Analysis of Codon Usage Bias of 30 Chloroplast Genomes in Ulva (Ulvophyceae, Chlorophyta)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Collection and Processing
2.2. Codon Usage Indices
2.3. Identification of Optimal Codon
2.4. ENC Plot Analyses
2.5. PR2 Plot Analyses
2.6. Neutrality Plot Analyses
2.7. Correspondence Analyses
3. Results
3.1. Nucleotide Composition
3.2. The Relative Synonymous Codon Usage and Optimal Codons
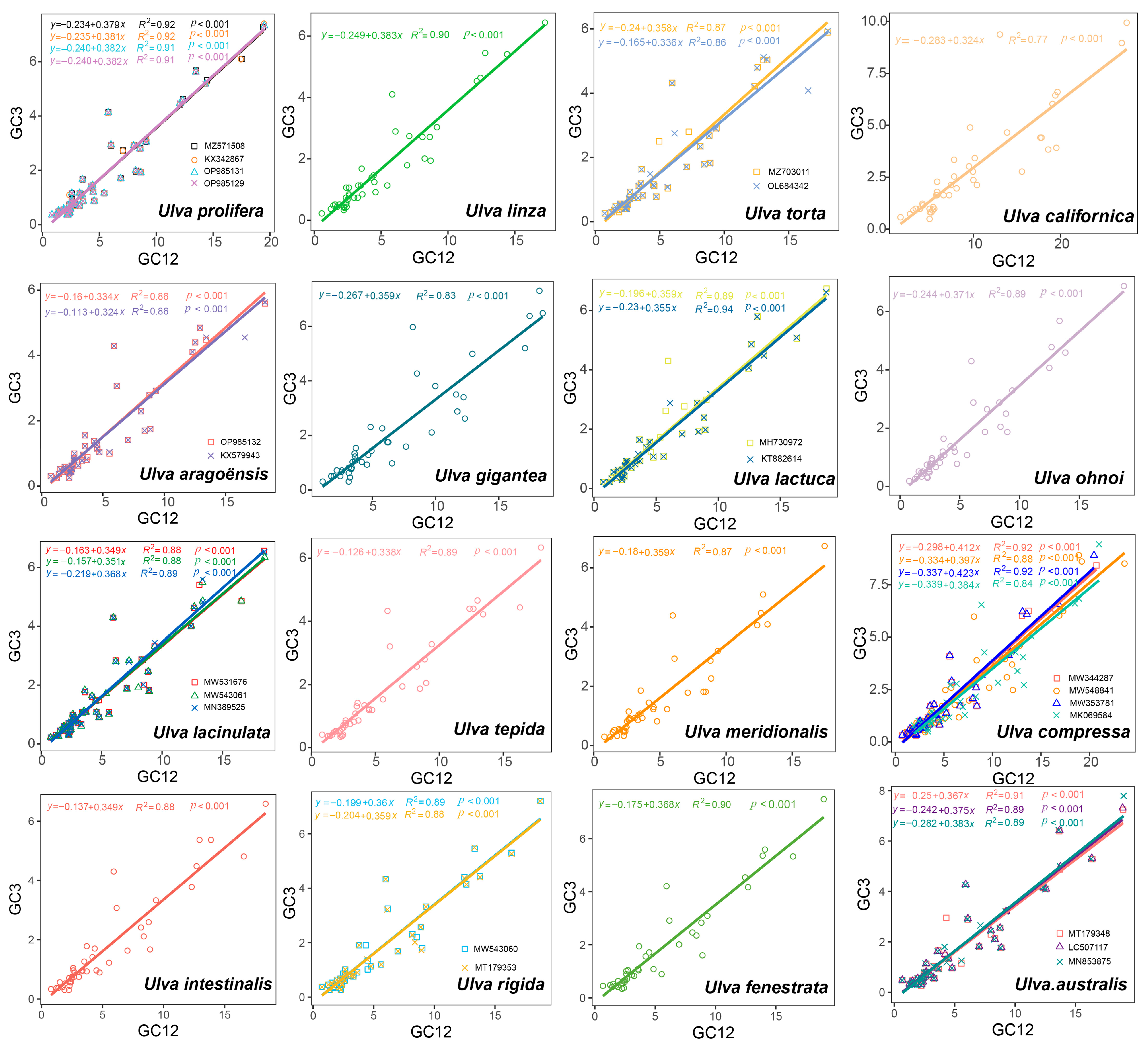
3.3. ENC Plot Analyses
3.4. PR2 Plot Analyses
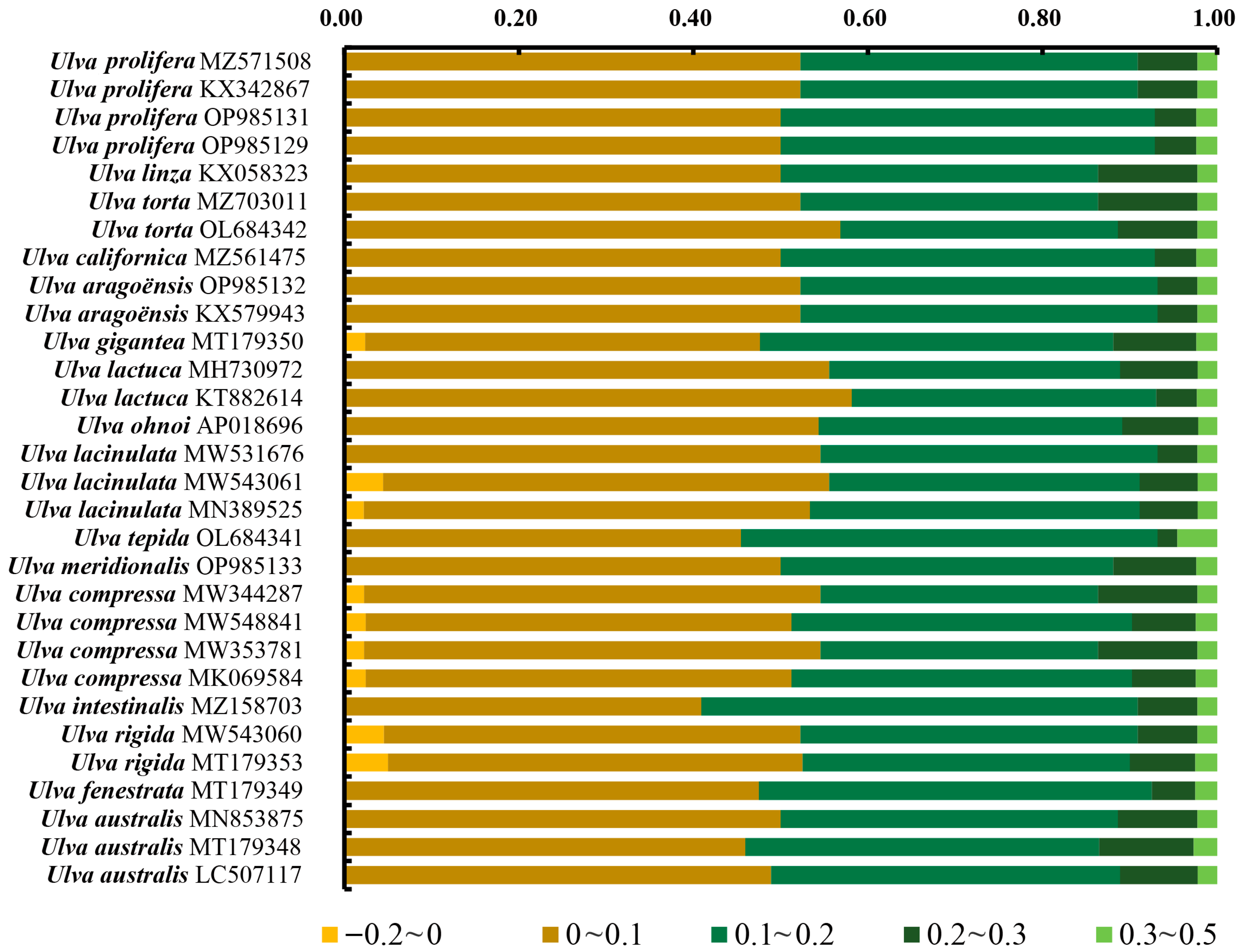
3.5. Neutrality Plot Analyses
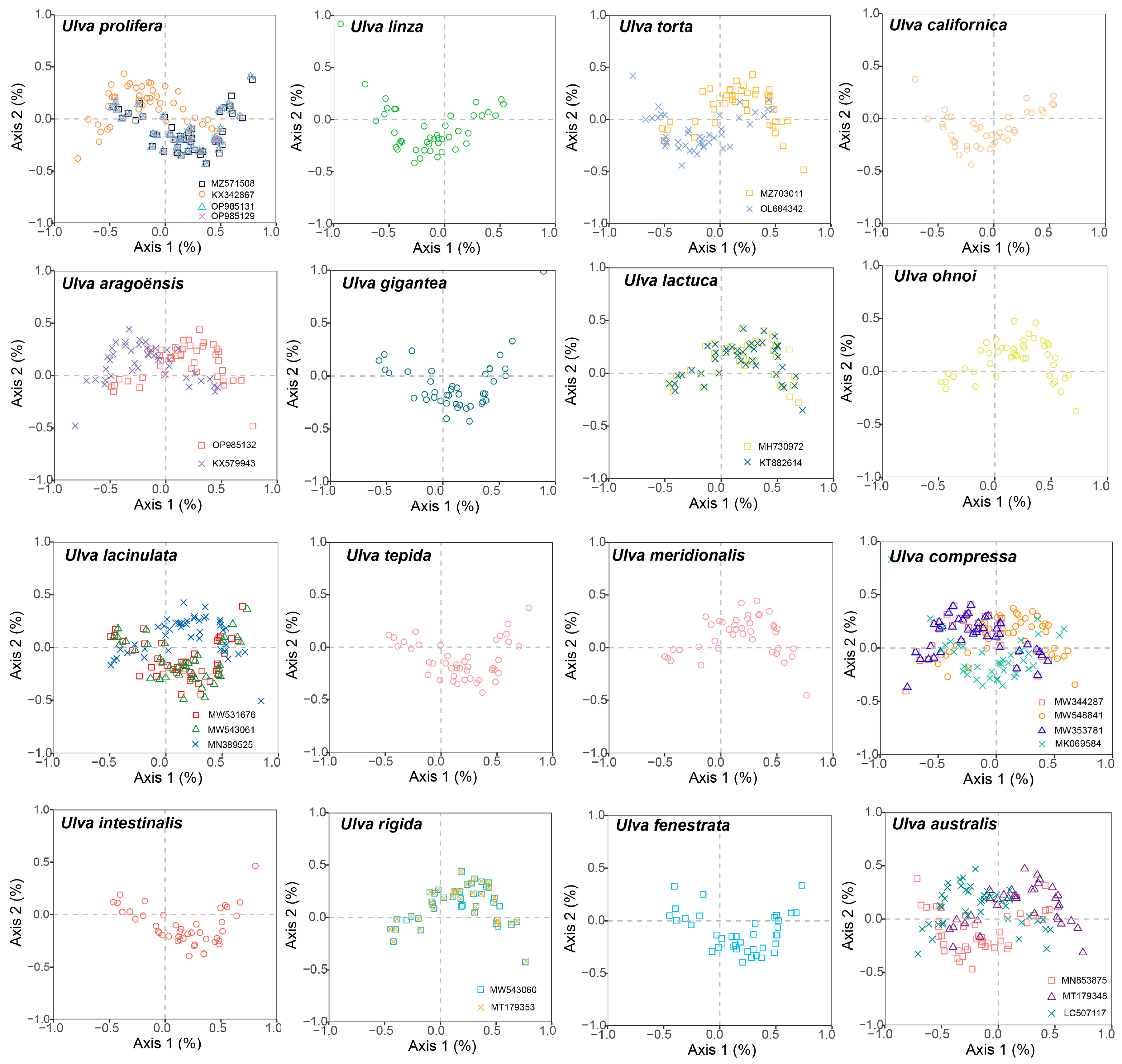
3.6. Correspondence Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ichihara, K.; Arai, S.; Uchimura, M.; Fay, E.J.; Ebata, H.; Hiraoka, M.; Shimada, S. New species of freshwater Ulva, Ulva limnetica (Ulvales, Ulvophyceae) from the Ryukyu Islands, Japan. Phycol. Res. 2009, 57, 94–103. [Google Scholar] [CrossRef]
- Liu, X.Q.; Wang, Z.L.; Zhang, X.L. A review of the green tides in the Yellow Sea, China. Mar. Environ. Res. 2016, 119, 189–196. [Google Scholar] [CrossRef]
- Tran, L.A.T.; Vieira, C.; Steinhagen, S.; Maggs, C.A.; Hiraoka, M.; Shimada, S.; Van Nguyen, T.; De Clerck, O.; Leliaert, F. An appraisal of Ulva (Ulvophyceae, Chlorophyta) taxonomy. J. Appl. Phycol. 2022, 34, 2689–2703. [Google Scholar] [CrossRef]
- Florez, J.Z.; Camus, C.; Hengst, M.B.; Buschmann, A.H. A Functional Perspective Analysis of Macroalgae and Epiphytic Bacterial Community Interaction. Front. Microbiol. 2017, 8, 2561. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Li, D.; Chen, C.; Ge, J.; Muller-Karger, F.E.; Liu, J.; Yu, F.; He, M.X. On the recurrent Ulva prolifera blooms in the Yellow Sea and East China Sea. J. Geophys. Res. Oceans 2010, 115, C05017. [Google Scholar] [CrossRef]
- Blomster, J.; Bäck, S.; Fewer, D.P.; Kiirikki, M.; Lehvo, A.; Maggs, C.A.; Stanhope, M.J. Novel morphology in Enteromorpha (Ulvophyceae) forming green tides. Am. J. Bot. 2002, 89, 1756–1763. [Google Scholar] [CrossRef]
- Hiraoka, M.; Ohno, M.; Kawaguchi, S.; Yoshida, G. Crossing test among floating Ulva thalli forming ‘green tide’ in Japan. Hydrobiologia 2004, 512, 239–245. [Google Scholar] [CrossRef]
- Lawton, R.J.; Mata, L.; de Nys, R.; Paul, N.A. Algal Bioremediation of Waste Waters from Land-Based Aquaculture Using Ulva: Selecting Target Species and Strains. PLoS ONE 2013, 8, e77344. [Google Scholar] [CrossRef]
- Cahill, P.L.; Hurd, C.L.; Lokman, M. Keeping the water clean—Seaweed biofiltration outperforms traditional bacterial biofilms in recirculating aquaculture. Aquaculture 2010, 306, 153–159. [Google Scholar] [CrossRef]
- Wichard, T. Exploring bacteria-induced growth and morphogenesis in the green macroalga order Ulvales (Chlorophyta). Front. Plant Sci. 2015, 6, 86. [Google Scholar] [CrossRef]
- Spoerner, M.; Wichard, T.; Bachhuber, T.; Stratmann, J.; Oertel, W. Growth and Thallus Morphogenesis of Ulva mutabilis (Chlorophyta) Depends on A Combination of Two Bacterial Species Excreting Regulatory Factors. J. Phycol. 2012, 48, 1433–1447. [Google Scholar] [CrossRef] [PubMed]
- Kessler, R.W.; Weiss, A.; Kuegler, S.; Hermes, C.; Wichard, T. Macroalgal–bacterial interactions: Role of dimethylsulfoniopropionate in microbial gardening by Ulva (Chlorophyta). Mol. Ecol. 2018, 27, 1808–1819. [Google Scholar] [CrossRef] [PubMed]
- Allouache, A.; Majda, A.; Toudert, A.Z.; Amrane, A.; Ballesteros, M. Cellulosic bioethanol production from Ulva Lactuca macroalgae. Cell. Chem. Technol. 2021, 55, 629–635. [Google Scholar] [CrossRef]
- Chemodanov, A.; Jinjikhashvily, G.; Habiby, O.; Liberzon, A.; Israel, A.; Yakhini, Z.; Golberg, A. Net primary productivity, biofuel production and CO2 emissions reduction potential of Ulva sp. (Chlorophyta) biomass in a coastal area of the Eastern Mediterranean. Energy Convers. Manag. 2017, 148, 1497–1507. [Google Scholar] [CrossRef]
- Puthiya Veettil, R.; Rabia; Mathew, D.K.; Gondi, R.; Sankarapandian, K.; Kannan, M.; Kumar, G.; Al-Qaradawi, S.Y.; Jeyakumar, R.B. Synergistic Effect of Surfactant on Disperser Energy and Liquefaction Potential of Macroalgae (Ulva intestinalis) for Biofuel Production. Fermentation 2023, 9, 55. [Google Scholar] [CrossRef]
- Gao, G.; Clare, A.S.; Rose, C.; Caldwell, G.S. Ulva rigida in the future ocean: Potential for carbon capture, bioremediation and biomethane production. GCB Bioenergy 2018, 10, 39–51. [Google Scholar] [CrossRef]
- Ramachandra, T.V.; Hebbale, D. Bioethanol from macroalgae: Prospects and challenges. Renew. Sust. Energ. Rev. 2020, 117, 109479. [Google Scholar] [CrossRef]
- Hofmann, L.C.; Strauss, S.; Shpigel, M.; Guttman, L.; Stengel, D.B.; Rebours, C.; Gjorgovska, N.; Turan, G.; Balina, K.; Zammit, G.; et al. The green seaweed Ulva: Tomorrow’s “wheat of the sea” in foods, feeds, nutrition, and biomaterials. Crit. Rev. Food Sci. Nutr. 2024, 64, 1–36. [Google Scholar] [CrossRef]
- Juul, L.; Nissen, S.H.; Bruhn, A.; Alexi, N.; Jensen, S.K.; Hammershøj, M.; Dalsgaard, T.K. Ulva species: A critical review on the green seaweed as a source of food protein. Trends Food Sci. Technol. 2024, 149, 104534. [Google Scholar] [CrossRef]
- Ning, L.; Yao, Z.; Zhu, B. Ulva (Enteromorpha) Polysaccharides and Oligosaccharides: A Potential Functional Food Source from Green-Tide-Forming Macroalgae. Mar. Drugs 2022, 20, 202. [Google Scholar] [CrossRef]
- Ou, J.Y.; Wei, Y.J.; Liu, F.L.; Huang, C.H. Anti-allergic effects of Ulva-derived polysaccharides, oligosaccharides and residues in a murine model of food allergy. Heliyon 2023, 9, e22840. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, X.; Shi, Z. Comparative Study on Codon Usage Patterns across Chloroplast Genomes of Eighteen Taraxacum Species. Horticulturae 2024, 10, 492. [Google Scholar] [CrossRef]
- Thomas, P.D.; Ferrer, M.F.; Lozano, M.J.; Gómez, R.M. A study on the codon usage bias of arenavirus common genes. Front. Microbiol. 2025, 15, 1490076. [Google Scholar] [CrossRef]
- Yang, X.; Wang, Y.; Gong, W.; Li, Y. Comparative Analysis of the Codon Usage Pattern in the Chloroplast Genomes of Gnetales Species. Int. J. Mol. Sci. 2024, 25, 10622. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Chen, S.; Liang, M.; Qu, S.; Feng, S.; Wang, D.; Wang, G. Comparative analysis of codon usage bias in chloroplast genomes of ten medicinal species of Rutaceae. BMC Plant Biol. 2024, 24, 424. [Google Scholar] [CrossRef]
- Parvathy, S.T.; Udayasuriyan, V.; Bhadana, V. Codon usage bias. Mol. Biol. Rep. 2022, 49, 539–565. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Cheng, Z.; Luo, L.; Cheng, K.; Gan, S.; Shi, Y.; Liu, C.; Wang, D. Comparative analysis of codon usage patterns of Plasmodium helical interspersed subtelomeric (PHIST) proteins. Front. Microbiol. 2023, 14, 1320060. [Google Scholar] [CrossRef]
- Athey, J.; Alexaki, A.; Osipova, E.; Rostovtsev, A.; Santana-Quintero, L.V.; Katneni, U.; Simonyan, V.; Kimchi-Sarfaty, C. A new and updated resource for codon usage tables. BMC Bioinform. 2017, 18, 391. [Google Scholar] [CrossRef]
- Wong, E.H.M.; Smith, D.K.; Rabadan, R.; Peiris, M.; Poon, L.L.M. Codon usage bias and the evolution of influenza A viruses. Codon Usage Biases of Influenza Virus. BMC Evol. Biol. 2010, 10, 253. [Google Scholar] [CrossRef]
- Anhlan, D.; Grundmann, N.; Makalowski, W.; Ludwig, S.; Scholtissek, C. Origin of the 1918 pandemic H1N1 influenza A virus as studied by codon usage patterns and phylogenetic analysis. RNA 2011, 17, 64–73. [Google Scholar] [CrossRef]
- Prat, Y.; Fromer, M.; Linial, N.; Linial, M. Codon usage is associated with the evolutionary age of genes in metazoan genomes. BMC Evol. Biol. 2009, 9, 285. [Google Scholar] [CrossRef] [PubMed]
- Kwon, K.C.; Chan, H.T.; León, I.R.; Williams-Carrier, R.; Barkan, A.; Daniell, H. Codon Optimization to Enhance Expression Yields Insights into Chloroplast Translation. Plant Physiol. 2016, 172, 62–77. [Google Scholar] [CrossRef] [PubMed]
- Bahir, I.; Fromer, M.; Prat, Y.; Linial, M. Viral adaptation to host: A proteome-based analysis of codon usage and amino acid preferences. Mol. Syst. Biol. 2009, 5, 311. [Google Scholar] [CrossRef]
- Pandit, A.; Sinha, S. Differential Trends in the Codon Usage Patterns in HIV-1 Genes. PLoS ONE 2011, 6, e28889. [Google Scholar] [CrossRef]
- Tao, J.; Yao, H. Comprehensive analysis of the codon usage patterns of polyprotein of Zika virus. Prog. Biophys. Mol. Biol. 2020, 150, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, T.H.; Alqahtani, A.M.; Alqahtani, T.; Emran, T.B.; Aldahish, A.A.; Uddin, A. Analysis of Codon Usage of Speech Gene FoxP2 among Animals. Biology 2021, 10, 111078. [Google Scholar] [CrossRef]
- Gao, W.; Chen, X.; He, J.; Sha, A.; Luo, Y.; Xiao, W.; Xiong, Z.; Li, Q. Intraspecific and interspecific variations in the synonymous codon usage in mitochondrial genomes of 8 pleurotus strains. BMC Genom. 2024, 25, 456. [Google Scholar] [CrossRef]
- Dobrogojski, J.; Adamiec, M.; Luciński, R. The chloroplast genome: A review. Acta Physiol. Plant. 2020, 42, 98. [Google Scholar] [CrossRef]
- HÄDer, D.-P. Photosynthesis in Plants and Algae. Anticancer Res. 2022, 42, 5035. [Google Scholar] [CrossRef]
- Zhang, Y.; Du, L.; Liu, A.; Chen, J.; Wu, L.; Hu, W.; Zhang, W.; Kim, K.; Lee, S.C.; Yang, T.J.; et al. The Complete Chloroplast Genome Sequences of Five Epimedium Species: Lights into Phylogenetic and Taxonomic Analyses. Front. Plant Sci. 2016, 7, 306. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.A.; Gan, Z.M.; Zhang, F.; Yi, X.Y.; Zhang, J.Z.; Wan, X.H. Analysis of codon usage patterns in citrus based on coding sequence data. BMC Genom. 2020, 21, 234. [Google Scholar] [CrossRef]
- Yang, M.H.; Liu, J.H.; Yang, W.Q.; Li, Z.; Hai, Y.L.; Duan, B.Z.; Zhang, H.Z.; Yang, X.L.; Xia, C.L. Analysis of codon usage patterns in 48 Aconitum species. BMC Genom. 2023, 24, 703. [Google Scholar] [CrossRef]
- Wang, D.; Yang, B. Analysis of codon usage bias of thioredoxin in apicomplexan protozoa. Parasites Vectors 2023, 16, 431. [Google Scholar] [CrossRef]
- Wright, F. The ‘effective number of codons’ used in a gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Fuglsang, A. The ‘effective number of codons’ revisited. Biochem. Biophys. Res. Commun. 2004, 317, 957–964. [Google Scholar] [CrossRef]
- Zeng, Y.; Shen, L.; Chen, S.; Qu, S.; Hou, N. Codon Usage Profiling of Chloroplast Genome in Juglandaceae. Forests 2023, 14, 020378. [Google Scholar] [CrossRef]
- Wang, Z.J.; Cai, Q.W.; Wang, Y.; Li, M.H.; Wang, C.C.; Wang, Z.X.; Jiao, C.Y.; Xu, C.C.; Wang, H.Y.; Zhang, Z.L. Comparative Analysis of Codon Bias in the Chloroplast Genomes of Theaceae Species. Front. Genet. 2022, 13, 824610. [Google Scholar] [CrossRef]
- Sueoka, N. Intrastrand parity rules of DNA base composition and usage biases of synonymous codons. J. Mol. Evol. 1995, 40, 318–325. [Google Scholar] [CrossRef]
- Yang, Q.; Xin, C.; Xiao, Q.-S.; Lin, Y.-T.; Li, L.; Zhao, J.-L. Codon usage bias in chloroplast genes implicate adaptive evolution of four ginger species. Front. Plant Sci. 2023, 14, 1304264. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Yengkhom, S.; Uddin, A. Analysis of codon usage bias of chloroplast genes in Oryza species Codon usage of chloroplast genes in Oryza species. Planta 2020, 252, 67. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Theory and Applications of Correspondence Analysis. Biometrics 1986, 42, 223. [Google Scholar] [CrossRef]
- Xue, C.; Zhang, Y.; Li, H.; Liu, Z.; Gao, W.; Liu, M.; Wang, H.; Liu, P.; Zhao, J. The genome of Candidatus phytoplasma ziziphi provides insights into their biological characteristics. BMC Plant Biol. 2023, 23, 251. [Google Scholar] [CrossRef] [PubMed]
- Long, S.; Yao, H.; Wu, Q.; Li, G. Analysis of compositional bias and codon usage pattern of the coding sequence in Banna virus genome. Virus Res. 2018, 258, 68–72. [Google Scholar] [CrossRef]
- Deb, B.; Uddin, A.; Chakraborty, S. Codon usage pattern and its influencing factors in different genomes of hepadnaviruses. Arch. Virol. 2020, 165, 557–570. [Google Scholar] [CrossRef]
- Wu, P.; Xiao, W.; Luo, Y.; Xiong, Z.; Chen, X.; He, J.; Sha, A.; Gui, M.; Li, Q. Comprehensive analysis of codon bias in 13 Ganoderma mitochondrial genomes. Front. Microbiol. 2023, 14, 1170790. [Google Scholar] [CrossRef]
- Kawabe, A.; Miyashita, N.T. Patterns of codon usage bias in three dicot and four monocot plant species. Genes Genet. Syst. 2003, 78, 343–352. [Google Scholar] [CrossRef]
- Li, N.; Li, Y.Y.; Zheng, C.C.; Huang, J.G.; Zhang, S.Z. Genome-wide comparative analysis of the codon usage patterns in plants. Genes Genom. 2016, 38, 723–731. [Google Scholar] [CrossRef]
- Quax, T.E.F.; Claassens, N.J.; Söll, D.; van der Oost, J. Codon Bias as a Means to Fine-Tune Gene Expression. Mol. Cell 2015, 59, 149–161. [Google Scholar] [CrossRef]
- Liu, Y. A code within the genetic code: Codon usage regulates co-translational protein folding. Cell. Commun. Signal. 2020, 18, 145. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, Q.; Zhao, F. Synonymous but Not Silent: The Codon Usage Code for Gene Expression and Protein Folding. Annu. Rev. Biochem. 2021, 90, 375–401. [Google Scholar] [CrossRef]
- Yang, C.; Zhao, Q.; Wang, Y.; Zhao, J.; Qiao, L.; Wu, B.; Yan, S.; Zheng, J.; Zheng, X. Comparative Analysis of Genomic and Transcriptome Sequences Reveals Divergent Patterns of Codon Bias in Wheat and Its Ancestor Species. Front. Genet. 2021, 12, 732432. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Gautam, T.; Singh, A.K.; Burma, P.K. Evaluating the effect of codon optimization on expression of bar gene in transgenic tobacco plants. J. Plant Biochem. Biotechnol. 2019, 28, 189–202. [Google Scholar] [CrossRef]
- Chen, J.; Ma, W.Q.; Hu, X.W.; Zhou, K.B. Synonymous Codon Usage Bias in the Chloroplast Genomes of 13 Oil-Tea Camellia Samples from South China. Forests 2023, 14, 794. [Google Scholar] [CrossRef]
- Song, Y.F.; Yang, Q.H.; Yi, X.G.; Zhu, Z.Q.; Wang, X.R.; Li, M. Comparative Analysis of Codon Usage Patterns in Chloroplast Genomes of Cherries. Forests 2022, 13, 1891. [Google Scholar] [CrossRef]
- Zhang, Y.; Shen, Z.; Meng, X.; Zhang, L.; Liu, Z.; Liu, M.; Zhang, F.; Zhao, J. Codon usage patterns across seven Rosales species. BMC Plant Biol. 2022, 22, 65. [Google Scholar] [CrossRef]
- Gao, D.; Sun, Z.; Bi, G.; Zhang, X. The complete plastome of Blidingia marginata and comparative analysis with the relative species in Ulvales. Aquat. Bot. 2022, 183, 103568. [Google Scholar] [CrossRef]
- Fang, J.; Zheng, L.; Liu, G.; Zhu, H. Comparative Analysis of Chloroplast Genomes in Cephaleuros and Its Related Genus (Trentepohlia): Insights into Adaptive Evolution. Genes 2024, 15, 839. [Google Scholar] [CrossRef]
- Fang, J.; Hu, Y.; Hu, Z. Comparative analysis of codon usage patterns in 16 chloroplast genomes of suborder Halimedineae. BMC Genom. 2024, 25, 945. [Google Scholar] [CrossRef]
- De Clerck, O.; Kao, S.M.; Bogaert, K.A.; Blomme, J.; Foflonker, F.; Kwantes, M.; Vancaester, E.; Vanderstraeten, L.; Aydogdu, E.; Boesger, J.; et al. Insights into the Evolution of Multicellularity from the Sea Lettuce Genome. Curr. Biol. 2018, 28, 2921–2933. [Google Scholar] [CrossRef] [PubMed]
- Osorio, H.; Tapia-Reyes, P.; Espinoza, D.; Laporte, D.; González, A.; Castro-Nallar, E.; Moenne, A. The Genome of the Marine Alga Ulva compressa (Chlorophyta) Reveals Protein-Coding Genes with Similarity to Plants and Green Microalgae, but Also to Animal, Bacterial, and Fungal Genes. Int. J. Mol. Sci. 2022, 23, 7279. [Google Scholar] [CrossRef]
- Powell, J.; Dion, K. Effects of Codon Usage on Gene Expression: Empirical Studies on Drosophila. J. Mol. Evol. 2015, 80, 219–226. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, T.; Ma, Z.; Ding, T.; Yang, Y.; Wang, F.; Wan, X.; Laing, F.; Chen, X.; Yao, H. Codon usage bias and phylogenetic analysis of chloroplast genome in 36 gracilariaceae species. Funct. Integr. Genom. 2024, 24, 45. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Luo, Y.; Sha, A.; Xiao, W.; Xiong, Z.; Chen, X.; He, J.; Peng, L.; Zou, L. Analysis of synonymous codon usage patterns in mitochondrial genomes of nine Amanita species. Front. Microbiol. 2023, 14, 1134228. [Google Scholar] [CrossRef]
- Begum, N.S.; Chakraborty, S. Influencing elements of codon usage bias in Birnaviridae and its evolutionary analysis. Virus Res. 2022, 310, 198672. [Google Scholar] [CrossRef]
- Wang, Z.J.; Xu, B.B.; Li, B.; Zhou, Q.Q.; Wang, G.Y.; Jiang, X.Z.; Wang, C.C.; Xu, Z.D. Comparative analysis of codon usage patterns in chloroplast genomes of six Euphorbiaceae species. PeerJ 2020, 8, e8251. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Uddin, A.; Das, S.; Chakraborty, S. Mutation pressure and natural selection on codon usage in chloroplast genes of two species in Pisum L. (Fabaceae: Faboideae). Mitochondrial DNA A 2019, 30, 664–673. [Google Scholar] [CrossRef]
- Yu, T.; Li, J.; Yang, Y.; Qi, L.; Chen, B.; Zhao, F.; Bao, Q.; Wu, J. Codon usage patterns and adaptive evolution of marine unicellular cyanobacteria Synechococcus and Prochlorococcus. Mol. Phylogen. Evol. 2012, 62, 206–213. [Google Scholar] [CrossRef]
- Rajneesh; Pathak, J.; Kannaujiya, V.K.; Singh, S.; Sinha, R. Codon usage analysis of photolyase encoding genes of cyanobacteria inhabiting diverse habitats. 3 Biotech 2017, 7, 192. [Google Scholar] [CrossRef]




| Species | Accession Number | A3% | T3% | C3% | G3% | GC% | GC1% | GC2% | GC3% |
|---|---|---|---|---|---|---|---|---|---|
| Ulva prolifera | MZ571508 | 42.41 | 49.25 | 5.10 | 3.23 | 24.60 | 35.46 | 30.00 | 8.33 |
| Ulva prolifera | KX342867 | 42.41 | 49.37 | 4.98 | 3.24 | 24.60 | 35.47 | 29.95 | 8.37 |
| Ulva prolifera | OP985131 | 42.21 | 49.48 | 5.17 | 3.13 | 24.75 | 35.87 | 30.07 | 8.30 |
| Ulva prolifera | OP985129 | 42.21 | 49.48 | 5.55 | 2.75 | 24.75 | 35.87 | 30.07 | 8.30 |
| Ulva linza | KX058323 | 42.83 | 48.50 | 5.39 | 3.28 | 26.13 | 38.09 | 31.63 | 8.67 |
| Ulva torta | MZ703011 | 42.46 | 50.08 | 4.88 | 2.58 | 24.79 | 36.45 | 30.47 | 7.46 |
| Ulva torta | OL684342 | 43.29 | 49.60 | 4.44 | 2.67 | 25.00 | 35.92 | 30.39 | 8.68 |
| Ulva californica | MZ561475 | 43.32 | 49.31 | 4.87 | 2.50 | 26.70 | 37.35 | 33.39 | 9.35 |
| Ulva aragoënsis | OP985132 | 42.88 | 49.96 | 4.53 | 2.64 | 24.24 | 35.78 | 29.77 | 7.16 |
| Ulva aragoënsis | KX579943 | 43.26 | 49.69 | 4.38 | 2.67 | 23.84 | 34.58 | 27.78 | 9.16 |
| Ulva gigantea | MT179350 | 42.79 | 49.06 | 5.40 | 2.75 | 26.59 | 39.28 | 30.77 | 9.73 |
| Ulva lactuca | MH730972 | 42.72 | 49.47 | 4.85 | 2.96 | 25.34 | 36.19 | 30.65 | 9.18 |
| Ulva lactuca | KT882614 | 42.93 | 49.34 | 4.81 | 2.91 | 23.91 | 34.75 | 29.25 | 7.73 |
| Ulva ohnoi | AP018696 | 42.66 | 49.54 | 4.82 | 2.98 | 24.37 | 35.55 | 29.74 | 7.80 |
| Ulva lacinulata | MW531676 | 43.03 | 49.14 | 4.94 | 2.90 | 24.81 | 36.17 | 30.42 | 7.84 |
| Ulva lacinulata | MW543061 | 42.78 | 49.48 | 4.85 | 2.89 | 24.18 | 35.23 | 29.58 | 7.74 |
| Ulva lacinulata | MN389525 | 42.75 | 49.46 | 4.88 | 2.91 | 24.20 | 35.25 | 29.56 | 7.79 |
| Ulva tepida | OL684341 | 43.09 | 49.46 | 4.71 | 2.74 | 23.79 | 34.67 | 29.26 | 7.45 |
| Ulva meridionalis | OP985133 | 42.60 | 49.63 | 4.94 | 2.84 | 24.28 | 35.40 | 29.67 | 7.77 |
| Ulva compressa | MW344287 | 42.12 | 48.06 | 6.06 | 3.75 | 25.22 | 36.08 | 29.77 | 9.81 |
| Ulva compressa | MW548841 | 43.08 | 47.85 | 5.84 | 3.23 | 25.91 | 37.56 | 31.11 | 9.07 |
| Ulva compressa | MW353781 | 42.33 | 48.65 | 5.50 | 3.52 | 24.72 | 35.56 | 29.58 | 9.02 |
| Ulva compressa | MK069584 | 42.56 | 47.94 | 6.37 | 3.13 | 28.95 | 42.77 | 34.58 | 9.50 |
| Ulva intestinalis | MZ158703 | 42.91 | 49.32 | 4.89 | 2.87 | 24.01 | 34.99 | 29.29 | 7.76 |
| Ulva rigida | MW543060 | 42.86 | 49.37 | 4.68 | 3.09 | 23.98 | 34.88 | 29.31 | 7.77 |
| Ulva rigida | MT179353 | 42.77 | 49.52 | 4.67 | 3.04 | 23.98 | 34.85 | 29.37 | 7.71 |
| Ulva fenestrata | MT179349 | 42.70 | 49.17 | 5.06 | 3.07 | 24.04 | 34.66 | 29.32 | 8.14 |
| Ulva australis | MN853875 | 42.90 | 49.38 | 4.77 | 2.94 | 24.05 | 35.11 | 29.34 | 7.71 |
| Ulva australis | MT179348 | 40.91 | 49.47 | 6.68 | 2.93 | 23.70 | 34.29 | 28.92 | 7.88 |
| Ulva australis | LC507117 | 43.28 | 48.64 | 5.06 | 3.02 | 25.58 | 35.51 | 33.14 | 8.08 |
| Species | Accession Number | Average ENC Value | Gene Number | |
|---|---|---|---|---|
| ENC < 35 | 35 ≤ ENC ≤ 61 | |||
| Ulva prolifera | MZ571508 | 32.22 | 41 | 3 |
| Ulva prolifera | KX342867 | 32.20 | 41 | 3 |
| Ulva prolifera | OP985131 | 32.11 | 38 | 4 |
| Ulva prolifera | OP985129 | 32.11 | 38 | 4 |
| Ulva linza | KX058323 | 32.01 | 40 | 4 |
| Ulva torta | MZ703011 | 31.43 | 44 | 0 |
| Ulva torta | OL684342 | 31.56 | 44 | 0 |
| Ulva californica | MZ561475 | 31.40 | 42 | 0 |
| Ulva aragoënsis | OP985132 | 31.58 | 43 | 1 |
| Ulva aragoënsis | KX579943 | 31.62 | 43 | 1 |
| Ulva gigantea | MT179350 | 31.84 | 40 | 2 |
| Ulva lactuca | MH730972 | 31.91 | 41 | 4 |
| Ulva lactuca | KT882614 | 32.02 | 39 | 4 |
| Ulva ohnoi | AP018696 | 31.83 | 43 | 3 |
| Ulva lacinulata | MW531676 | 31.94 | 41 | 3 |
| Ulva lacinulata | MW543061 | 31.95 | 42 | 3 |
| Ulva lacinulata | MN389525 | 31.94 | 42 | 3 |
| Ulva tepida | OL684341 | 31.84 | 43 | 1 |
| Ulva meridionalis | OP985133 | 32.10 | 40 | 2 |
| Ulva compressa | MW344287 | 32.74 | 36 | 8 |
| Ulva compressa | MW548841 | 32.57 | 34 | 7 |
| Ulva compressa | MW353781 | 32.76 | 37 | 7 |
| Ulva compressa | MK069584 | 32.71 | 33 | 8 |
| Ulva intestinalis | MZ158703 | 31.78 | 41 | 3 |
| Ulva rigida | MW543060 | 32.22 | 39 | 5 |
| Ulva rigida | MT179353 | 32.11 | 36 | 4 |
| Ulva fenestrata | MT179349 | 32.19 | 37 | 3 |
| Ulva australis | MN853875 | 31.81 | 42 | 2 |
| Ulva australis | MT179348 | 31.43 | 37 | 0 |
| Ulva australis | LC507117 | 31.80 | 44 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, J.; Qin, L.; Liu, H.; Hu, Z. Analysis of Codon Usage Bias of 30 Chloroplast Genomes in Ulva (Ulvophyceae, Chlorophyta). Genes 2025, 16, 608. https://doi.org/10.3390/genes16050608
Fang J, Qin L, Liu H, Hu Z. Analysis of Codon Usage Bias of 30 Chloroplast Genomes in Ulva (Ulvophyceae, Chlorophyta). Genes. 2025; 16(5):608. https://doi.org/10.3390/genes16050608
Chicago/Turabian StyleFang, Jiao, Liming Qin, Hongni Liu, and Zhangfeng Hu. 2025. "Analysis of Codon Usage Bias of 30 Chloroplast Genomes in Ulva (Ulvophyceae, Chlorophyta)" Genes 16, no. 5: 608. https://doi.org/10.3390/genes16050608
APA StyleFang, J., Qin, L., Liu, H., & Hu, Z. (2025). Analysis of Codon Usage Bias of 30 Chloroplast Genomes in Ulva (Ulvophyceae, Chlorophyta). Genes, 16(5), 608. https://doi.org/10.3390/genes16050608





