Comparative Analysis of the Genetic Composition of Minorities in the Carpathian Basin Through Genome-Wide Autosomal Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets
2.2. Inferring Population Structure and Ancestry
2.3. DNA Segment Analyses
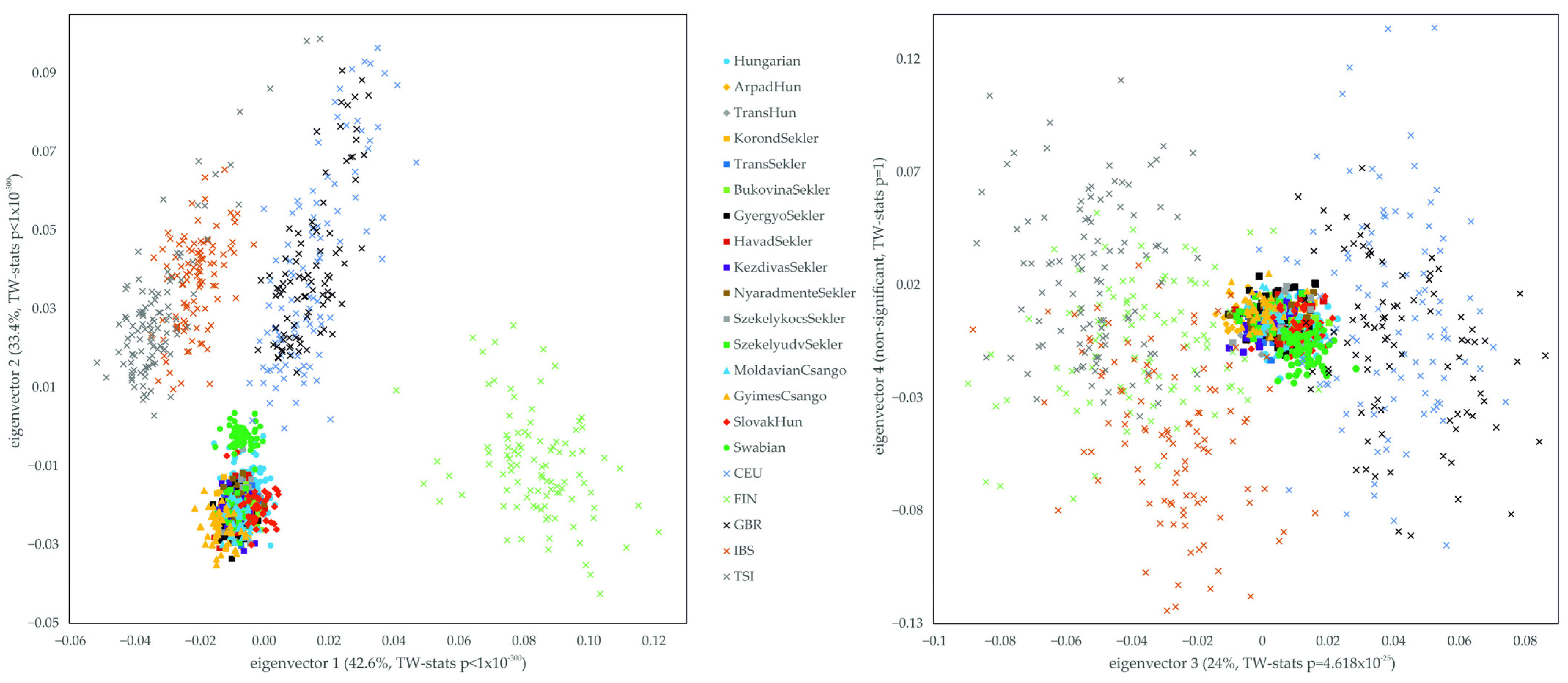
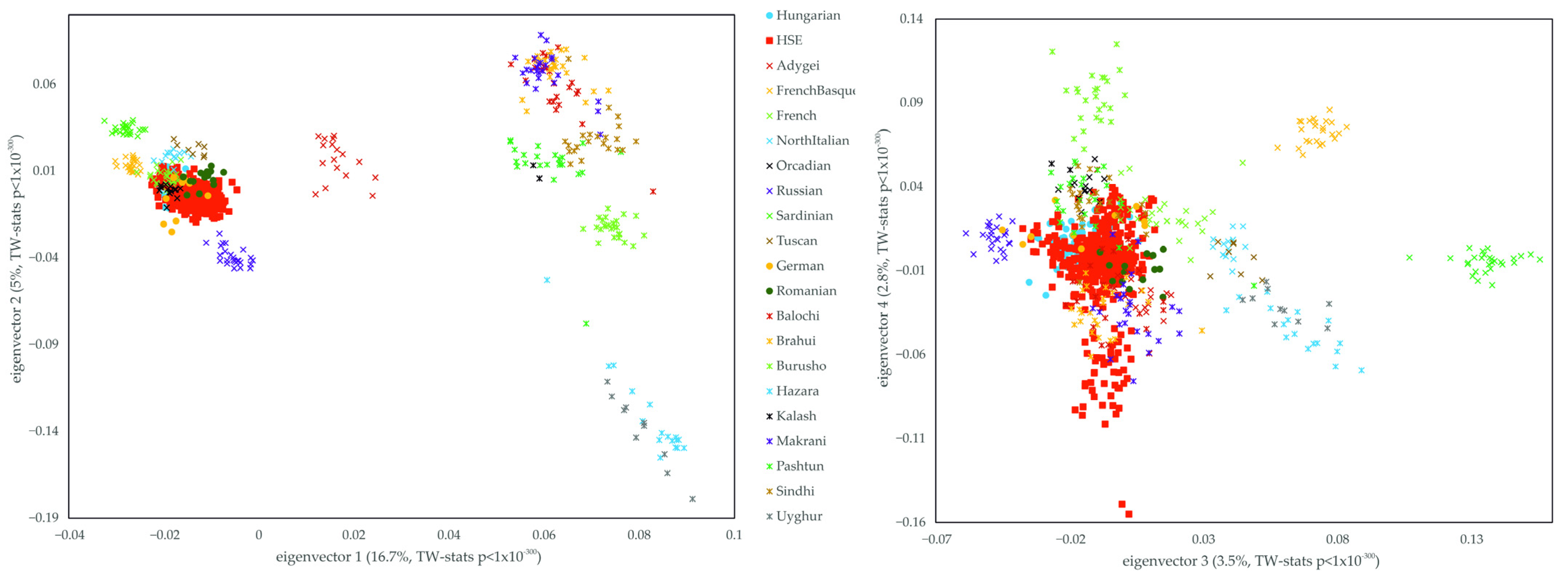
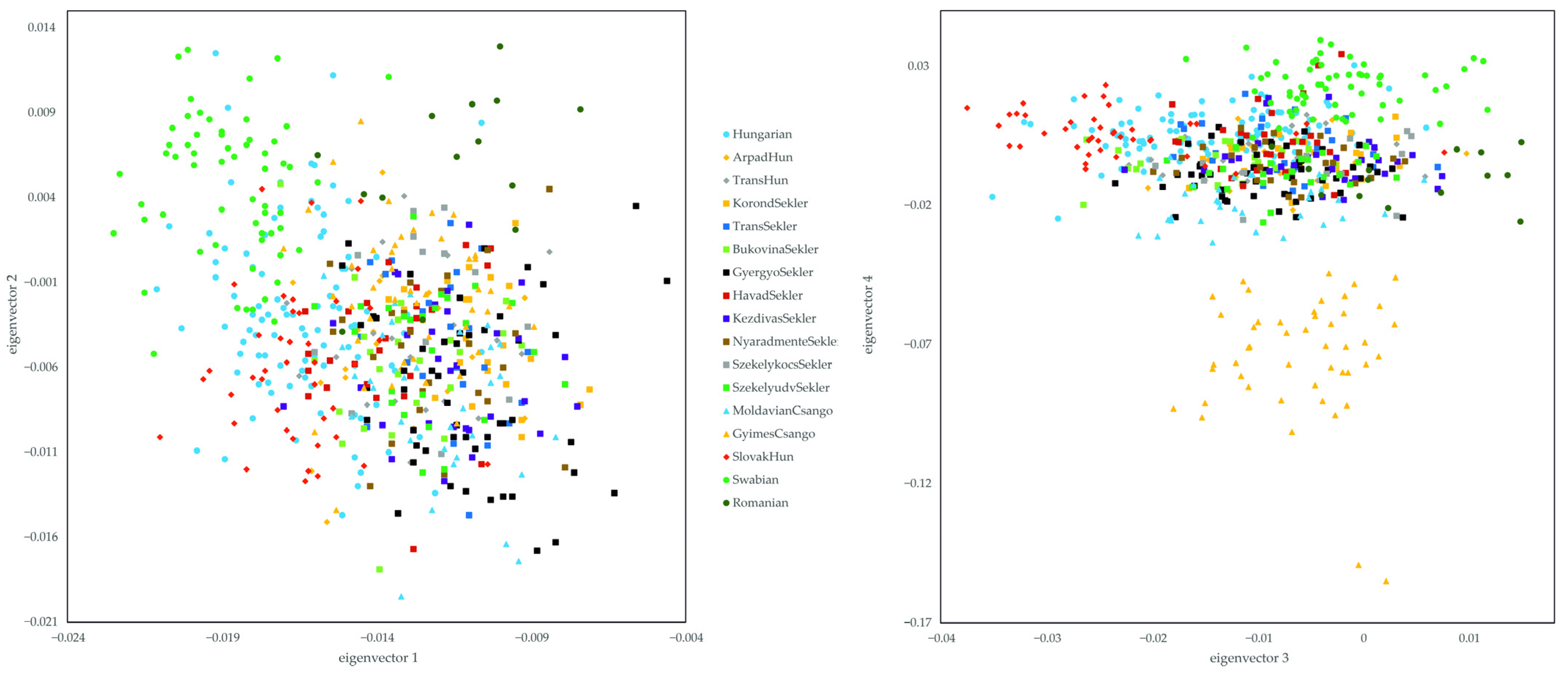
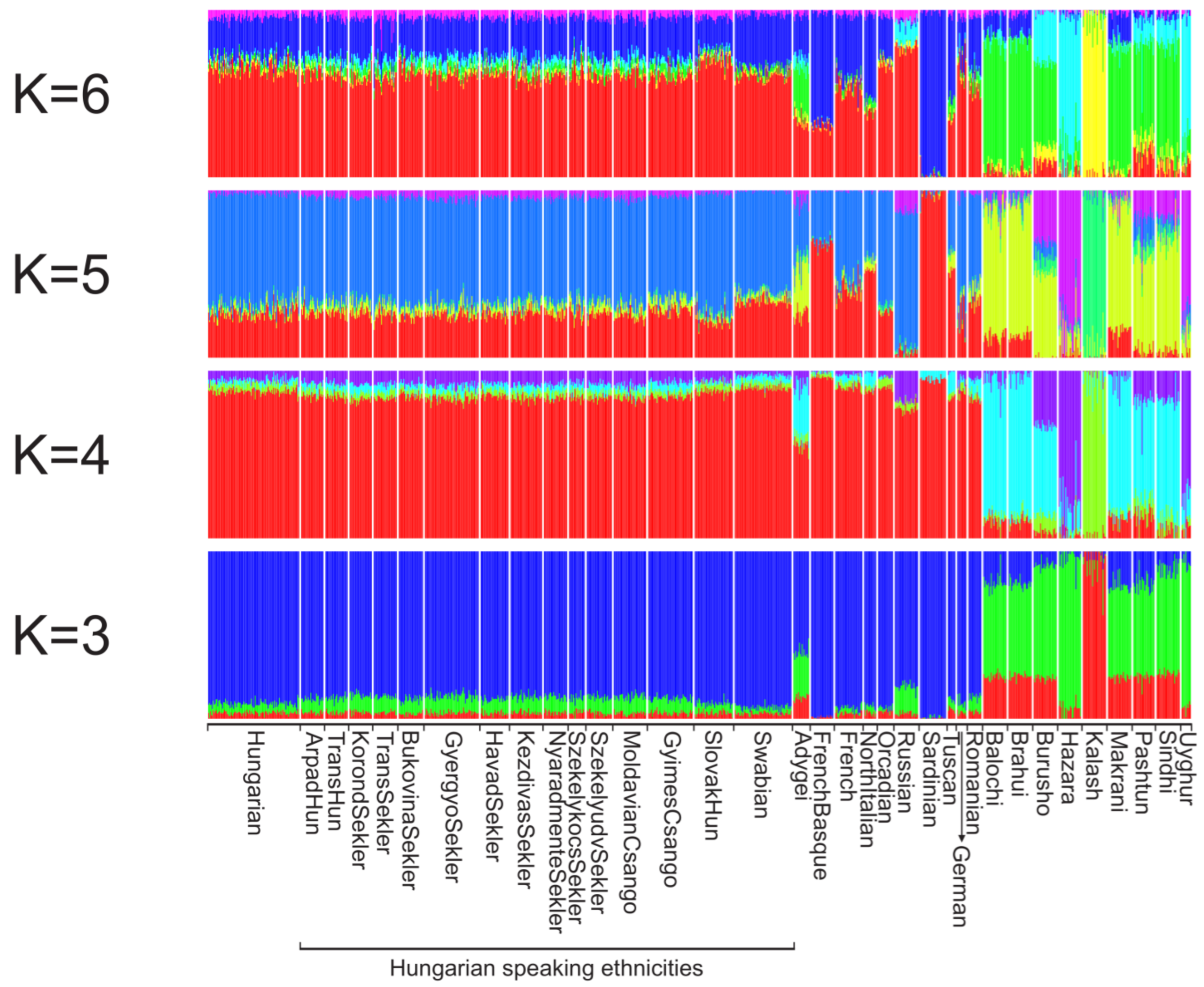
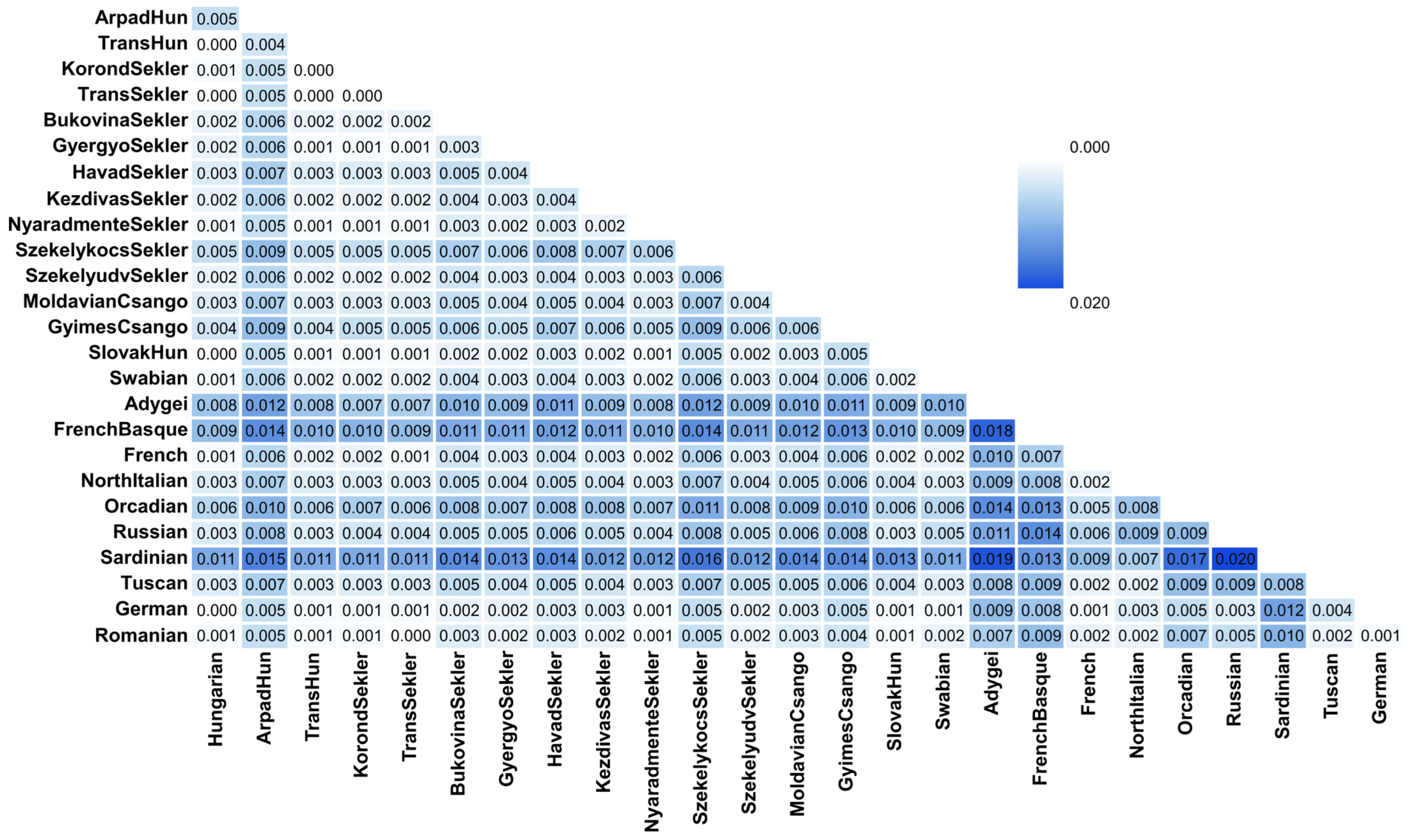
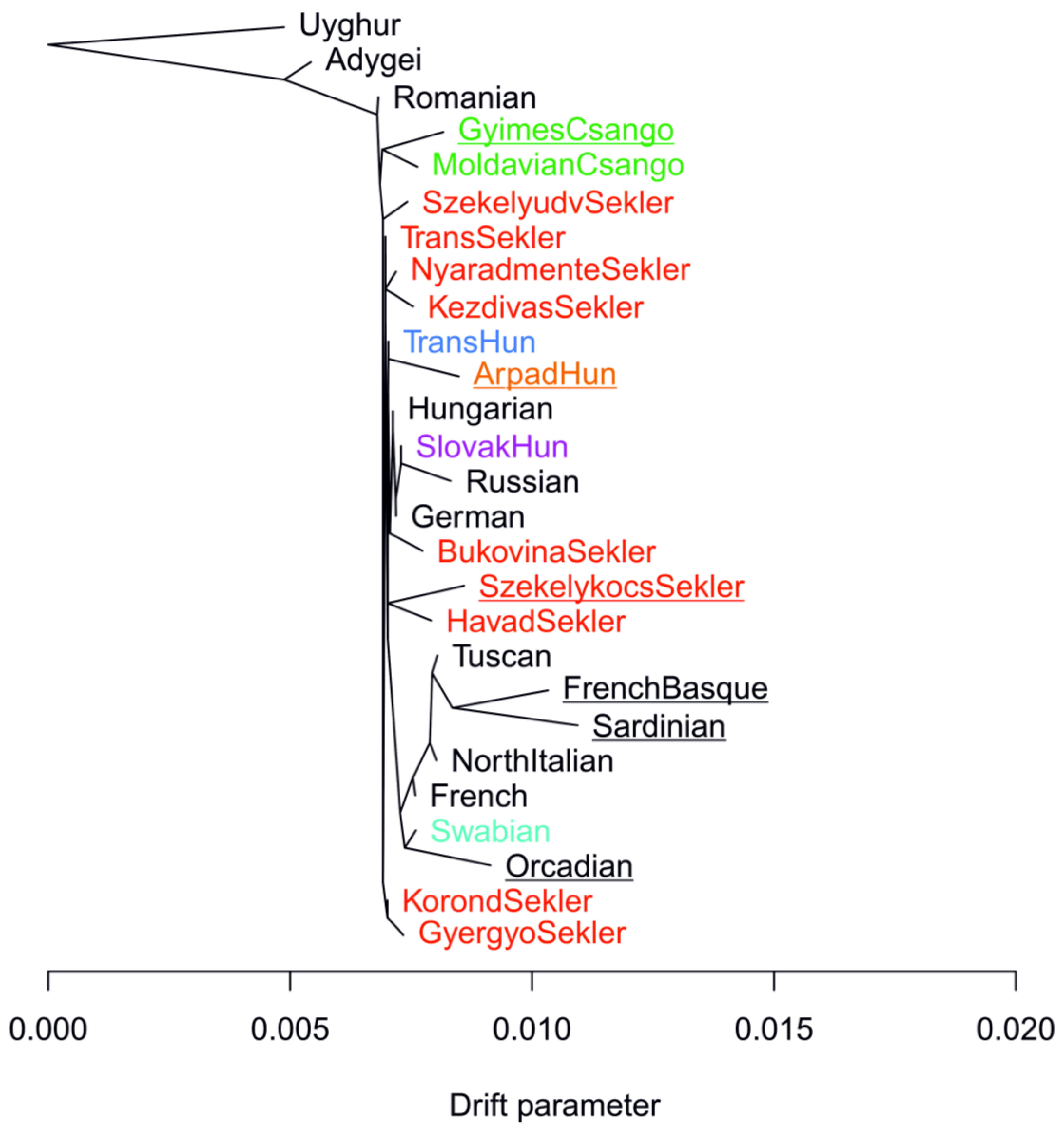
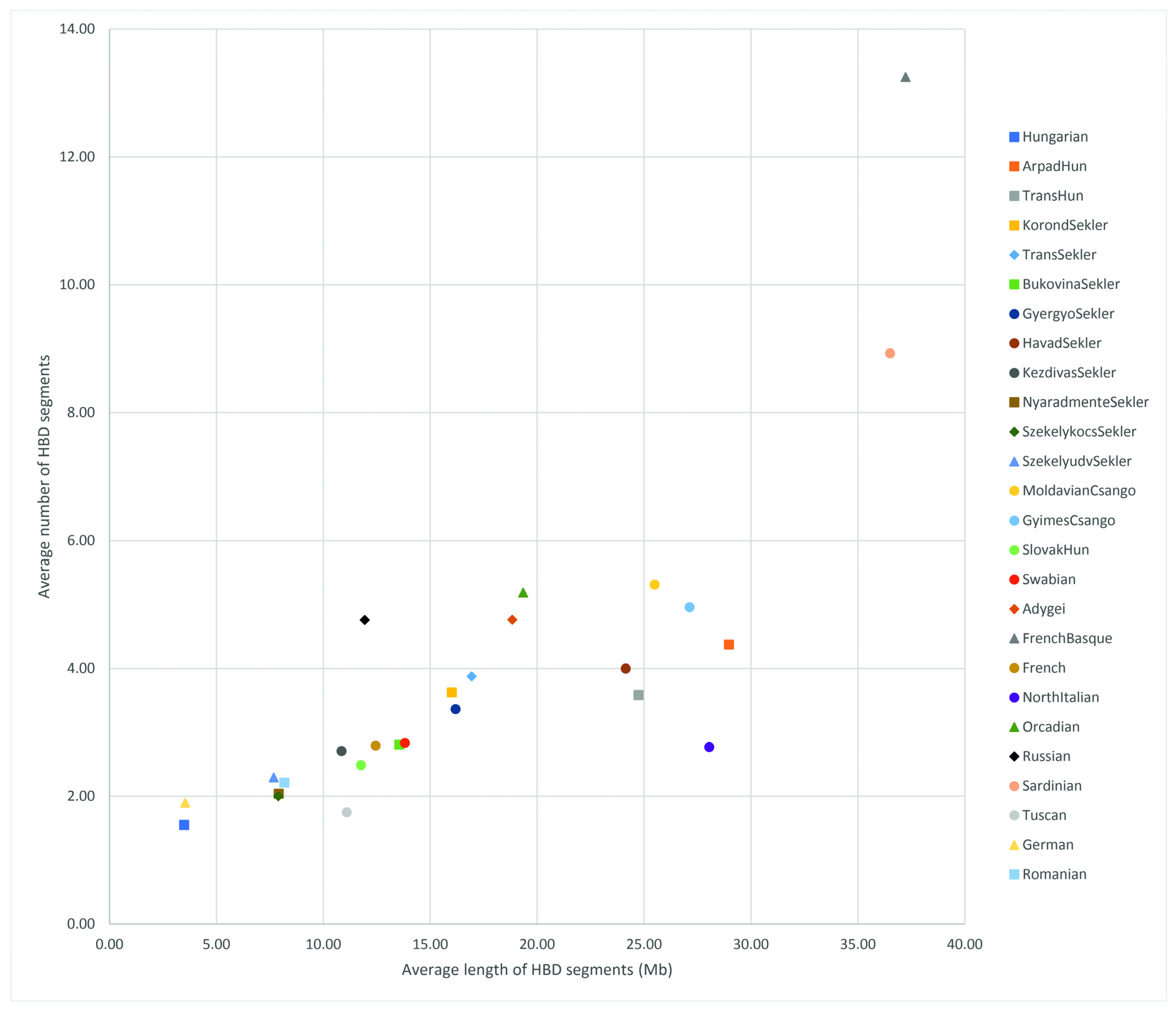
3. Results
3.1. Relationship of the Hungarian, Slovakia Living Hungarian, Hungary Living Swabian and Transylvanian Samples to European Populations
3.2. Haplotype-Based Analysis Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HWE | Hardy–Weinberg Equilibrium |
| MAF | Minor Allele Frequency |
| HSE | Hungarian-speaking Ethnicities |
| HapMap | International Haplotype Mapping Project |
| HGDP | Human Genome Diversity Project |
| 1KGP | 1000 Genomes Project |
| CEU | HGDP Central European Americans |
| FIN | HGDP Finnish |
| GBR | HGDP British |
| IBS | HGDP Iberian Peninsula |
| TSI | HGDP Tuscans |
| PCA | Principal Component Analysis |
| FST | Fixation Index or Pairwise Average Allele Frequency Differentiation |
| ML | Maximum Likelihood Estimation |
| CV | Cross-Validation |
| SNP | Single Nucleotide Polymorphism |
| IBD | Identity By Descent |
| HBD | Homozigosity By Descent |
| VCF | Variant Call Format |
References
- Moores, E.M.; Fairbridge, R.W. (Eds.) Encyclopedia of European and Asian Regional Geology; Encyclopedia of Earth Sciences Series; Springer: Houston, TX, USA, 1998. [Google Scholar]
- Jordan, P.A.; Adamič, M.O.; Woodman, P. (Eds.) Exonyms and the International Standardisation of Geographical Names: Approaches Towards the Resolution of an Apparent Contradiction; LIT Verlag: Wien, Austria, 2007. [Google Scholar]
- Hoffmann, G.W. (Ed.) A Geography of Europe: Problems and Prospects; John Wiley & Sons Inc.: Hoboken, NJ, USA, 1983. [Google Scholar]
- Róna-Tas, A. Hungarians and Europe in the Early Middle Ages: An Introduction to Early Hungarian History; Central European University Press: Budapest, Hungary, 1999. [Google Scholar]
- Walter, P. Conceptions of Ethnicity in Early Medieval Studies; Austrian Academy of Sciences Press: Vienna, Austria, 1998. [Google Scholar]
- Curta, F. The Making of the Slavs: History and Archaeology of the Lower Danube Region; Cambridge University Press: New York, NY, USA, 2008. [Google Scholar]
- Hungarian Central Statistical Office. Population Census 2011 in Hungary. Available online: https://www.ksh.hu/docs/eng/xftp/idoszaki/nepsz2011/enepszelo2011.pdf (accessed on 24 October 2024).
- Statistical Office of the Republic of Serbia. 2011 Census of Population, Households and Dwellings in the Republic of Serbia. Available online: https://publikacije.stat.gov.rs/G2014/PdfE/G20144012.pdf (accessed on 24 October 2024).
- The State Statistics Service of Ukraine. The All-Ukrainian Population Census 2001. Available online: https://km.ukrstat.gov.ua/ukr/statinf/dem/nas_r.htm (accessed on 24 October 2024).
- Croatian Bureau of Statistics. 2011 Census of Population, Households, and Dwellings in Croatia. Available online: https://web.dzs.hr/eng/censuses/census2011/results/censustabshtm.htm (accessed on 24 October 2024).
- Austrian Federal Statistical Office. Census 2011 in Austria. Available online: https://www.statistik.at/fileadmin/publications/Census_2011_-_Austria.pdf (accessed on 24 October 2024).
- Statistical Office of the Slovak Republic. Census of Population, Dwellings and Apartments 2011 in the Slovak Republic. Available online: https://susr.statistics.sk/wps/wcm/connect/d79eb13d-fe8b-4939-8c1b-ca2a45e3f47e/Table_9_Resident_Population_by_nationality_by_regions_2001_2011_Census.pdf?MOD=AJPERES&CVID=kojGKwI&CVID=kojGKwI&CVID=kojGKwI&CVID=kojGKwI&CVID=kojGKwI&CVID=kojGKwI (accessed on 28 October 2024).
- Statistical Office of the Republic of Slovenia. 2011 Census of Population, Households, and Dwellings in the Republic of Slovenia. Available online: https://www.stat.si/doc/pub/Ljudje_druzine_stanovanja.pdf (accessed on 24 October 2024).
- National Institute of Statistics of Romania. 2011 Census of Population and Dwellings in Romania. Available online: https://view.officeapps.live.com/op/view.aspx?src=https%3A%2F%2Fwww.recensamantromania.ro%2Fwp-content%2Fuploads%2F2021%2F11%2Fvol2_t1.xls&wdOrigin=BROWSELINK (accessed on 24 October 2024).
- Balaresque, P.; Bowden, G.R.; Adams, S.M.; Leung, H.Y.; King, T.E.; Rosser, Z.H.; Goodwin, J.; Moisan, J.P.; Richard, C.; Millward, A.; et al. A predominantly neolithic origin for European paternal lineages. PLoS Biol. 2010, 8, e1000285. [Google Scholar] [CrossRef] [PubMed]
- Cruciani, F.; Trombetta, B.; Antonelli, C.; Pascone, R.; Valesini, G.; Scalzi, V.; Vona, G.; Melegh, B.; Zagradisnik, B.; Assum, G.; et al. Strong intra- and inter-continental differentiation revealed by Y chromosome SNPs M269, U106 and U152. Forensic Sci. Int. Genet. 2011, 5, e49–e52. [Google Scholar] [CrossRef] [PubMed]
- Myres, N.M.; Rootsi, S.; Lin, A.A.; Jarve, M.; King, R.J.; Kutuev, I.; Cabrera, V.M.; Khusnutdinova, E.K.; Pshenichnov, A.; Yunusbayev, B.; et al. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur. J. Hum. Genet. 2011, 19, 95–101. [Google Scholar] [CrossRef]
- Rocca, R.A.; Magoon, G.; Reynolds, D.F.; Krahn, T.; Tilroe, V.O.; Op den Velde Boots, P.M.; Grierson, A.J. Discovery of Western European R1b1a2 Y chromosome variants in 1000 genomes project data: An online community approach. PLoS ONE 2012, 7, e41634. [Google Scholar] [CrossRef]
- Underhill, P.A.; Poznik, G.D.; Rootsi, S.; Jarve, M.; Lin, A.A.; Wang, J.; Passarelli, B.; Kanbar, J.; Myres, N.M.; King, R.J.; et al. The phylogenetic and geographic structure of Y-chromosome haplogroup R1a. Eur. J. Hum. Genet. 2015, 23, 124–131. [Google Scholar] [CrossRef]
- Kayser, M.; Lao, O.; Anslinger, K.; Augustin, C.; Bargel, G.; Edelmann, J.; Elias, S.; Heinrich, M.; Henke, J.; Henke, L.; et al. Significant genetic differentiation between Poland and Germany follows present-day political borders, as revealed by Y-chromosome analysis. Hum. Genet. 2005, 117, 428–443. [Google Scholar] [CrossRef]
- Battaglia, V.; Fornarino, S.; Al-Zahery, N.; Olivieri, A.; Pala, M.; Myres, N.M.; King, R.J.; Rootsi, S.; Marjanovic, D.; Primorac, D.; et al. Y-chromosomal evidence of the cultural diffusion of agriculture in Southeast Europe. Eur. J. Hum. Genet. 2009, 17, 820–830. [Google Scholar] [CrossRef]
- Cruciani, F.; La Fratta, R.; Santolamazza, P.; Sellitto, D.; Pascone, R.; Moral, P.; Watson, E.; Guida, V.; Colomb, E.B.; Zaharova, B.; et al. Phylogeographic analysis of haplogroup E3b (E-M215) y chromosomes reveals multiple migratory events within and out of Africa. Am. J. Hum. Genet. 2004, 74, 1014–1022. [Google Scholar] [CrossRef]
- Semino, O.; Magri, C.; Benuzzi, G.; Lin, A.A.; Al-Zahery, N.; Battaglia, V.; Maccioni, L.; Triantaphyllidis, C.; Shen, P.; Oefner, P.J.; et al. Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: Inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am. J. Hum. Genet. 2004, 74, 1023–1034. [Google Scholar] [CrossRef]
- Torroni, A.; Richards, M.; Macaulay, V.; Forster, P.; Villems, R.; Norby, S.; Savontaus, M.L.; Huoponen, K.; Scozzari, R.; Bandelt, H.J. mtDNA haplogroups and frequency patterns in Europe. Am. J. Hum. Genet. 2000, 66, 1173–1177. [Google Scholar] [CrossRef]
- Pereira, L.; Richards, M.; Goios, A.; Alonso, A.; Albarran, C.; Garcia, O.; Behar, D.M.; Golge, M.; Hatina, J.; Al-Gazali, L.; et al. High-resolution mtDNA evidence for the late-glacial resettlement of Europe from an Iberian refugium. Genome Res. 2005, 15, 19–24. [Google Scholar] [CrossRef]
- Achilli, A.; Rengo, C.; Magri, C.; Battaglia, V.; Olivieri, A.; Scozzari, R.; Cruciani, F.; Zeviani, M.; Briem, E.; Carelli, V.; et al. The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am. J. Hum. Genet. 2004, 75, 910–918. [Google Scholar] [CrossRef]
- Guglielmino, C.R.; De Silvestri, A.; Beres, J. Probable ancestors of Hungarian ethnic groups: An admixture analysis. Ann. Hum. Genet. 2000, 64, 145–159. [Google Scholar] [CrossRef]
- Biro, A.; Feher, T.; Barany, G.; Pamjav, H. Testing Central and Inner Asian admixture among contemporary Hungarians. Forensic Sci. Int. Genet. 2015, 15, 121–126. [Google Scholar] [CrossRef]
- Hellenthal, G.; Busby, G.B.J.; Band, G.; Wilson, J.F.; Capelli, C.; Falush, D.; Myers, S. A genetic atlas of human admixture history. Science 2014, 343, 747–751. [Google Scholar] [CrossRef]
- Brandstatter, A.; Egyed, B.; Zimmermann, B.; Duftner, N.; Padar, Z.; Parson, W. Migration rates and genetic structure of two Hungarian ethnic groups in Transylvania, Romania. Ann. Hum. Genet. 2007, 71, 791–803. [Google Scholar] [CrossRef]
- Adam, V.; Banfai, Z.; Maasz, A.; Sumegi, K.; Miseta, A.; Melegh, B. Investigating the genetic characteristics of the Csangos, a traditionally Hungarian speaking ethnic group residing in Romania. J. Hum. Genet. 2020, 65, 1093–1103. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- International HapMap, C.; Frazer, K.A.; Ballinger, D.G.; Cox, D.R.; Hinds, D.A.; Stuve, L.L.; Gibbs, R.A.; Belmont, J.W.; Boudreau, A.; Hardenbol, P.; et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 2007, 449, 851–861. [Google Scholar] [CrossRef]
- Cavalli-Sforza, L.L. The Human Genome Diversity Project: Past, present and future. Nat. Rev. Genet. 2005, 6, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Cann, H.M.; de Toma, C.; Cazes, L.; Legrand, M.F.; Morel, V.; Piouffre, L.; Bodmer, J.; Bodmer, W.F.; Bonne-Tamir, B.; Cambon-Thomsen, A.; et al. A human genome diversity cell line panel. Science 2002, 296, 261–262. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.A.; Mahajan, S.; Ramachandran, S.; Zhao, C.; Pritchard, J.K.; Feldman, M.W. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 2005, 1, e70. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.A.; Pritchard, J.K.; Weber, J.L.; Cann, H.M.; Kidd, K.K.; Zhivotovsky, L.A.; Feldman, M.W. Genetic structure of human populations. Science 2002, 298, 2381–2385. [Google Scholar] [CrossRef]
- Bergstrom, A.; McCarthy, S.A.; Hui, R.; Almarri, M.A.; Ayub, Q.; Danecek, P.; Chen, Y.; Felkel, S.; Hallast, P.; Kamm, J.; et al. Insights into human genetic variation and population history from 929 diverse genomes. Science 2020, 367, eaay5012. [Google Scholar] [CrossRef]
- Yunusbayev, B.; Metspalu, M.; Metspalu, E.; Valeev, A.; Litvinov, S.; Valiev, R.; Akhmetova, V.; Balanovska, E.; Balanovsky, O.; Turdikulova, S.; et al. The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia. PLoS Genet. 2015, 11, e1005068. [Google Scholar] [CrossRef]
- Behar, D.M.; Metspalu, M.; Baran, Y.; Kopelman, N.M.; Yunusbayev, B.; Gladstein, A.; Tzur, S.; Sahakyan, H.; Bahmanimehr, A.; Yepiskoposyan, L.; et al. No evidence from genome-wide data of a Khazar origin for the Ashkenazi Jews. Hum. Biol. 2013, 85, 859–900. [Google Scholar] [CrossRef]
- Genomes Project, C.; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Patterson, N.; Price, A.L.; Reich, D. Population structure and eigenanalysis. PLoS Genet. 2006, 2, e190. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Browning, B.L.; Browning, S.R. A fast, powerful method for detecting identity by descent. Am. J. Hum. Genet. 2011, 88, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S. PLINK/SEQ: A Library for the Analysis of Genetic Variation Data. Available online: https://zzz.bwh.harvard.edu/plinkseq/start-pseq.shtml (accessed on 24 October 2024).
- Browning, B.L.; Browning, S.R. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 2013, 194, 459–471. [Google Scholar] [CrossRef]
- Atzmon, G.; Hao, L.; Pe’er, I.; Velez, C.; Pearlman, A.; Palamara, P.F.; Morrow, B.; Friedman, E.; Oddoux, C.; Burns, E.; et al. Abraham’s children in the genome era: Major Jewish diaspora populations comprise distinct genetic clusters with shared Middle Eastern Ancestry. Am. J. Hum. Genet. 2010, 86, 850–859. [Google Scholar] [CrossRef]
- Green, R.E.; Krause, J.; Briggs, A.W.; Maricic, T.; Stenzel, U.; Kircher, M.; Patterson, N.; Li, H.; Zhai, W.; Fritz, M.H.; et al. A draft sequence of the Neandertal genome. Science 2010, 328, 710–722. [Google Scholar] [CrossRef]
- Patterson, N.; Moorjani, P.; Luo, Y.; Mallick, S.; Rohland, N.; Zhan, Y.; Genschoreck, T.; Webster, T.; Reich, D. Ancient admixture in human history. Genetics 2012, 192, 1065–1093. [Google Scholar] [CrossRef]
- Hermann, G.M. Székely Történeti Kistükör 1848-ig [Small Mirror of Székely History Until 1848]; Litera könyvkiadó: Székelyudvarhely, Romania, 2004. [Google Scholar]
- Eberhardt, P. Ethnic Groups and Population Changes in Twentieth Century Eastern Europe; Routledge: London, UK, 2003. [Google Scholar]
- Benkő, E. The Szeklers in the Middle Ages. In Archaeology of the Middle Ages and the Early Modern Period in Hungary; Hungarian Academy of Sciences, Institute of Archaeology: Budapest, Hungary, 2010. [Google Scholar]



| W Population 1 | X Population 2 | Y Population 3 | Z Population 4 | D-Statistics | Z-Score |
|---|---|---|---|---|---|
| Yoruba | Hungarian | Romanian | ArpadHun | 0.000303 | 2.316 |
| Yoruba | Hungarian | Romanian | BukovinaSekler | 0.000262 | 2.122 |
| Yoruba | Hungarian | Romanian | HavadSekler | 0.000266 | 2.165 |
| Yoruba | MoldavianCsango | Romanian | ArpadHun | 0.000323 | 2.445 |
| Yoruba | MoldavianCsango | Romanian | GyimesCsango | 0.000278 | 2.405 |
| Yoruba | Hungarian | German | Swabian | 0.000073 | 0.609 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szabó, A.; Sümegi, K.; Bánfai, Z.; Hadzsiev, K.; Gallyas, F.; Miseta, A.; Kásler, M.; Melegh, B. Comparative Analysis of the Genetic Composition of Minorities in the Carpathian Basin Through Genome-Wide Autosomal Data. Genes 2025, 16, 607. https://doi.org/10.3390/genes16050607
Szabó A, Sümegi K, Bánfai Z, Hadzsiev K, Gallyas F, Miseta A, Kásler M, Melegh B. Comparative Analysis of the Genetic Composition of Minorities in the Carpathian Basin Through Genome-Wide Autosomal Data. Genes. 2025; 16(5):607. https://doi.org/10.3390/genes16050607
Chicago/Turabian StyleSzabó, András, Katalin Sümegi, Zsolt Bánfai, Kinga Hadzsiev, Ferenc Gallyas, Attila Miseta, Miklós Kásler, and Béla Melegh. 2025. "Comparative Analysis of the Genetic Composition of Minorities in the Carpathian Basin Through Genome-Wide Autosomal Data" Genes 16, no. 5: 607. https://doi.org/10.3390/genes16050607
APA StyleSzabó, A., Sümegi, K., Bánfai, Z., Hadzsiev, K., Gallyas, F., Miseta, A., Kásler, M., & Melegh, B. (2025). Comparative Analysis of the Genetic Composition of Minorities in the Carpathian Basin Through Genome-Wide Autosomal Data. Genes, 16(5), 607. https://doi.org/10.3390/genes16050607






