Genome-Wide Identification of the Polygalacturonase Gene Family and Its Potential Association with Abscission Zone in Capsicum annuum L.
Abstract
1. Introduction
2. Results
2.1. Identification and Characteristics of CaPGs
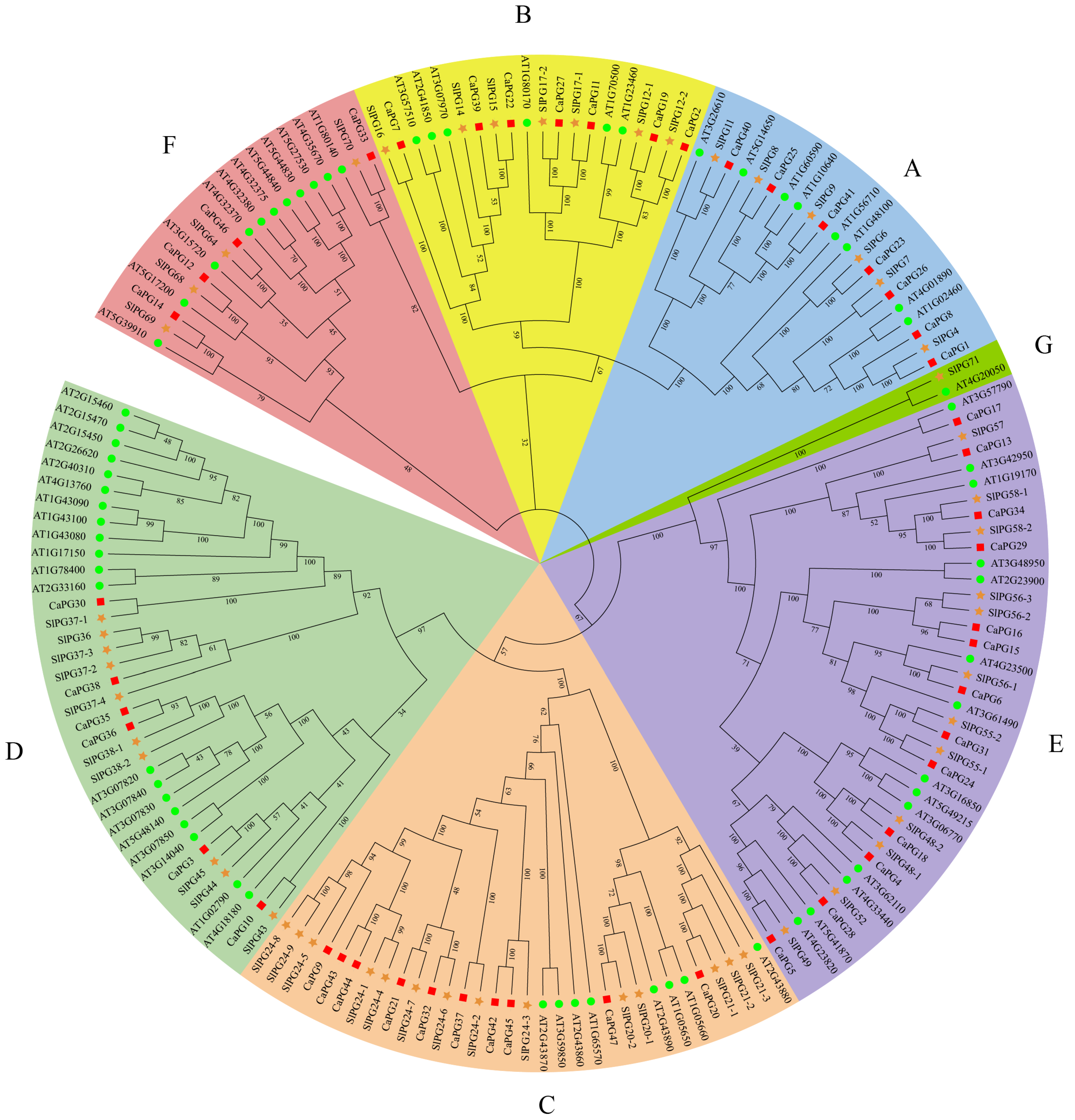
2.2. Phylogenetic Analysis and Classification of CaPGs
2.3. Distribution of CaPG Genes Across Chromosomes
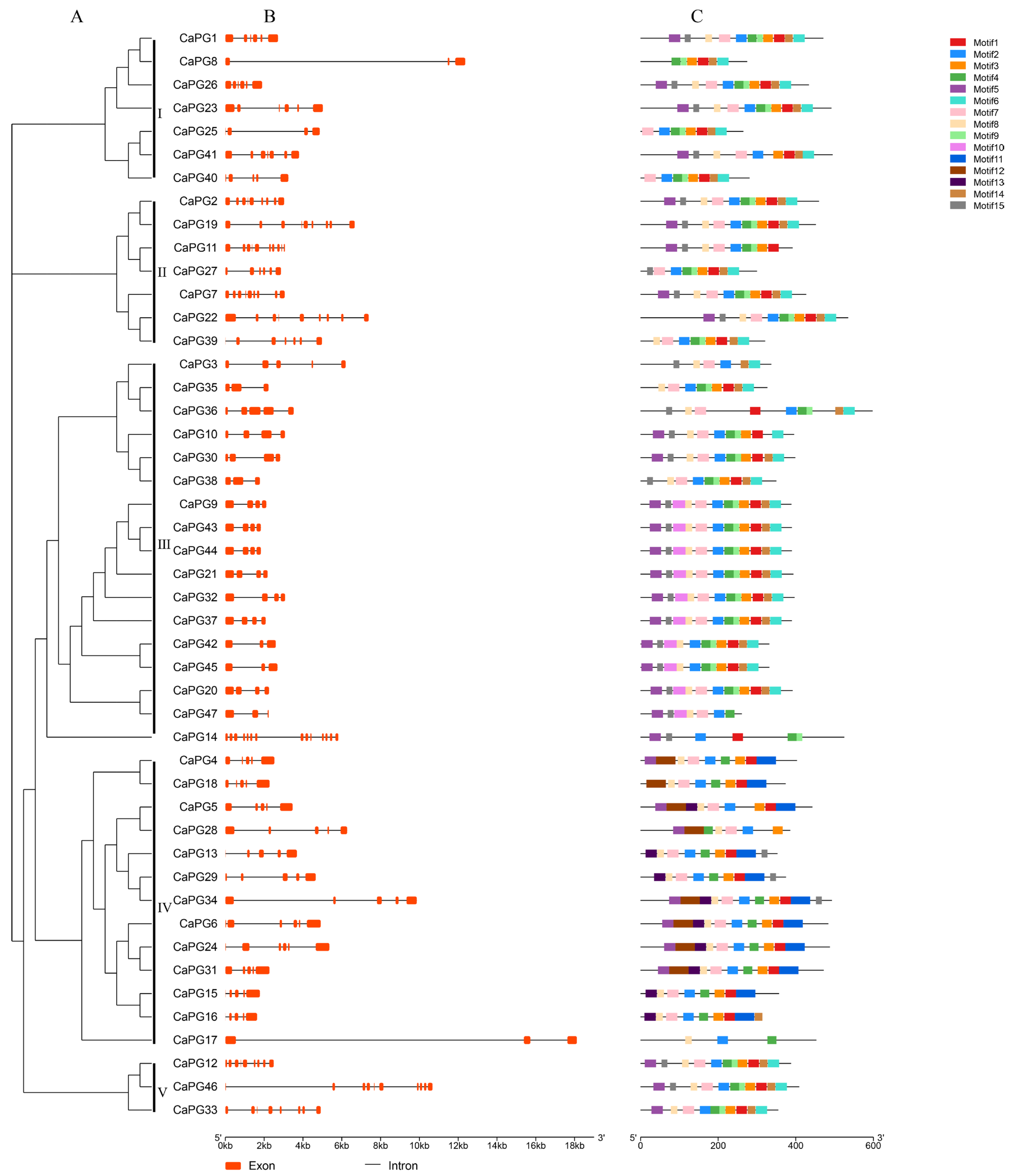
2.4. Gene Structure and Conserved Motif Composition
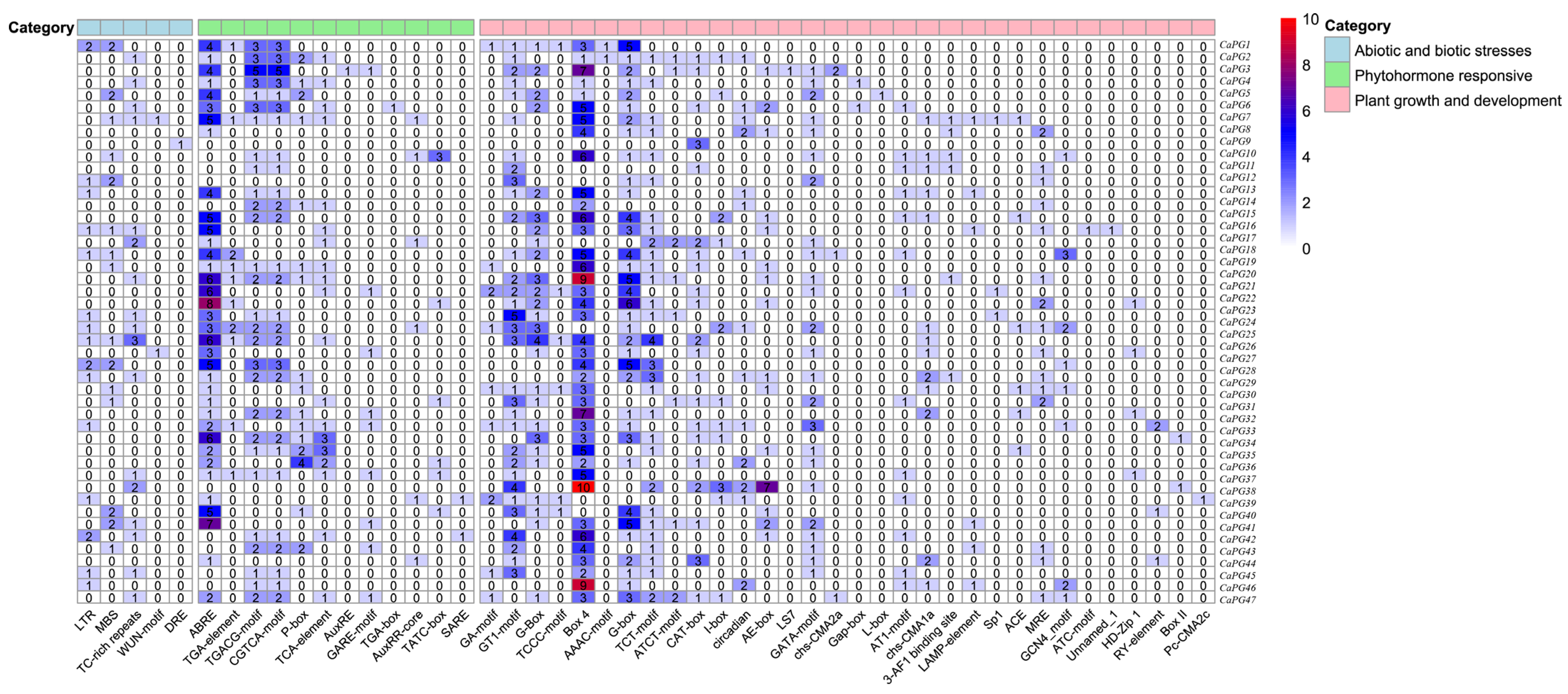
2.5. Analysis of Cis-Acting Elements
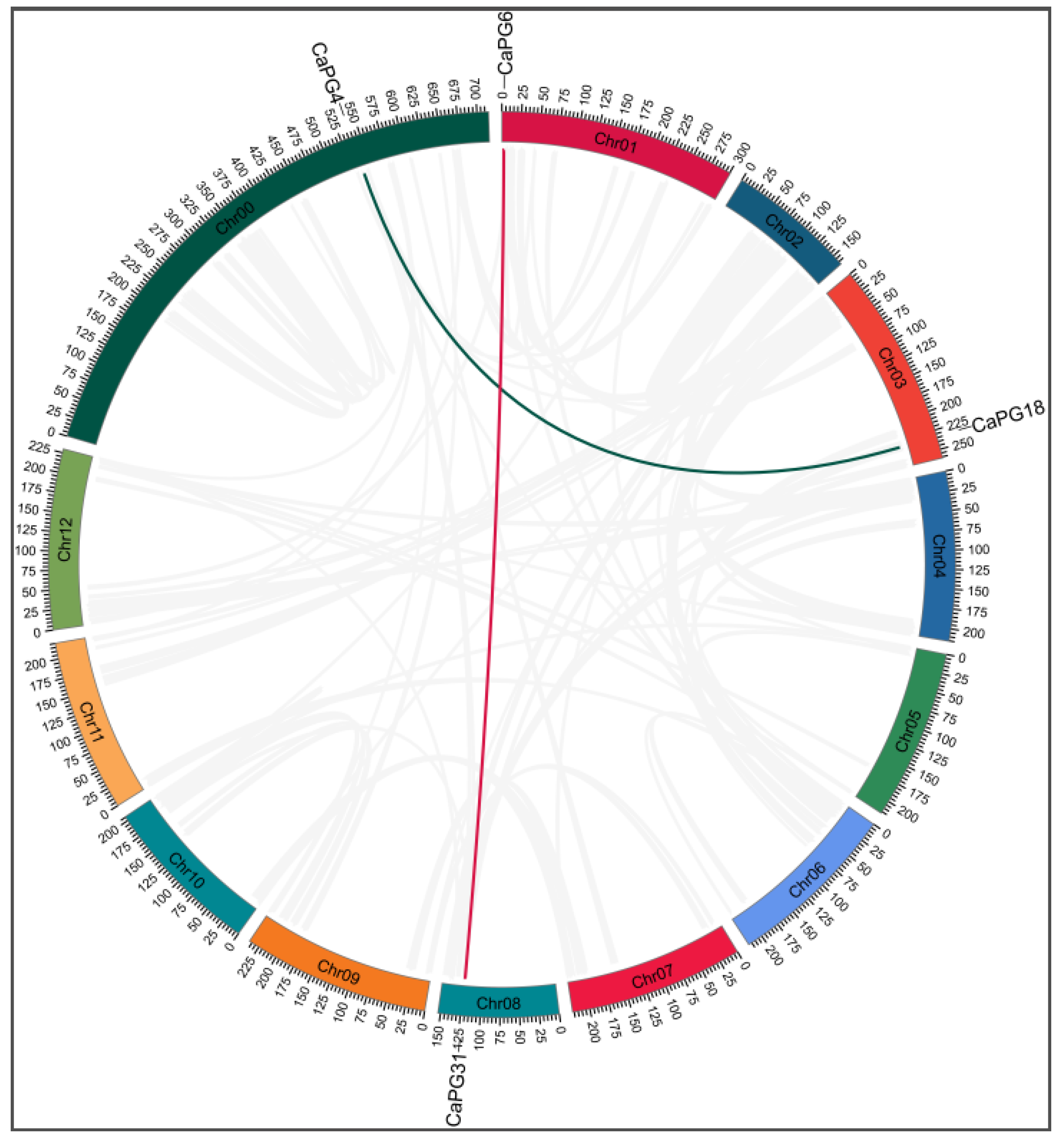
2.6. Intraspecific Collinearity Analysis of CaPG Genes
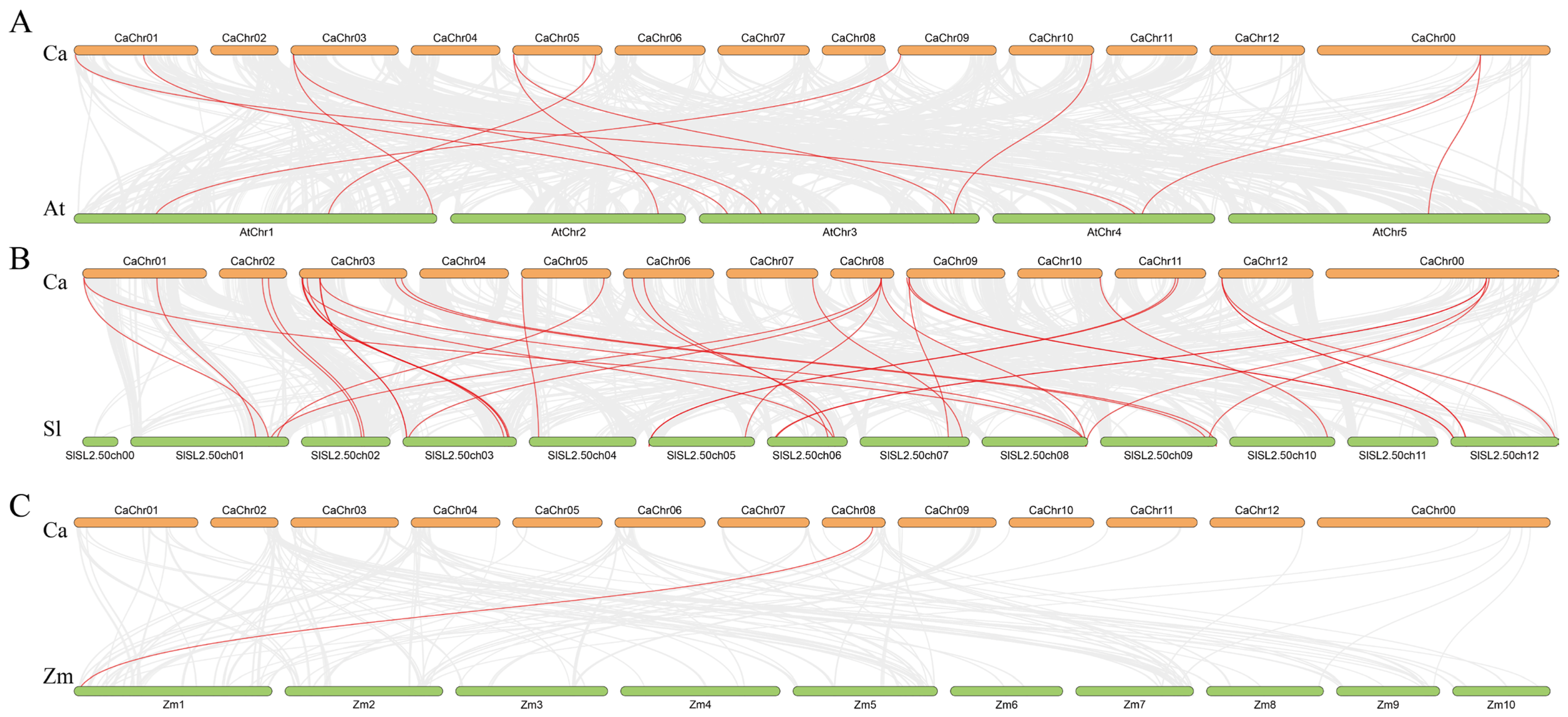
2.7. Synteny Analysis Among CaPG Genes
2.8. Expression Profiles of CaPG Genes in Different Tissues and Fruit Developmental Stages
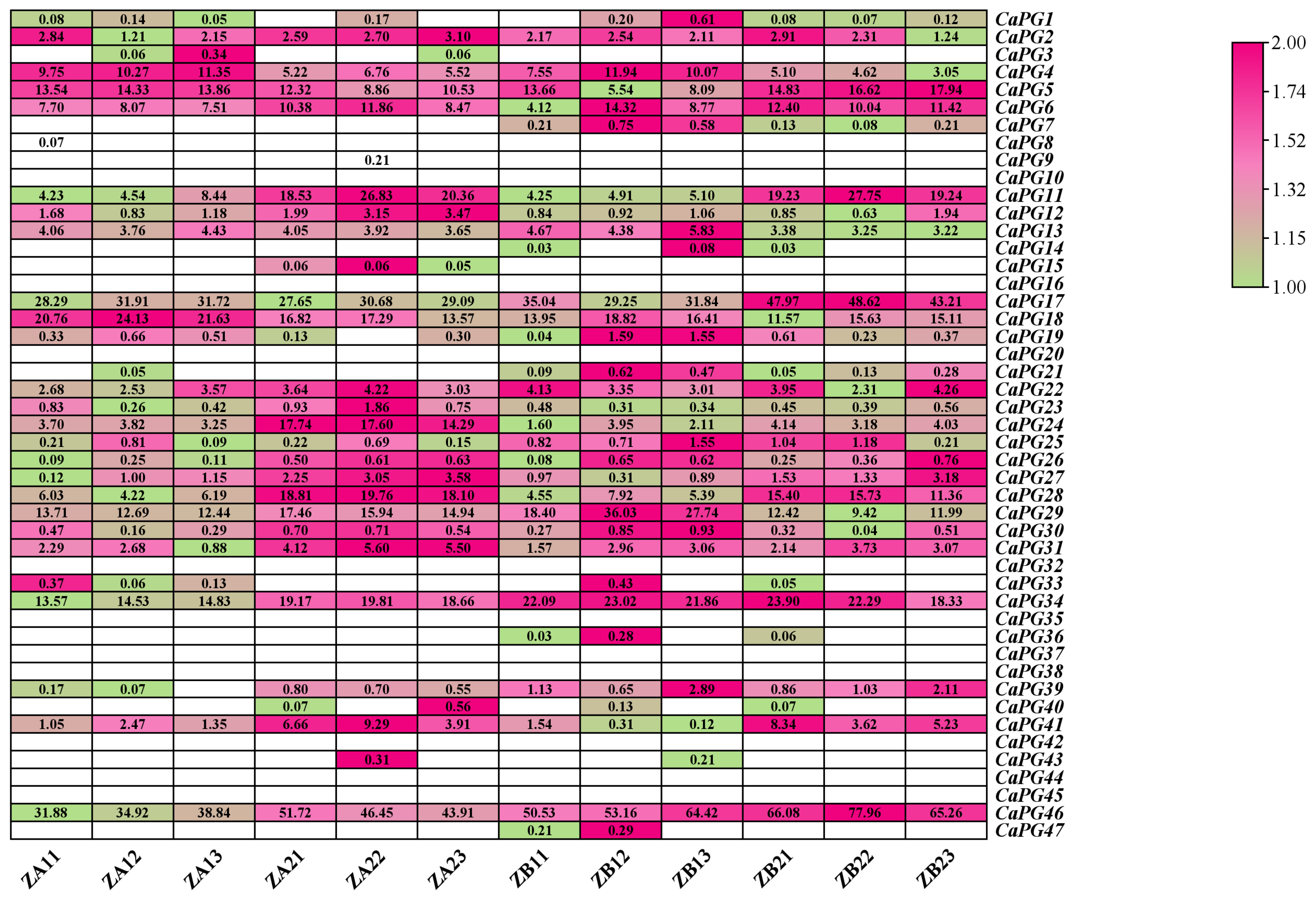
2.9. Expression Patterns of CaPG Genes Related to Pedicel Abscission Susceptibility
2.10. qRT-PCR Validation of CaPG Genes Involvement in Ethylene-Induced Pedicel Abscission
3. Discussion
4. Materials and Methods
4.1. Plant Growth and Treatments
4.2. Identification and Chromosomal Distribution of CaPG Genes
4.3. Physicochemical Properties of CaPG Proteins
4.4. Subcellular Localization Prediction and Signal Peptide Analysis of CaPG Proteins
4.5. Analysis of Conserved Motifs and Gene Structures
4.6. Analysis of Promoters and Phylogeny of CaPG Genes
4.7. Collinearity Analysis and Visualization of CaPG Genes
4.8. Tissue-Specific Expression Profiles of CaPG Genes
4.9. RT-qPCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, H.; Zhang, Y.; Zhang, L.; Wang, X.; Zhang, K.; Cheng, F.; Chen, S. Genetic regulation of chlorophyll biosynthesis in Pepper Fruit: Roles of CaAPRR2 and CaGLK2. Genes 2025, 16, 219. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yang, C.; He, J.; Zhang, L.; Xing, X.; Zhang, K.; Yu, H.; Zhang, Z.; Wu, H.; Cheng, F.; et al. Characterization of metabolites and transcriptome of Pepper accessions from four southern provinces of China. Genes 2025, 16, 137. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, S.; Kilian, B.; Koepke, T.; Abarca, J.; Whiting, M.; Dhingra, A. Transcriptome analysis reveals potential mechanisms for ethylene-inducible pedicel-Fruit abscission zone activation in non-climacteric sweet cherry (Prunus avium L.). Horticulturae 2022, 7, 270. [Google Scholar] [CrossRef]
- Hasan, A.M.; Lakhwani, D.; Pathak, S.; Gupta, P.; Bag, S.K.; Nath, P.; Trivedi, P. K Transcriptome analysis of ripe and unripe fruit tissue of banana identifies major metabolic networks involved in fruit ripening process. BMC Plant Biol. 2014, 14, 316. [Google Scholar] [CrossRef]
- Meir, S.; Philosoph-Hadas, S.; Sundaresan, S.; Selvaraj, K.S.V.; Burd, S.; Ophir, R.; Kochanek, B.; Reid, M.S.; Jiang, C.Z.; Lers, A. Microarray analysis of the abscission-related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiol. 2010, 154, 785–797. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, L.; Li, R.; Cai, Y.; Wang, X.; Fu, X.; Dong, X.; Qi, M.; Jiang, C.-Z.; Xu, T.; et al. The HD-Zip transcription factor SlHB15A regulates abscission by modulating jasmonoyl-isoleucine biosynthesis. Plant Physiol. 2022, 190, 1800–1814. [Google Scholar] [CrossRef]
- Qi, X.X.; Hu, S.; Zhou, H.S.; Liu, X.; Wang, L.F.; Zhao, B.Y.; Huang, X.S.; Zhang, S.L. A MADS-box transcription factor of Kuerle xiangli (Pyrus sinkiangensis Yu) PsJOINTLESS gene functions in floral organ abscission. Gene 2017, 621, 13–20. [Google Scholar] [CrossRef]
- Yi, J.W.; Wang, Y.; Ma, X.S.; Zhang, J.Q.; Zhao, M.L.; Huang, X.M.; Li, J.G.; Hu, G.B.; Wang, H.C. LcERF2 modulates cell wall metabolism by directly targeting a UDP-glucose-4-epimerase gene to regulate pedicel development and fruit abscission of litchi. Plant J. 2021, 106, 620–635. [Google Scholar] [CrossRef]
- Lee, Y.S.; Do, V.G.; Kim, S.; Kweon, H.; McGhie, T.K. Cold stress triggers premature fruit abscission through ABA-dependent signal transduction in early developing apple. PLoS ONE 2021, 16, e0248887. [Google Scholar] [CrossRef]
- Yang, Z.Q.; Zhong, X.M.; Fan, Y.; Wang, H.C.; Li, J.G.; Huang, X.M. Burst of reactive oxygen species in pedicel-mediated fruit abscission after carbohydrate supply was cut off in longan (Dimocarpus longan). Front. Plant Sci. 2015, 6, 833. [Google Scholar] [CrossRef]
- Costa, F.; Peace, C.P.; Stella, S.; Musacchi, S.; Bazzani, M.; Sansavini, S.; Weg, W.E.V.D. QTL dynamics for fruit firmness and softening around an ethylene-dependent polygalacturonase gene in apple (Malus × domestica Borkh.). J. Exp. Bot. 2010, 61, 4301–4313. [Google Scholar] [CrossRef] [PubMed]
- Paniagua, C.; Ric-Varas, P.; A García-Gago, J.; López-Casado, G.; Blanco-Portales, R.; Muñoz-Blanco, J.; Schückel, J.; Knox, J.P.; Matas, A.J.; A Quesada, M.; et al. Elucidating the role of polygalacturonase genes in strawberry fruit softening. J. Exp. Bot. 2020, 71, 5107–5121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.L.; Ma, M.; Zhang, H.P.; Zhang, S.L.; Qian, M.; Zhang, Z.; Luo, W.Q.; Fan, J.B.; Liu, Z.Q.; Wang, L.B. Genome-wide analysis of polygalacturonase gene family from pear genome and identification of the member involved in pear softening. BMC Plant Biol. 2019, 19, 587. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.M.; Shi, Y.; Geng, X.Q.; Xing, G.M. CRISPR/Cas9-mediated targeted mutagenesis of tomato polygalacturonase gene (SlPG) delays fruit softening. Front. Plant Sci. 2022, 13, 834018. [Google Scholar] [CrossRef]
- Roongsattham, P.; Morcillo, F.; Jantasuriyarat, C.; Pizot, M.; Moussu, S.; Jayaweera, D.; Collin, M.; Gonzalez-Carranza, Z.H.; Amblard, P.; Tregear, J.W.; et al. Temporal and spatial expression of polygalacturonase gene family members reveals divergent regulation during fleshy fruit ripening and abscission in the monocot species oil palm. BMC Plant Biol. 2012, 12, 150. [Google Scholar] [CrossRef]
- Kim, J.; Patterson, S.E. Expression divergence and functional redundancy of polygalacturonases in floral organ abscission. Plant Signal. Behav. 2009, 4, 1180–1182. [Google Scholar] [CrossRef]
- Ge, T.; Huang, X.; Pan, X.T.; Zhang, J.; Xie, R.J. Genome-wide identification and expression analysis of citrus fruitlet abscission-related polygalacturonase genes. 3 Biotech 2019, 9, 250. [Google Scholar] [CrossRef]
- Taylor, J.E.; Tucker, G.A.; Lasslett, Y.; Smith, C.J.; Arnold, C.M.; Watson, C.F.; Schuch, W.; Grierson, D.; Roberts, J.A. Polygalacturonase expression during leaf abscission of normal and transgenic tomato plants. Planta 2013, 238, 743–754. [Google Scholar] [CrossRef]
- Ahmed, S.S.; Gong, Z.H.; Khan, M.A.; Yin, Y.X.; Guo, W.L.; Imran, J. Activity and expression of polygalacturonase vary at different fruit ripening stages of sweet pepper cultivars. Genet. Mol. Res. 2011, 10, 3275–3290. [Google Scholar] [CrossRef]
- Li, P.P.; Zhang, R.H.; Zhou, H.D.; Mo, Y.R.; Wu, S.; Zhang, X.; Xie, Z.H.; Zhang, T.Y.; Zhao, K.; LV, J.H.; et al. Melatonin delays softening of postharvest pepper fruits (Capsicum annuum L.) by regulating cell wall degradation, membrane stability and antioxidant systems. Postharvest Biol. Technol. 2024, 212, 112852. [Google Scholar] [CrossRef]
- Li, Z.D.; Zhao, W.T.; Wang, P.; Zhao, S.; Wang, D.; Zhao, X.Y. Transcriptome analysis integrated with changes in cell wall polysaccharides of different fresh-cut chili pepper cultivars during storage reveals the softening mechanism. Food Chem. 2024, 452, 139445. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Dong, J.C.; Zhang, S.B.; Song, Z.; Guan, W.D.; Yuan, F.C.; Zhong, J.; Liu, J.; Hu, K.L.; Cheng, J.W. Fine mapping and gene silencing pinpoint Capana10g002229 as a strong candidate gene regulating the deciduous character of ripe pepper fruit (Capsicum spp.). Theor. Appl. Genet. 2023, 136, 107. [Google Scholar] [CrossRef]
- Jiang, F.L.; Lopez, A.; Jeon, S.; Freitas, S.T.D.; Yu, Q.H.; Wu, Z.; Labavitch, J.M.; Tian, S.; Powell, A.L.T.; Mitcham, E. Disassembly of the fruit cell wall by the ripening-associated polygalacturonase and expansin influences tomato cracking. Hortic. Res. 2019, 6, 17. [Google Scholar] [CrossRef]
- Jiang, C.Z.; Lu, F.; Imsabai, W.; Meir, S.; Reid, M.S. Silencing polygalacturonase expression inhibits tomato petiole abscission. J. Exp. Bot. 2008, 59, 973–979. [Google Scholar] [CrossRef]
- Ferrari, S.; Galletti, R.; Pontiggia, D.; Manfredini, C.; Lionetti, V.; Bellincampi, D.; Cervone, F.; Lorenzo, G.D.G. Transgenic expression of a fungal endo-polygalacturonase increases plant resistance to pathogens and reduces auxin sensitivity. Plant Physiol. 2008, 146, 669–681. [Google Scholar] [CrossRef]
- González-Carranza, Z.H.; Elliott, K.A.; Roberts, J.A. Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J. Exp. Bot. 2007, 58, 3719–3730. [Google Scholar] [CrossRef]
- Ke, X.B.; Wang, H.S.; Li, Y.; Zhu, B.; Zang, Y.X.; He, Y.; Cao, J.S.; Zhu, Z.J.; Yu, Y.J. Genome-wide identification and analysis of polygalacturonase genes in Solanum lycopersicum. Int. J. Mol. Sci. 2018, 19, 2290. [Google Scholar] [CrossRef]
- Kozlowski, L.P. Proteome-pI 2.0: Proteome isoelectric point database update. Nucleic Acids Res. 2021, 49, D407–D412. [Google Scholar] [CrossRef]
- Loell, K.; Nanda, V. Marginal protein stability drives subcellular proteome isoelectric point. Proc. Natl. Acad. Sci. USA 2018, 115, E7704–E7713. [Google Scholar] [CrossRef]
- Lu, L.; Hou, Q.C.; Wang, L.L.; Zhang, T.Y.; Zhao, W.; Yan, T.W.; Zhao, L.N.; Li, J.P.; Wan, X.Y. Genome-wide identification and characterization of polygalacturonase gene family in maize (Zea mays L.). Int. J. Mol. Sci. 2021, 22, 10722. [Google Scholar] [CrossRef]
- He, P.W.; Zhang, J.Z.; Lv, Z.F.; Cui, P.; Xu, X.M.; George, M.S.; Lu, G.Q. Genome-wide identification and expression analysis of the polygalacturonase gene family in sweetpotato. BMC Plant Biol. 2023, 23, 300. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.T.; Hu, R.X.; Lin, Y.X.; Yang, Y.Q.; Chen, Q.; Li, M.Y.; Zhang, Y.; Zhang, Y.T.; Wang, Y.; He, W.; et al. Genome-wide analysis of polygalacturonase gene family reveals its role in strawberry softening. Plants 2024, 13, 1838. [Google Scholar] [CrossRef] [PubMed]
- Parra, G.; Bradnam, K.; Rose, A.B.; Korf, I. Comparative and functional analysis of intron-mediated enhancement signals reveals conserved features among plants. Nucleic Acids Res. 2011, 39, 8403–8415. [Google Scholar] [CrossRef] [PubMed]
- Ressayre, A.; Glémin, S.; Montalent, P.; Serre-Giardi, L.; Dillmann, C.; Joets, J. Introns structure patterns of variation in nucleotide composition in Arabidopsis thaliana and rice protein-coding genes. Genome Biol. Evol. 2015, 7, 3263–3276. [Google Scholar] [CrossRef]
- Godoy Herz, M.A.; Kornblihtt, A.R. Alternative splicing and transcription elongation in plants. Front. Plant Sci. 2019, 10, 1123. [Google Scholar] [CrossRef]
- Reddy, A.S.N.; Marquez, Y.; Kalyna, M.; Barta, A. Complexity of the alternative splicing landscape in plants. Plant Cell 2013, 25, 1630–1653. [Google Scholar] [CrossRef]
- Meir, S.; Philosoph-Hadas, S.; Riov, J.; Tucker, M.L.; Patterson, S.E.; Roberts, J.A. Re-evaluation of the ethylene-dependent and -independent pathways in the regulation of floral and organ abscission. J. Exp. Bot. 2019, 70, 2887–2899. [Google Scholar] [CrossRef]
- Wang, G.Q.; Wei, P.C.; Tan, F.; Yu, M.; Zhang, X.Y.; Chen, Q.J.; Wang, X.C. The Transcription factor AtDOF4.7 is involved in ethylene- and IDA-mediated organ abscission in Arabidopsis. Front. Plant Sci. 2016, 7, 1833. [Google Scholar] [CrossRef]
- Qin, C.; Yu, C.S.; Shen, Y.O.; Fang, X.D.; Chen, L.; Min, J.M.; Cheng, J.W.; Zhao, S.C.; Xu, M.; Luo, Y.; et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135–5140. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Chao, J.M.; Li, Z.Y.; Sun, Y.H.; Aluko, O.O.; Wu, X.R.; Wang, Q.; Liu, G.S. MG2C: A user-friendly online tool for drawing genetic maps. Mol. Hortic. 2021, 1, 16. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Deng, M.; Zhao, K.; Zhou, H.; Wu, R.; Li, M.; Cheng, H.; Li, P.; Zhang, R.; Lv, J. Characterization of plant homeodomain transcription factor genes involved in flower development and multiple abiotic stress response in pepper. Genes 2023, 14, 1737. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Chen, H.; Zhang, D.; Ma, J.J. Ectopic expression of the apple cytokinin response regulator MdRR9 gene in tomatoes promotes shoot branching. Scientia Hortic. 2023, 321, 112228. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Gu, H.J.; Feng, W.X.; Mehari, T.G.; Wang, Y.F.; Wang, Z.Y.; Xu, Y.F.; Zhao, Y.Z.; Tang, J.F.; Zhang, K.; Zhou, Z.T.; et al. Genome-wide analysis and functional validation of the cotton FAH gene family for salt stress. BMC Genom. 2025, 26, 271. [Google Scholar] [CrossRef]
- Chen, C.J.; Wu, Y.; Li, J.W.; Wang, X.; Zeng, Z.H.; Xu, J.; Liu, Y.L.; Feng, J.T.; Chen, H.; He, Y.H.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-DataMining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; Wang, X.Y.; Sun, Y.; Joseph, P.V.; Paterson, A.H. Detection of colinear blocks and synteny and evolutionary analyses based on utilization of MCScanX. Nat. Protoc. 2024, 19, 2206–2229. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



| Gene Name | Gene ID | Amino Acid Number | Molecular Weight (Da) | Theoretical pI | Instability Index | Aliphatic Index | Average of Hydropathicity (GRAVY) | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| CaPG1 | Capana00g000318 | 471 | 51,283.65 | 5.66 | 55.72 | 78.43 | −0.179 | chloroplast |
| CaPG2 | Capana00g001601 | 460 | 49,752.13 | 5.43 | 32.55 | 85.17 | −0.162 | cytoplasm |
| CaPG3 | Capana00g003267 | 337 | 35,659.78 | 6.52 | 39.79 | 89.38 | −0.059 | extracellular matrix |
| CaPG4 | Capana00g003270 | 403 | 43,430.89 | 5.54 | 38.86 | 85.78 | 0.049 | cytoplasm |
| CaPG5 | Capana00g003407 | 443 | 48,502.59 | 8.92 | 34.18 | 90.61 | −0.095 | tonoplast |
| CaPG6 | Capana01g000186 | 484 | 53,203.5 | 5.5 | 38.59 | 88.82 | −0.095 | chloroplast |
| CaPG7 | Capana01g002618 | 427 | 47,497.98 | 9.43 | 26.25 | 85.36 | −0.353 | chloroplast |
| CaPG8 | Capana01g002826 | 275 | 29,680.37 | 5.2 | 46.98 | 82.22 | −0.167 | cytoplasm |
| CaPG9 | Capana02g000966 | 389 | 41,790.44 | 8.61 | 34.05 | 86.68 | −0.125 | nucleus |
| CaPG10 | Capana02g001324 | 396 | 42,839.4 | 7.18 | 35.38 | 85.13 | −0.067 | extracellular matrix |
| CaPG11 | Capana03g000428 | 392 | 43,288.71 | 8.82 | 42.73 | 85.48 | 0.026 | chloroplast |
| CaPG12 | Capana03g000435 | 388 | 41,598.44 | 8.59 | 38.97 | 90.67 | −0.059 | nucleus |
| CaPG13 | Capana03g000564 | 353 | 39,730.85 | 6.66 | 41.45 | 87.99 | −0.399 | cytoskeleton |
| CaPG14 | Capana03g001086 | 525 | 57,077.33 | 7.8 | 38.98 | 93.33 | 0.022 | cytoplasm |
| CaPG15 | Capana03g002144 | 357 | 38,892.43 | 5.35 | 36.79 | 88.99 | −0.045 | cytoplasm |
| CaPG16 | Capana03g002171 | 313 | 34,204.04 | 5.35 | 36.4 | 89.07 | −0.02 | cytoplasm |
| CaPG17 | Capana03g003120 | 453 | 50,599.63 | 8.83 | 48.51 | 86.47 | −0.201 | cytoplasm |
| CaPG18 | Capana03g003665 | 374 | 40,615.51 | 5.39 | 39.73 | 87.51 | −0.079 | chloroplast |
| CaPG19 | Capana03g004218 | 452 | 49,398.24 | 4.97 | 38.09 | 78.45 | −0.312 | extracellular matrix |
| CaPG20 | Capana04g002230 | 392 | 42,882.99 | 9.8 | 31.47 | 74.82 | −0.215 | nucleus |
| CaPG21 | Capana04g002231 | 394 | 42,274.84 | 8.71 | 34.46 | 80.66 | −0.124 | nucleus |
| CaPG22 | Capana05g000108 | 535 | 59,457.05 | 5.62 | 25.29 | 73.57 | −0.741 | nucleus |
| CaPG23 | Capana05g000884 | 492 | 53,276.09 | 4.96 | 53.51 | 77.83 | −0.211 | nucleus |
| CaPG24 | Capana05g001033 | 488 | 53,375.7 | 5.41 | 37.8 | 90.7 | −0.096 | endoplasmic reticulum |
| CaPG25 | Capana05g002022 | 265 | 28,432.09 | 8.19 | 47 | 73.51 | −0.217 | chloroplast |
| CaPG26 | Capana05g002171 | 434 | 46,785.2 | 6.69 | 51.14 | 75.74 | −0.149 | nucleus |
| CaPG27 | Capana06g001141 | 300 | 33,023.9 | 9.28 | 40.01 | 84.1 | −0.065 | nucleus |
| CaPG28 | Capana06g001746 | 386 | 42,390.74 | 9.08 | 41.27 | 98.42 | 0.003 | cytoplasm |
| CaPG29 | Capana07g000744 | 375 | 42,090.84 | 7.23 | 36.51 | 86.69 | −0.332 | cytoplasm |
| CaPG30 | Capana07g001965 | 399 | 43,410.72 | 9.03 | 36.37 | 83.01 | −0.033 | nucleus |
| CaPG31 | Capana08g000965 | 472 | 51,887.04 | 8.26 | 38.85 | 89.64 | −0.187 | tonoplast |
| CaPG32 | Capana08g001207 | 397 | 42,209.5 | 5.52 | 42.32 | 88.14 | −0.038 | extracellular matrix |
| CaPG33 | Capana09g000012 | 355 | 38,489.07 | 5.45 | 33.72 | 82.08 | −0.155 | extracellular matrix |
| CaPG34 | Capana09g000145 | 493 | 55,367.46 | 8.9 | 40.58 | 92.45 | −0.145 | chloroplast |
| CaPG35 | Capana09g000459 | 327 | 34,860.55 | 7.47 | 29.79 | 80.67 | −0.296 | nucleus |
| CaPG36 | Capana09g000460 | 598 | 63,472.85 | 6.69 | 30.57 | 83.04 | −0.177 | nucleus |
| CaPG37 | Capana09g000516 | 390 | 41,929.81 | 8.87 | 29.01 | 79.72 | −0.12 | nucleus |
| CaPG38 | Capana10g000004 | 350 | 37,695.18 | 4.95 | 45.59 | 83.6 | −0.163 | nucleus |
| CaPG39 | Capana10g002229 | 321 | 35,227.22 | 5.69 | 27.29 | 73.77 | −0.355 | nucleus |
| CaPG40 | Capana11g001239 | 281 | 30,660.88 | 8.41 | 37.33 | 86.62 | −0.19 | nucleus |
| CaPG41 | Capana11g001285 | 495 | 54,293.94 | 8.97 | 39.4 | 75.96 | −0.245 | chloroplast |
| CaPG42 | Capana12g000384 | 332 | 35,479.08 | 7.49 | 26.59 | 81.54 | −0.152 | nucleus |
| CaPG43 | Capana12g000385 | 390 | 41,382.38 | 6.74 | 37 | 78.74 | −0.105 | nucleus |
| CaPG44 | Capana12g000386 | 390 | 41,441.45 | 7.07 | 36.02 | 78.74 | −0.113 | nucleus |
| CaPG45 | Capana12g000387 | 332 | 35,479.08 | 7.49 | 26.59 | 81.54 | −0.152 | nucleus |
| CaPG46 | Capana12g001281 | 409 | 44,343.14 | 5.98 | 40.42 | 87.43 | −0.048 | nucleus |
| CaPG47 | Capana12g002138 | 261 | 28,032.14 | 8.8 | 27.91 | 96.32 | 0.207 | extracellular matrix |
| Duplicated Pairs | Ka 1 | Ks 2 | Ka/Ks | Purifying Selection | Duplicate Type |
|---|---|---|---|---|---|
| CaPG4–CaPG18 | 0.1123 | 0.7115 | 0.1578 | Yes | Segmental |
| CaPG6–CaPG31 | 0.2493 | 2.2193 | 0.1123 | Yes | Segmental |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, L.; Lu, C.; Yan, X.; Yang, S.; Zhou, P.; Lai, W.; He, J. Genome-Wide Identification of the Polygalacturonase Gene Family and Its Potential Association with Abscission Zone in Capsicum annuum L. Genes 2025, 16, 579. https://doi.org/10.3390/genes16050579
He L, Lu C, Yan X, Yang S, Zhou P, Lai W, He J. Genome-Wide Identification of the Polygalacturonase Gene Family and Its Potential Association with Abscission Zone in Capsicum annuum L. Genes. 2025; 16(5):579. https://doi.org/10.3390/genes16050579
Chicago/Turabian StyleHe, Lei, Chen Lu, Xi Yan, Sha Yang, Peng Zhou, Wei Lai, and Jianwen He. 2025. "Genome-Wide Identification of the Polygalacturonase Gene Family and Its Potential Association with Abscission Zone in Capsicum annuum L." Genes 16, no. 5: 579. https://doi.org/10.3390/genes16050579
APA StyleHe, L., Lu, C., Yan, X., Yang, S., Zhou, P., Lai, W., & He, J. (2025). Genome-Wide Identification of the Polygalacturonase Gene Family and Its Potential Association with Abscission Zone in Capsicum annuum L. Genes, 16(5), 579. https://doi.org/10.3390/genes16050579





