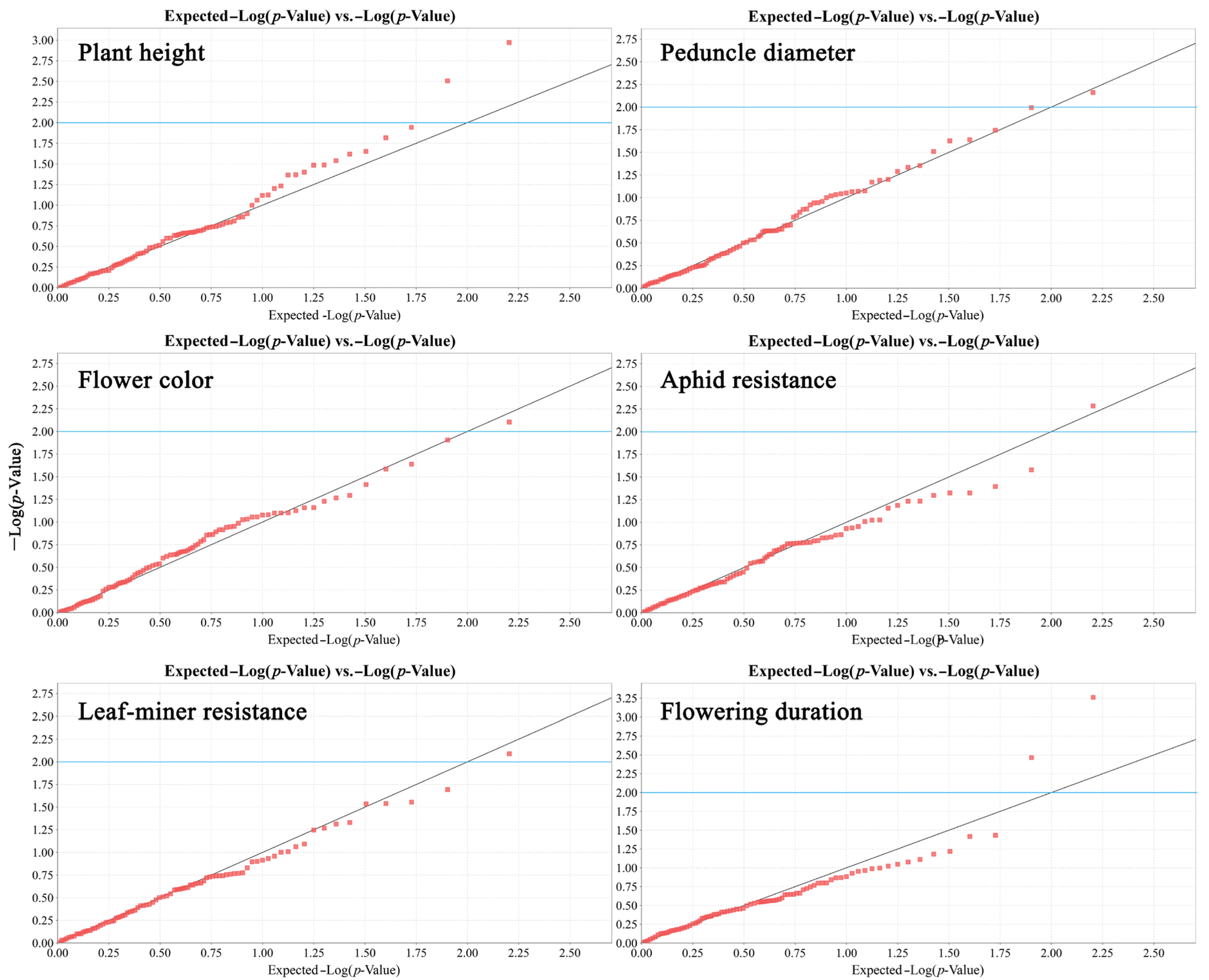
3.3. Marker–Trait Association Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, S.L.; Dai, S.L. The Chinese Chrysanthemum Book; China Forestry Publishing House: Beijing, China, 2013. [Google Scholar]
- Su, J.; Jiang, J.; Zhang, F.; Liu, Y.; Ding, L.; Chen, S.; Chen, F. Current achievements and future prospects in the genetic breeding of chrysanthemum: A review. Hortic. Res. 2019, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Teixeira Da Silva, J.A.; Shinoyama, H.; Aida, R.; Matsushita, Y.; Raj, S.K.; Chen, F. Chrysanthemum biotechnology: Quo vadis? Crit. Rev. Plant Sci. 2013, 32, 21–52. [Google Scholar] [CrossRef]
- Pan, B.; Pan, B.; Du, Y.; Chen, Q.; Chen, Q.; Wang, Y.; Chen, L.; Chen, L.; Li, H.; Huang, C.; et al. China’s chrysanthemum in the global market: Evaluating the international competitiveness and influencing factors. Front. Sustain. Food Syst. 2025, 9, 1521709. [Google Scholar] [CrossRef]
- Zhang, X.; Su, J.; Jia, F.; He, Y.; Liao, Y.; Wang, Z.; Jiang, J.; Guan, Z.; Fang, W.; Chen, F.; et al. Genetic architecture and genomic prediction of plant height-related traits in chrysanthemum. Hortic. Res. 2024, 11, uhad236. [Google Scholar] [CrossRef]
- Sun, J.; Wen, C. Phenotypic variation and inheritance of leaf weight in cut chrysanthemum: Phenotypic variation and inheritance of leaf weight in cut chrysanthemum. Euphytica 2025, 221, 27. [Google Scholar] [CrossRef]
- Miler, N.; Jedrzejczyk, I. Chrysanthemum plants regenerated from ovaries: A study on genetic and phenotypic variation. Turk. J. Bot. 2018, 42, 289–297. [Google Scholar]
- Lu, C.; Li, Y.; Wang, J.; Qu, J.; Chen, Y.; Chen, X.; Huang, H.; Dai, S. Flower color classification and correlation between color space values with pigments in potted multiflora chrysanthemum. Sci. Hortic. 2021, 283, 110082. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Mackill, D.J. Start codon targeted (SCoT) polymorphism: A simple, novel dna marker technique for generating gene-targeted markers in plants. Plant Mol. Biol. Rep. 2009, 27, 86–93. [Google Scholar] [CrossRef]
- Al-Khayri, J.M.; Mahdy, E.; Taha, H.; Eldomiaty, A.S.; Abd-Elfattah, M.A.; Abdel, L.A.; Rezk, A.A.; Shehata, W.F.; Almaghasla, M.I.; Shalaby, T.A.; et al. Genetic and morphological diversity assessment of five kalanchoe genotypes by SCoT, ISSR and RAPD-PCR markers. Plants 2022, 11, 1722. [Google Scholar] [CrossRef]
- Deng, L.; Liang, Q.; He, X.; Luo, C.; Chen, H.; Qin, Z. Investigation and analysis of genetic diversity of diospyros germplasms using SCoT molecular markers in guangxi. PLoS ONE 2015, 10, e136510. [Google Scholar] [CrossRef]
- Rai, M.K. Start codon targeted (SCoT) polymorphism marker in plant genome analysis: Current status and prospects. Planta 2023, 257, 34. [Google Scholar] [CrossRef] [PubMed]
- Shaban, A.S.; Arab, S.A.; Basuoni, M.M.; Abozahra, M.S.; Abdelkawy, A.M.; Mohamed, M.M.; Merah, O. SCoT, ISSR, and SDS-PAGE investigation of genetic diversity in several egyptian wheat genotypes under normal and drought conditions. Int. J. Agron. 2022, 2022, 7024028. [Google Scholar] [CrossRef]
- Poczai, P.; Varga, I.; Laos, M.; Cseh, A.; Bell, N.; Valkonen, J.P.; Hyvönen, J. Advances in plant gene-targeted and functional markers: A review. Plant Methods 2013, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Sumitomo, K.; Shirasawa, K.; Isobe, S.; Hirakawa, H.; Hisamatsu, T.; Nakano, Y.; Yagi, M.; Ohmiya, A. Genome-wide association study overcomes the genome complexity in autohexaploid chrysanthemum and tags snp markers onto the flower color genes. Sci. Rep. 2019, 9, 13947–13949. [Google Scholar] [CrossRef]
- Nguyen, T.K.; Ha, S.T.T.; Lim, J.H. Analysis of chrysanthemum genetic diversity by genotyping-by-sequencing. Hortic. Environ. Biotechnol. 2020, 61, 903–913. [Google Scholar] [CrossRef]
- Song, X.; Xu, Y.; Gao, K.; Fan, G.; Zhang, F.; Deng, C.; Dai, S.; Huang, H.; Xin, H.; Li, Y. High-density genetic map construction and identification of loci controlling flower-type traits in chrysanthemum (Chrysanthemum × morifolium Ramat.). Hortic. Res. 2020, 7, 108. [Google Scholar] [CrossRef]
- Li, P.; Zhang, F.; Chen, S.; Jiang, J.; Wang, H.; Su, J.; Fang, W.; Guan, Z.; Chen, F. Genetic diversity, population structure and association analysis in cut chrysanthemum (Chrysanthemum morifolium Ramat.). Mol. Genet. Genom. 2016, 291, 1117–1125. [Google Scholar] [CrossRef]
- Feng, S.; He, R.; Jiang, M.; Lu, J.; Shen, X.; Liu, J.; Wang, Z.; Wang, H. Genetic diversity and relationships of medicinal chrysanthemum morifolium revealed by start codon targeted (SCoT) markers. Sci. Hortic. 2016, 201, 118–123. [Google Scholar] [CrossRef]
- Samarina, L.S.; Malyarovskaya, V.I.; Reim, S.; Yakushina, L.G.; Koninskaya, N.G.; Klemeshova, K.V.; Shkhalakhova, R.M.; Matskiv, A.O.; Shurkina, E.S.; Gabueva, T.Y.; et al. Transferability of ISSR, SCoT and SSR markers for Chrysanthemum × morifolium ramat and genetic relationships among commercial russian cultivars. Plants 2021, 10, 1302. [Google Scholar] [CrossRef]
- Kulus, D.; Tymoszuk, A.; Gościnna, K.; Osial, M. Enhancing germination and growth of chrysanthemum synthetic seeds through iron oxide nanoparticles and indole-3-acetic acid: Impact of treatment duration on metabolic activity and genetic stability. Nanotechnol. Sci. Appl. 2025, 18, 139–155. [Google Scholar] [CrossRef]
- Tymoszuk, A.; Szałaj, U.; Wojnarowicz, J.; Kowalska, J.; Antkowiak, M.; Kulus, D. Zinc oxide and silver effects on the growth, pigment content and genetic stability of chrysanthemums propagated by the node culture method. Folia Hortic. 2024, 36, 35–66. [Google Scholar] [CrossRef]
- NY/T 2228-2012; Guidelines for the Conduct of Tests for Distinctness, Uniformity, and Stability—Chrysanthemum. Ministry of Agriculture of the People’s Republic of China: Beijing, China, 2012.
- R Core Team. R: A Language and Environment for Statistical Computing. Version 4.3.1, R Foundation for Statistical Computing. 2023. Available online: http://www.R-project.org/ (accessed on 25 March 2025).
- Nagy, S.; Poczai, P.; Cernák, I.; Gorji, A.M.; Hegedűs, G.; Taller, J. PICcalc: An online program to calculate polymorphic information content for molecular genetic studies. Biochem. Genet. 2012, 50, 670–672. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Muse, S.V. Powermarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Earl, D.A.; Vonholdt, B.M. Structure harvester: A website and program for visualizing structure output and implementing the evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. Genalex 6.5: Genetic analysis in excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. Tassel: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Hodaei, M.; Rahimmalek, M.; Arzani, A. Genetic diversity of iranian Chrysanthemum morifolium cultivars using morphological traits and sequence-related amplified polymorphism (SRAP) markers. Hortic. Environ. Biotechnol. 2019, 60, 753–765. [Google Scholar] [CrossRef]
- Roein, Z.; Hassanpour Asil, M.; Sabouri, A.; Dadras, A.R. Genetic structure of chrysanthemum genotypes from iran assessed by AFLP markers and phenotypic traits. Plant Syst. Evol. 2014, 300, 493–503. [Google Scholar] [CrossRef]
- Fu, X.; Su, J.; Yu, K.; Cai, Y.; Zhang, F.; Chen, S.; Fang, W.; Fadi, C.; Guan, Z. Genetic variation and association mapping of aphid (Macrosiphoniella sanbourni) resistance in chrysanthemum (Chrysanthemum morifolium ramat.). Euphytica 2018, 214, 21. [Google Scholar] [CrossRef]
- Yang, X.; Su, J.; Qu, Y.; Jiang, J.; Guan, Z.; Fang, W.; Chen, F.; Zhang, F. Dissecting the inheritance pattern of the anemone flower type and tubular floral traits of chrysanthemum in segregating f1 populations. Euphytica 2023, 219, 16. [Google Scholar] [CrossRef]
- Zhang, F.; Chen, S.; Chen, F.; Fang, W.; Deng, Y.; Chang, Q.; Liu, P. Genetic analysis and associated SRAP markers for flowering traits of chrysanthemum (Chrysanthemum morifolium). Euphytica 2011, 177, 15–24. [Google Scholar] [CrossRef]
- Zhou, Y.; Wei, X.; Abbas, F.; Yu, Y.; Yu, R.; Fan, Y. Genome-wide identification of simple sequence repeats and assessment of genetic diversity in Hedychium. J. Appl. Res. Med. Aromat. Plants 2021, 24, 100312. [Google Scholar] [CrossRef]
- Su, J.; Zhang, F.; Li, P.; Guan, Z.; Fang, W.; Chen, F. Genetic variation and association mapping of waterlogging tolerance in chrysanthemum. Planta 2016, 244, 1241–1252. [Google Scholar] [CrossRef]
- Su, J.; Zhang, F.; Chong, X.; Song, A.; Guan, Z.; Fang, W.; Chen, F. Genome-wide association study identifies favorable snp alleles and candidate genes for waterlogging tolerance in chrysanthemums. Hortic. Res. 2019, 6, 21. [Google Scholar] [CrossRef]
- Li, P.; Su, J.; Guan, Z.; Fang, W.; Chen, F.; Zhang, F. Association analysis of drought tolerance in cut chrysanthemum (Chrysanthemum morifolium Ramat.) At seedling stage. 3 Biotech 2018, 8, 226. [Google Scholar] [CrossRef]
- Wan, W.; Jia, F.; Liu, Z.; Sun, W.; Zhang, X.; Su, J.; Guan, Z.; Chen, F.; Zhang, F.; Fang, W. Quantitative evaluation and genome-wide association studies of chrysanthemum flower color. Sci. Hortic 2024, 338, 113561. [Google Scholar] [CrossRef]
- Su, J.; Zeng, J.; Wang, S.; Zhang, X.; Zhao, L.; Wen, S.; Zhang, F.; Jiang, J.; Chen, F. Multi-locus genome-wide association studies reveal the dynamic genetic architecture of flowering time in chrysanthemum. Plant Cell Rep. 2024, 43, 84. [Google Scholar] [CrossRef]
- Zhang, X.; Ning, X.; He, Y.; Su, J.; Wen, S.; Lu, Z.; Sun, W.; Wang, H.; Guan, Z.; Fang, W.; et al. GWAS reveals the genetic basis and genomic regions underlying four active compounds in chrysanthemum. Hortic. Plant J. 2025; in press. [Google Scholar] [CrossRef]

| Marker | Sequence (5′-3′) | OAT1 °C | Amplified Bands | Polymorphic Bands | Polymorphism Ratio | PIC2 |
|---|---|---|---|---|---|---|
| SCoT1 | CAACAATGGCTACCACCA | 55.4 | 5 | 5 | 100.00% | 0.769 |
| SCoT2 | CAACAATGGCTACCACCC | 53.4 | 6 | 6 | 100.00% | 0.767 |
| SCoT3 | CAACAATGGCTACCACCG | 54.1 | 3 | 3 | 100.00% | 0.408 |
| SCoT4 | CAACAATGGCTACCACCT | 52.1 | 4 | 4 | 100.00% | 0.708 |
| SCoT7 | CAACAATGGCTACCACGG | 54.1 | 5 | 5 | 100.00% | 0.539 |
| SCoT8 | CAACAATGGCTACCACGT | 52.6 | 5 | 5 | 100.00% | 0.714 |
| SCoT9 | CAACAATGGCTACCAGCA | 52.7 | 5 | 5 | 100.00% | 0.724 |
| SCoT11 | AAGCAATGGCTACCACCA | 58.1 | 9 | 9 | 100.00% | 0.774 |
| SCoT14 | ACGACATGGCGACCACGC | 66.3 | 12 | 12 | 100.00% | 0.896 |
| SCoT16 | ACCATGGCTACCACCGAC | 63 | 5 | 5 | 100.00% | 0.573 |
| SCoT18 | ACCATGGCTACCACCGCC | 66.5 | 9 | 9 | 100.00% | 0.721 |
| SCoT20 | ACCATGGCTACCACCGCG | 61.8 | 7 | 7 | 100.00% | 0.829 |
| SCoT21 | ACGACATGGCGACCCACA | 69.3 | 8 | 8 | 100.00% | 0.822 |
| SCoT22 | AACCATGGCTACCACCAC | 59.2 | 5 | 5 | 100.00% | 0.772 |
| SCoT23 | CACCATGGCTACCACCAG | 61 | 12 | 12 | 100.00% | 0.868 |
| SCoT24 | CACCATGGCTACCACCAT | 61 | 8 | 7 | 87.50% | 0.855 |
| SCoT27 | ACCATGGCTACCACCGTG | 62 | 9 | 9 | 100.00% | 0.879 |
| SCoT28 | CCATGGCTACCACCGCCA | 66.6 | 5 | 5 | 100.00% | 0.729 |
| SCoT30 | CCATGGCTACCACCGGCG | 64.2 | 10 | 10 | 100.00% | 0.772 |
| SCoT31 | CCATGGCTACCACCGCCT | 64.5 | 5 | 5 | 100.00% | 0.743 |
| SCoT34 | ACCATGGCTACCACCGCA | 65 | 4 | 4 | 100.00% | 0.478 |
| SCoT35 | CATGGCTACCACCGGCCC | 63.1 | 12 | 12 | 100.00% | 0.825 |
| SCoT36 | GCAACAATGGCTACCACC | 60 | 7 | 7 | 100.00% | 0.793 |
| Mean | 6.957 | 6.913 | 99.46% | 0.737 | ||
| Trait | Primer | Band | p-Value | R2 |
|---|---|---|---|---|
| Flowering duration | SCoT28 | band1 | 0.0005 | 20.92% |
| Plant height | SCoT3 | band3 | 0.0011 | 18.53% |
| Plant height | SCoT30 | band8 | 0.0031 | 14.89% |
| Flowering duration | SCoT31 | band1 | 0.0034 | 14.58% |
| Aphid resistance | SCoT35 | band10 | 0.0052 | 13.32% |
| Peduncle diameter | SCoT20 | band1 | 0.0069 | 12.31% |
| Flower color | SCoT14 | band10 | 0.0079 | 11.81% |
| Leaf-miner resistance | SCoT36 | band1 | 0.0082 | 11.18% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Chen, X.; Huang, Z.; Wei, L.; Wang, J.; Wen, S.; Liu, Y.; Zhou, Y. Phenotypic Characterization and Marker–Trait Association Analysis Using SCoT Markers in Chrysanthemum (Chrysanthemum morifolium Ramat.) Germplasm. Genes 2025, 16, 664. https://doi.org/10.3390/genes16060664
Wang F, Chen X, Huang Z, Wei L, Wang J, Wen S, Liu Y, Zhou Y. Phenotypic Characterization and Marker–Trait Association Analysis Using SCoT Markers in Chrysanthemum (Chrysanthemum morifolium Ramat.) Germplasm. Genes. 2025; 16(6):664. https://doi.org/10.3390/genes16060664
Chicago/Turabian StyleWang, Fenglan, Xiuzhe Chen, Zifeng Huang, Lisha Wei, Jun Wang, Shuang Wen, Yang Liu, and Yiwei Zhou. 2025. "Phenotypic Characterization and Marker–Trait Association Analysis Using SCoT Markers in Chrysanthemum (Chrysanthemum morifolium Ramat.) Germplasm" Genes 16, no. 6: 664. https://doi.org/10.3390/genes16060664
APA StyleWang, F., Chen, X., Huang, Z., Wei, L., Wang, J., Wen, S., Liu, Y., & Zhou, Y. (2025). Phenotypic Characterization and Marker–Trait Association Analysis Using SCoT Markers in Chrysanthemum (Chrysanthemum morifolium Ramat.) Germplasm. Genes, 16(6), 664. https://doi.org/10.3390/genes16060664






