PAX3 Regulatory Signatures and Gene Targets in Melanoma Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Harvest
2.2. Cleavage Under Targets and Release Using Nuclease (CUT&RUN)
2.3. CUT&RUN Library Preparation and Data Analysis
2.4. ATAC-Seq Processing
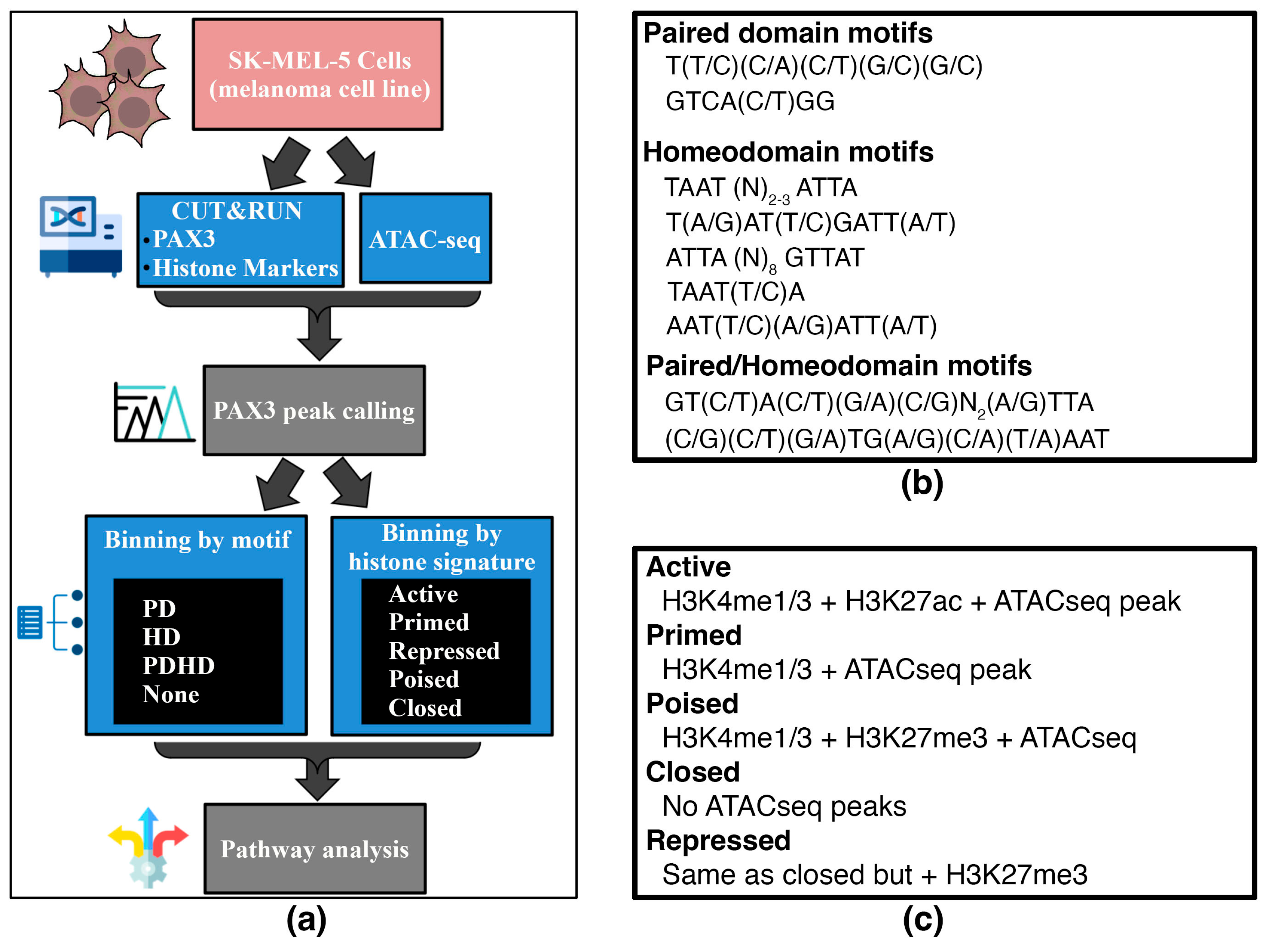
2.5. Identification and Classification of PAX3 Binding Motifs
2.6. Gene State Assessment and Classification
2.7. Data Processing and Analysis
3. Results
3.1. PAX3-Bound Cistromic Regions in Melanoma Cells
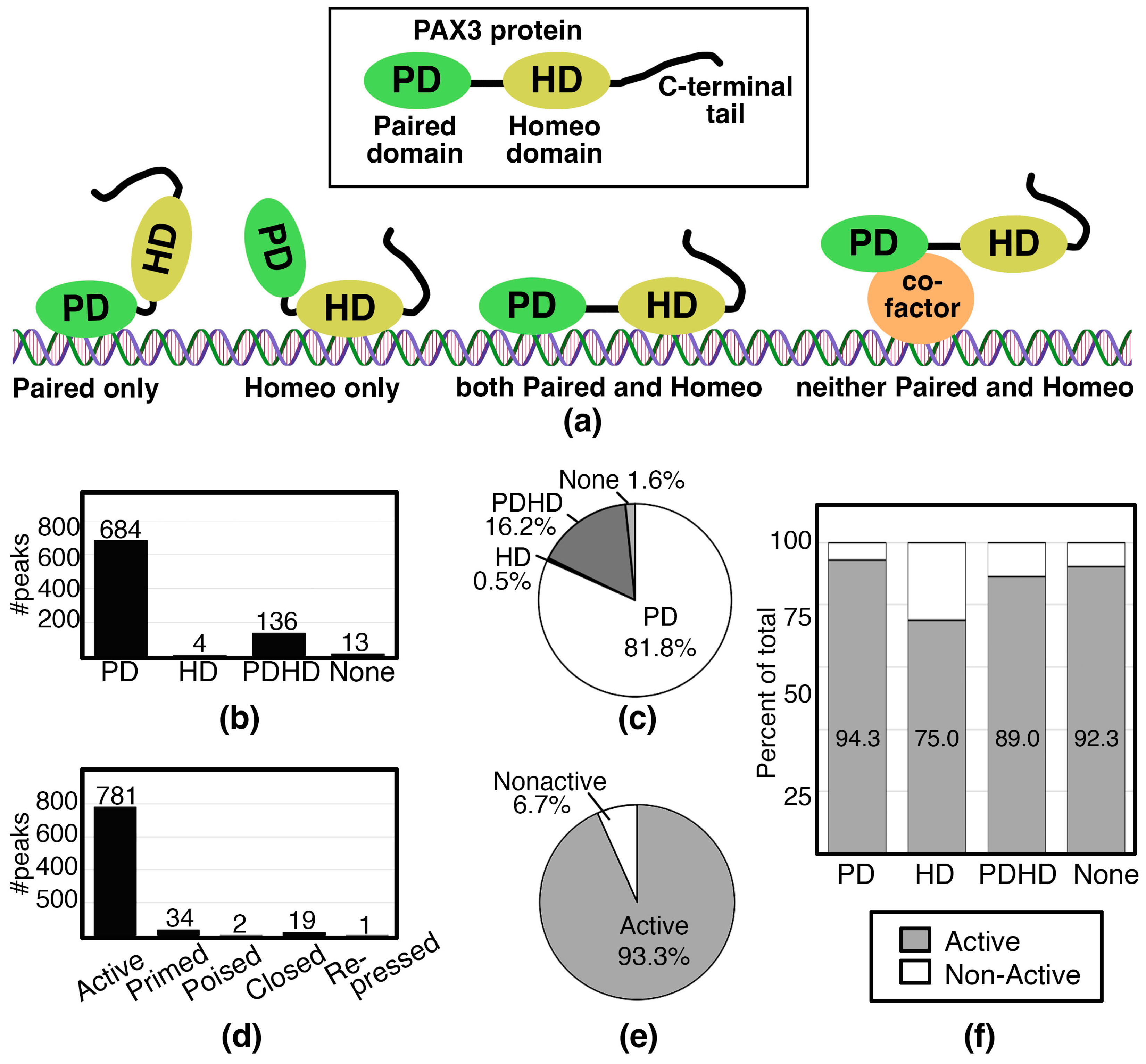
3.2. Binning of PAX3 Peaks with Paired and/or Homeodomain Binding Sites and with Histone Signatures
3.3. PAX3 Commonly Binds to Enhancers of Active Genes Through the Paired Domain in SK-MEL-5 Cells
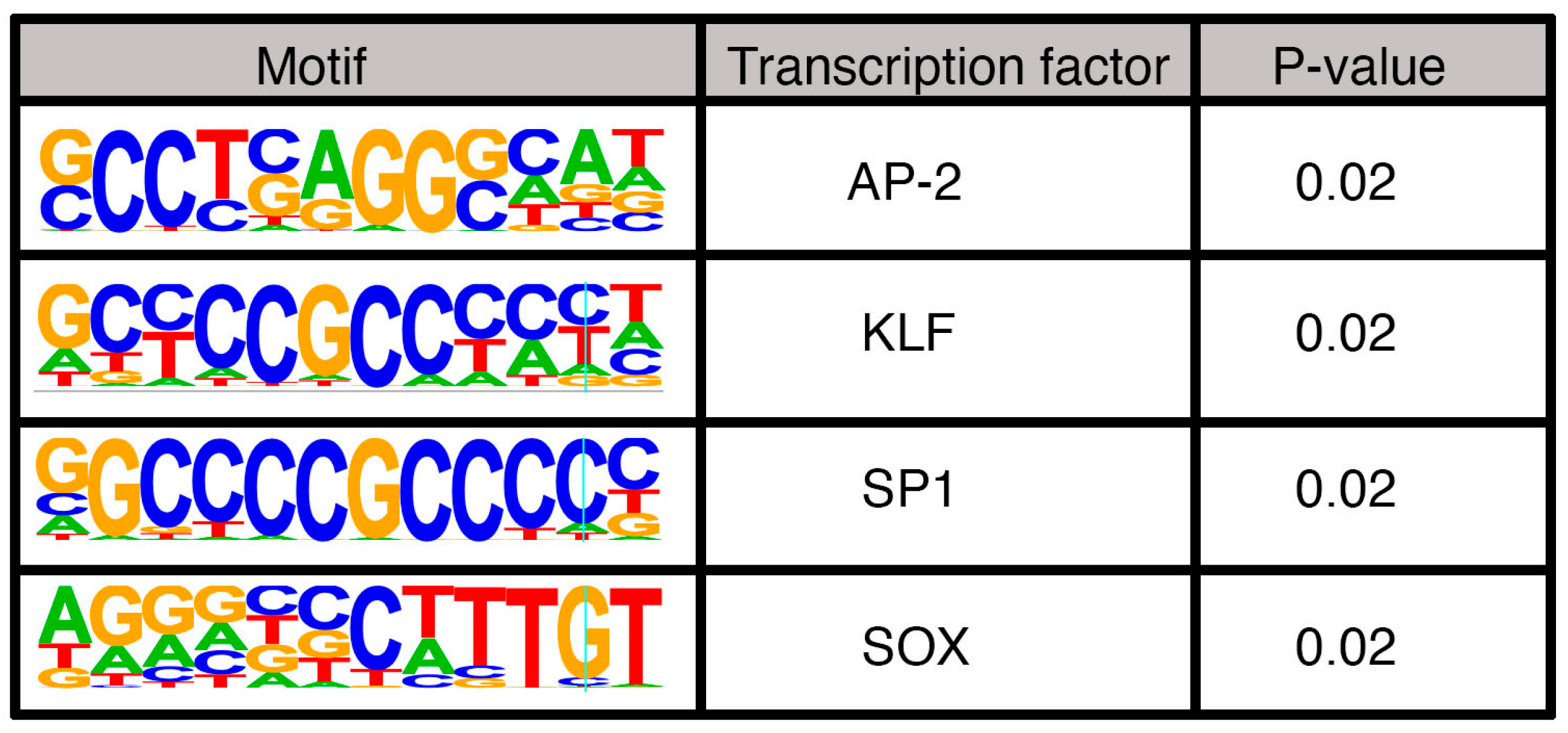
3.4. Pathway Analysis of Potential PAX3 Downstream Genes in SK-MEL-5 Melanoma Cells
3.5. Genes Associated with Distal PAX3 Enhancers Have a Neuronal Signature
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BP | Biological Processes |
| CNS | Central Nervous System |
| CUT&RUN | Cleavage Under Targets & Release Using Nuclease |
| FP-RMS | Fusion Positive Rhabdomyosarcoma |
| GO | Gene Ontology |
| HD | Homeodomain |
| MF | Molecular Function |
| NCI | National Cancer Institute |
| PD | Paired Domain |
| PDHD | Homeodomain and Paired Domain |
| PTM | Post-Translational Modification |
| TSS | Transcriptional Start Site |
References
- Kubic, J.D.; Young, K.P.; Plummer, R.S.; Ludvik, A.E.; Lang, D. Pigmentation PAX-ways: The role of Pax3 in melanogenesis, melanocyte stem cell maintenance, and disease. Pigment Cell Melanoma Res. 2008, 21, 627–645. [Google Scholar] [CrossRef] [PubMed]
- Tajbakhsh, S.; Rocancourt, D.; Cossu, G.; Buckingham, M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell 1997, 89, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Geles, K.G.; Paik, J.H.; DePinho, R.A.; Tjian, R. Codependent activators direct myoblast-specific MyoD transcription. Dev. Cell 2008, 15, 534–546. [Google Scholar] [CrossRef]
- Smith, M.P.; Rana, S.; Ferguson, J.; Rowling, E.J.; Flaherty, K.T.; Wargo, J.A.; Marais, R.; Wellbrock, C. A PAX3/BRN2 rheostat controls the dynamics of BRAF mediated MITF regulation in MITF(high)/AXL(low) melanoma. Pigment Cell Melanoma Res. 2019, 32, 280–291. [Google Scholar] [CrossRef]
- Watanabe, A.; Takeda, K.; Ploplis, B.; Tachibana, M. Epistatic relationship between Waardenburg syndrome genes MITF and PAX3. Nat. Genet. 1998, 18, 283–286. [Google Scholar] [CrossRef]
- Corry, G.N.; Underhill, D.A. Pax3 target gene recognition occurs through distinct modes that are differentially affected by disease-associated mutations. Pigment Cell Res. 2005, 18, 427–438. [Google Scholar] [CrossRef]
- Tassabehji, M.; Read, A.P.; Newton, V.E.; Harris, R.; Balling, R.; Gruss, P.; Strachan, T. Waardenburg’s syndrome patients have mutations in the human homologue of the Pax-3 paired box gene. Nature 1992, 355, 635–636. [Google Scholar] [CrossRef]
- Bonvin, E.; Falletta, P.; Shaw, H.; Delmas, V.; Goding, C.R. A phosphatidylinositol 3-kinase-Pax3 axis regulates Brn-2 expression in melanoma. Mol. Cell Biol. 2012, 32, 4674–4683. [Google Scholar] [CrossRef]
- Kubic, J.D.; Little, E.C.; Lui, J.W.; Iizuka, T.; Lang, D. PAX3 and ETS1 synergistically activate MET expression in melanoma cells. Oncogene 2015, 34, 4964–4974. [Google Scholar] [CrossRef]
- Kubic, J.D.; Little, E.C.; Kaiser, R.S.; Young, K.P.; Lang, D. FOXD3 Promotes PAX3 Expression in Melanoma Cells. J. Cell. Biochem. 2016, 117, 533–541. [Google Scholar] [CrossRef]
- Mascarenhas, J.B.; Littlejohn, E.L.; Wolsky, R.J.; Young, K.P.; Nelson, M.; Salgia, R.; Lang, D. PAX3 and SOX10 activate MET receptor expression in melanoma. Pigment Cell Melanoma Res. 2010, 23, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Magli, A.; Baik, J.; Mills, L.J.; Kwak, I.Y.; Dillon, B.S.; Mondragon Gonzalez, R.; Stafford, D.A.; Swanson, S.A.; Stewart, R.; Thomson, J.A.; et al. Time-dependent Pax3-mediated chromatin remodeling and cooperation with Six4 and Tead2 specify the skeletal myogenic lineage in developing mesoderm. PLoS Biol. 2019, 17, e3000153. [Google Scholar] [CrossRef] [PubMed]
- Magli, A.; Baik, J.; Pota, P.; Cordero, C.O.; Kwak, I.Y.; Garry, D.J.; Love, P.E.; Dynlacht, B.D.; Perlingeiro, R.C.R. Pax3 cooperates with Ldb1 to direct local chromosome architecture during myogenic lineage specification. Nat. Commun. 2019, 10, 2316. [Google Scholar] [CrossRef] [PubMed]
- Soleimani, V.D.; Punch, V.G.; Kawabe, Y.; Jones, A.E.; Palidwor, G.A.; Porter, C.J.; Cross, J.W.; Carvajal, J.J.; Kockx, C.E.; van IJcken, W.F.J.; et al. Transcriptional dominance of Pax7 in adult myogenesis is due to high-affinity recognition of homeodomain motifs. Dev. Cell 2012, 22, 1208–1220. [Google Scholar] [CrossRef]
- Kotov, A.; Seal, S.; Alkobtawi, M.; Kappes, V.; Ruiz, S.M.; Arbes, H.; Harland, R.M.; Peshkin, L.; Monsoro-Burq, A.H. A time-resolved single-cell roadmap of the logic driving anterior neural crest diversification from neural border to migration stages. Proc. Natl. Acad. Sci. USA 2024, 121, e2311685121. [Google Scholar] [CrossRef]
- Asante, Y.; Benischke, K.; Osman, I.; Ngo, Q.A.; Wurth, J.; Laubscher, D.; Kim, H.; Udhayakumar, B.; Khan, M.I.H.; Chin, D.H.; et al. PAX3-FOXO1 uses its activation domain to recruit CBP/P300 and shape RNA Pol2 cluster distribution. Nat. Commun. 2023, 14, 8361. [Google Scholar] [CrossRef]
- Cao, L.; Yu, Y.; Bilke, S.; Walker, R.L.; Mayeenuddin, L.H.; Azorsa, D.O.; Yang, F.; Pineda, M.; Helman, L.J.; Meltzer, P.S. Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer Res. 2010, 70, 6497–6508. [Google Scholar] [CrossRef]
- Gryder, B.E.; Yohe, M.E.; Chou, H.C.; Zhang, X.; Marques, J.; Wachtel, M.; Schaefer, B.; Sen, N.; Song, Y.; Gualtieri, A.; et al. PAX3-FOXO1 Establishes Myogenic Super Enhancers and Confers BET Bromodomain Vulnerability. Cancer Discov. 2017, 7, 884–899. [Google Scholar] [CrossRef]
- Hsieh, J.; Danis, E.P.; Owens, C.R.; Parrish, J.K.; Nowling, N.L.; Wolin, A.R.; Purdy, S.C.; Rosenbaum, S.R.; Ivancevic, A.M.; Chuong, E.B.; et al. Dependence of PAX3-FOXO1 chromatin occupancy on ETS1 at important disease-promoting genes exposes new targetable vulnerability in Fusion-Positive Rhabdomyosarcoma. Oncogene 2024, 14, 19–29. [Google Scholar] [CrossRef]
- Kim, Y.Y.; Gryder, B.E.; Sinniah, R.; Peach, M.L.; Shern, J.F.; Abdelmaksoud, A.; Pomella, S.; Woldemichael, G.M.; Stanton, B.Z.; Milewski, D.; et al. KDM3B inhibitors disrupt the oncogenic activity of PAX3-FOXO1 in fusion-positive rhabdomyosarcoma. Nat. Commun. 2024, 15, 1703. [Google Scholar] [CrossRef]
- Manceau, L.; Richard Albert, J.; Lollini, P.L.; Greenberg, M.V.C.; Gilardi-Hebenstreit, P.; Ribes, V. Divergent transcriptional and transforming properties of PAX3-FOXO1 and PAX7-FOXO1 paralogs. PLoS Genet. 2022, 18, e1009782. [Google Scholar] [CrossRef] [PubMed]
- Searcy, M.B.; Larsen, R.K.t.; Stevens, B.T.; Zhang, Y.; Jin, H.; Drummond, C.J.; Langdon, C.G.; Gadek, K.E.; Vuong, K.; Reed, K.B.; et al. PAX3-FOXO1 dictates myogenic reprogramming and rhabdomyosarcoma identity in endothelial progenitors. Nat. Commun. 2023, 14, 7291. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Abu-Zaid, A.; Jin, H.; Fang, J.; Wu, Q.; Wang, T.; Feng, H.; Quarni, W.; Shao, Y.; Maxham, L.; et al. Targeting KDM4 for treating PAX3-FOXO1-driven alveolar rhabdomyosarcoma. Sci. Transl. Med. 2022, 14, eabq2096. [Google Scholar] [CrossRef]
- Sunkel, B.D.; Wang, M.; LaHaye, S.; Kelly, B.J.; Fitch, J.R.; Barr, F.G.; White, P.; Stanton, B.Z. Evidence of pioneer factor activity of an oncogenic fusion transcription factor. iScience 2021, 24, 102867. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, J.; Liu, Q.; McDonald, W.H.; Bomber, M.L.; Layden, H.M.; Ellis, J.; Borinstein, S.C.; Hiebert, S.W.; Stengel, K.R. PAX3-FOXO1 coordinates enhancer architecture, eRNA transcription, and RNA polymerase pause release at select gene targets. Mol. Cell 2022, 82, 4428–4442.e4427. [Google Scholar] [CrossRef]
- Galibert, M.D.; Yavuzer, U.; Dexter, T.J.; Goding, C.R. Pax3 and regulation of the melanocyte-specific tyrosinase-related protein-1 promoter. J. Biol. Chem. 1999, 274, 26894–26900. [Google Scholar] [CrossRef]
- Kwang, S.J.; Brugger, S.M.; Lazik, A.; Merrill, A.E.; Wu, L.Y.; Liu, Y.H.; Ishii, M.; Sangiorgi, F.O.; Rauchman, M.; Sucov, H.M.; et al. Msx2 is an immediate downstream effector of Pax3 in the development of the murine cardiac neural crest. Development 2002, 129, 527–538. [Google Scholar] [CrossRef]
- Lang, D.; Chen, F.; Milewski, R.; Li, J.; Lu, M.M.; Epstein, J.A. Pax3 is required for enteric ganglia formation and functions with Sox10 to modulate expression of c-ret. J. Clin. Investig. 2000, 106, 963–971. [Google Scholar] [CrossRef]
- Lang, D.; Lu, M.M.; Huang, L.; Engleka, K.A.; Zhang, M.; Chu, E.Y.; Lipner, S.; Skoultchi, A.; Millar, S.E.; Epstein, J.A. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature 2005, 433, 884–887. [Google Scholar] [CrossRef]
- Nakazaki, H.; Reddy, A.C.; Mania-Farnell, B.L.; Shen, Y.W.; Ichi, S.; McCabe, C.; George, D.; McLone, D.G.; Tomita, T.; Mayanil, C.S. Key basic helix-loop-helix transcription factor genes Hes1 and Ngn2 are regulated by Pax3 during mouse embryonic development. Dev. Biol. 2008, 316, 510–523. [Google Scholar] [CrossRef]
- Smith, C.K., 2nd; Janney, M.J.; Allen, R.E. Temporal expression of myogenic regulatory genes during activation, proliferation, and differentiation of rat skeletal muscle satellite cells. J. Cell Physiol. 1994, 159, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Li, J.; Engleka, K.A.; Zhou, B.; Lu, M.M.; Plotkin, J.B.; Epstein, J.A. Persistent expression of Pax3 in the neural crest causes cleft palate and defective osteogenesis in mice. J. Clin. Investig. 2008, 118, 2076–2087. [Google Scholar] [CrossRef]
- Epstein, J.A.; Cai, J.X.; Maas, R.M. Pax3 recognizes a sequence within the 3′utr of the murine neurofibromatosis gene Nf1. Circulation 1994, 90, 635. [Google Scholar]
- Epstein, J.A.; Shapiro, D.N.; Cheng, J.; Lam, P.Y.; Maas, R.L. Pax3 modulates expression of the c-Met receptor during limb muscle development. Proc. Natl. Acad. Sci. USA 1996, 93, 4213–4218. [Google Scholar] [CrossRef]
- Fenby, B.T.; Fotaki, V.; Mason, J.O. Pax3 regulates Wnt1 expression via a conserved binding site in the 5′ proximal promoter. Biochim. Biophys. Acta 2008, 1779, 115–121. [Google Scholar] [CrossRef]
- Kubic, J.D.; Lui, J.W.; Little, E.C.; Ludvik, A.E.; Konda, S.; Salgia, R.; Aplin, A.E.; Lang, D. PAX3 and FOXD3 Promote CXCR4 Expression in Melanoma. J. Biol. Chem. 2015, 290, 21901–21914. [Google Scholar] [CrossRef]
- Lagha, M.; Kormish, J.D.; Rocancourt, D.; Manceau, M.; Epstein, J.A.; Zaret, K.S.; Relaix, F.; Buckingham, M.E. Pax3 regulation of FGF signaling affects the progression of embryonic progenitor cells into the myogenic program. Genes. Dev. 2008, 22, 1828–1837. [Google Scholar] [CrossRef]
- Lakkis, M.M.; Golden, J.A.; O’Shea, K.S.; Epstein, J.A. Neurofibromin deficiency in mice causes exencephaly and is a modifier for Splotch neural tube defects. Dev. Biol. 1999, 212, 80–92. [Google Scholar] [CrossRef][Green Version]
- Margue, C.M.; Bernasconi, M.; Barr, F.G.; Schafer, B.W. Transcriptional modulation of the anti-apoptotic protein BCL-XL by the paired box transcription factors PAX3 and PAX3/FKHR. Oncogene 2000, 19, 2921–2929. [Google Scholar] [CrossRef][Green Version]
- Mayanil, C.S.; Pool, A.; Nakazaki, H.; Reddy, A.C.; Mania-Farnell, B.; Yun, B.; George, D.; McLone, D.G.; Bremer, E.G. Regulation of murine TGFbeta2 by Pax3 during early embryonic development. J. Biol. Chem. 2006, 281, 24544–24552. [Google Scholar] [CrossRef]
- Relaix, F.; Polimeni, M.; Rocancourt, D.; Ponzetto, C.; Schafer, B.W.; Buckingham, M. The transcriptional activator PAX3-FKHR rescues the defects of Pax3 mutant mice but induces a myogenic gain-of-function phenotype with ligand-independent activation of Met signaling in vivo. Genes. Dev. 2003, 17, 2950–2965. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Li, Y.; Du, J.; Lin, H.; Cao, S.; Mao, Z.; Wu, R.; Liu, M.; Liu, Y.; Yin, Q. PAX3 Promotes Cell Migration and CXCR4 Gene Expression in Neural Crest Cells. J. Mol. Neurosci. 2018, 64, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tanami, H.; Imoto, I.; Hirasawa, A.; Yuki, Y.; Sonoda, I.; Inoue, J.; Yasui, K.; Misawa-Furihata, A.; Kawakami, Y.; Inazawa, J. Involvement of overexpressed wild-type BRAF in the growth of malignant melanoma cell lines. Oncogene 2004, 23, 8796–8804. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Persaud, Y.; Pratilas, C.A.; Taylor, B.S.; Janakiraman, M.; She, Q.B.; Gallardo, H.; Liu, C.; Merghoub, T.; Hefter, B.; et al. Concurrent loss of the PTEN and RB1 tumor suppressors attenuates RAF dependence in melanomas harboring (V600E)BRAF. Oncogene 2012, 31, 446–457. [Google Scholar] [CrossRef]
- Meers, M.P.; Bryson, T.D.; Henikoff, J.G.; Henikoff, S. Improved CUT&RUN chromatin profiling tools. Elife 2019, 8, e46314. [Google Scholar] [CrossRef]
- Skene, P.J.; Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 2017, 6, e21856. [Google Scholar] [CrossRef]
- Liu, N. Library Prep for CUT&RUN with NEBNext® UltraTM II DNA Library Prep Kit for Illumina® (E7645) V2; Protocols.io: Berkeley, CA, USA, 2018. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdottir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Donohue, L.K.H.; Guo, M.G.; Zhao, Y.; Jung, N.; Bussat, R.T.; Kim, D.S.; Neela, P.H.; Kellman, L.N.; Garcia, O.S.; Meyers, R.M.; et al. A cis-regulatory lexicon of DNA motif combinations mediating cell-type-specific gene regulation. Cell Genom. 2022, 2, 100191. [Google Scholar] [CrossRef]
- Mölder, F.; Jablonski, K.P.; Letcher, B.; Hall, M.B.; Tomkins-Tinch, C.H.; Sochat, V.; Forster, J.; Lee, S.; Twardziok, S.O.; Kanitz, A.; et al. Sustainable data analysis with Snakemake. F1000Res 2021, 10, 33. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 29 March 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Ramirez, F.; Ryan, D.P.; Gruning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dundar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; He, Q.Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef]
- Lawrence, M.; Huber, W.; Pages, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef]
- Lawrence, M.; Gentleman, R.; Carey, V. rtracklayer: An R package for interfacing with genome browsers. Bioinformatics 2009, 25, 1841–1842. [Google Scholar] [CrossRef]
- Team-TBD. BSgenome.Hsapiens.UCSC.hg19: Full Genome Sequences for Homo Sapiens (UCSC Version hg19, Based on GRCh37.p13); R Package Version 1.4.3; Bioconductor. 2020. Available online: https://bioconductor.org/packages/release/data/annotation/html/BSgenome.Hsapiens.UCSC.hg19.html (accessed on 29 March 2024).
- Pagés, H.; Aboyoun, P.; Gentleman, R.; DebRoy, S. Biostrings: Efficient Manipulation of Biological Strings; Bioconductor. 2024. Available online: https://bioconductor.org/packages/release/bioc/manuals/Biostrings/man/Biostrings.pdf (accessed on 29 March 2024).
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Chalepakis, G.; Goulding, M.; Read, A.; Strachan, T.; Gruss, P. Molecular basis of splotch and Waardenburg Pax-3 mutations. Proc. Natl. Acad. Sci. USA 1994, 91, 3685–3689. [Google Scholar] [CrossRef]
- Chalepakis, G.; Gruss, P. Identification of DNA recognition sequences for the Pax3 paired domain. Gene 1995, 162, 267–270. [Google Scholar] [CrossRef]
- Jun, S.; Desplan, C. Cooperative interactions between paired domain and homeodomain. Development 1996, 122, 2639–2650. [Google Scholar] [CrossRef]
- Epstein, J.A.; Song, B.; Lakkis, M.; Wang, C. Tumor-specific PAX3-FKHR transcription factor, but not PAX3, activates the platelet-derived growth factor alpha receptor. Mol. Cell Biol. 1998, 18, 4118–4130. [Google Scholar] [CrossRef]
- Chalepakis, G.; Wijnholds, J.; Gruss, P. Pax-3-DNA interaction: Flexibility in the DNA binding and induction of DNA conformational changes by paired domains. Nucleic Acids Res. 1994, 22, 3131–3137. [Google Scholar] [CrossRef]
- Pelletier, A.; Mayran, A.; Gouhier, A.; Omichinski, J.G.; Balsalobre, A.; Drouin, J. Pax7 pioneer factor action requires both paired and homeo DNA binding domains. Nucleic Acids Res. 2021, 49, 7424–7436. [Google Scholar] [CrossRef]
- Wilson, D.S.; Guenther, B.; Desplan, C.; Kuriyan, J. High resolution crystal structure of a paired (Pax) class cooperative homeodomain dimer on DNA. Cell 1995, 82, 709–719. [Google Scholar] [CrossRef]
- Phelan, S.A.; Loeken, M.R. Identification of a new binding motif for the paired domain of Pax-3 and unusual characteristics of spacing of bipartite recognition elements on binding and transcription activation. J. Biol. Chem. 1998, 273, 19153–19159. [Google Scholar] [CrossRef]
- Yin, Y.; Morgunova, E.; Jolma, A.; Kaasinen, E.; Sahu, B.; Khund-Sayeed, S.; Das, P.K.; Kivioja, T.; Dave, K.; Zhong, F.; et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 2017, 356, eaaj2239. [Google Scholar] [CrossRef]
- Chalepakis, G.; Jones, F.S.; Edelman, G.M.; Gruss, P. Pax-3 contains domains for transcription activation and transcription inhibition. Proc. Natl. Acad. Sci. USA 1994, 91, 12745–12749. [Google Scholar] [CrossRef]
- Barral, A.; Zaret, K.S. Pioneer factors: Roles and their regulation in development. Trends Genet. 2024, 40, 134–148. [Google Scholar] [CrossRef]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef]
- Crispatzu, G.; Rehimi, R.; Pachano, T.; Bleckwehl, T.; Cruz-Molina, S.; Xiao, C.; Mahabir, E.; Bazzi, H.; Rada-Iglesias, A. The chromatin, topological and regulatory properties of pluripotency-associated poised enhancers are conserved in vivo. Nat. Commun. 2021, 12, 4344. [Google Scholar] [CrossRef]
- Gates, L.A.; Foulds, C.E.; O’Malley, B.W. Histone Marks in the ’Driver’s Seat’: Functional Roles in Steering the Transcription Cycle. Trends Biochem. Sci. 2017, 42, 977–989. [Google Scholar] [CrossRef]
- Bondurand, N.; Pingault, V.; Goerich, D.E.; Lemort, N.; Sock, E.; Caignec, C.L.; Wegner, M.; Goossens, M. Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum. Mol. Genet. 2000, 9, 1907–1917. [Google Scholar] [CrossRef]
- Potterf, S.B.; Furumura, M.; Dunn, K.J.; Arnheiter, H.; Pavan, W.J. Transcription factor hierarchy in Waardenburg syndrome: Regulation of MITF expression by SOX10 and PAX3. Hum. Genet. 2000, 107, 1–6. [Google Scholar] [CrossRef]
- Smith, M.P.; Brunton, H.; Rowling, E.J.; Ferguson, J.; Arozarena, I.; Miskolczi, Z.; Lee, J.L.; Girotti, M.R.; Marais, R.; Levesque, M.P.; et al. Inhibiting Drivers of Non-mutational Drug Tolerance Is a Salvage Strategy for Targeted Melanoma Therapy. Cancer Cell 2016, 29, 270–284. [Google Scholar] [CrossRef]
- Eccles, M.R.; He, S.; Ahn, A.; Slobbe, L.J.; Jeffs, A.R.; Yoon, H.S.; Baguley, B.C. MITF and PAX3 Play Distinct Roles in Melanoma Cell Migration; Outline of a “Genetic Switch” Theory Involving MITF and PAX3 in Proliferative and Invasive Phenotypes of Melanoma. Front. Oncol. 2013, 3, 229. [Google Scholar] [CrossRef] [PubMed]
- Gruis, N.A.; Weaver-Feldhaus, J.; Liu, Q.; Frye, C.; Eeles, R.; Orlow, I.; Lacombe, L.; Ponce-Castaneda, V.; Lianes, P.; Latres, E.; et al. Genetic evidence in melanoma and bladder cancers that p16 and p53 function in separate pathways of tumor suppression. Am. J. Pathol. 1995, 146, 1199–1206. [Google Scholar] [PubMed]
- Dasen, J.S.; Martinez Barbera, J.P.; Herman, T.S.; Connell, S.O.; Olson, L.; Ju, B.; Tollkuhn, J.; Baek, S.H.; Rose, D.W.; Rosenfeld, M.G. Temporal regulation of a paired-like homeodomain repressor/TLE corepressor complex and a related activator is required for pituitary organogenesis. Genes. Dev. 2001, 15, 3193–3207. [Google Scholar] [CrossRef]
- Kioussi, C.; Gross, M.K.; Gruss, P. Pax3: A paired domain gene as a regulator in PNS myelination. Neuron 1995, 15, 553–562. [Google Scholar] [CrossRef]
- Lang, D.; Epstein, J.A. Sox10 and Pax3 physically interact to mediate activation of a conserved c-RET enhancer. Hum. Mol. Genet. 2003, 12, 937–945. [Google Scholar] [CrossRef]
- Seberg, H.E.; Van Otterloo, E.; Loftus, S.K.; Liu, H.; Bonde, G.; Sompallae, R.; Gildea, D.E.; Santana, J.F.; Manak, J.R.; Pavan, W.J.; et al. TFAP2 paralogs regulate melanocyte differentiation in parallel with MITF. PLoS Genet. 2017, 13, e1006636. [Google Scholar] [CrossRef]
- Milunsky, J.M.; Maher, T.M.; Zhao, G.; Wang, Z.; Mulliken, J.B.; Chitayat, D.; Clemens, M.; Stalker, H.J.; Bauer, M.; Burch, M.; et al. Genotype-phenotype analysis of the branchio-oculo-facial syndrome. Am. J. Med. Genet. A 2011, 155A, 22–32. [Google Scholar] [CrossRef]
- Kenny, C.; Dilshat, R.; Seberg, H.E.; Van Otterloo, E.; Bonde, G.; Helverson, A.; Franke, C.M.; Steingrimsson, E.; Cornell, R.A. TFAP2 paralogs facilitate chromatin access for MITF at pigmentation and cell proliferation genes. PLoS Genet. 2022, 18, e1010207. [Google Scholar] [CrossRef]
- Hong, C.S.; Devotta, A.; Lee, Y.H.; Park, B.Y.; Saint-Jeannet, J.P. Transcription factor AP2 epsilon (Tfap2e) regulates neural crest specification in Xenopus. Dev. Neurobiol. 2014, 74, 894–906. [Google Scholar] [CrossRef]
- Campbell, N.R.; Rao, A.; Hunter, M.V.; Sznurkowska, M.K.; Briker, L.; Zhang, M.; Baron, M.; Heilmann, S.; Deforet, M.; Kenny, C.; et al. Cooperation between melanoma cell states promotes metastasis through heterotypic cluster formation. Dev. Cell 2021, 56, 2808–2825.e10. [Google Scholar] [CrossRef]
- Black, A.R.; Black, J.D.; Azizkhan-Clifford, J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell Physiol. 2001, 188, 143–160. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.T.; Wu, R.L.; Castro-Munozledo, F.; Sun, T.T. Regulation of K3 keratin gene transcription by Sp1 and AP-2 in differentiating rabbit corneal epithelial cells. Mol. Cell Biol. 1997, 17, 3056–3064. [Google Scholar] [CrossRef] [PubMed]
- Bajard, L.; Relaix, F.; Lagha, M.; Rocancourt, D.; Daubas, P.; Buckingham, M.E. A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes. Dev. 2006, 20, 2450–2464. [Google Scholar] [CrossRef] [PubMed]
- Daubas, P.; Buckingham, M.E. Direct molecular regulation of the myogenic determination gene Myf5 by Pax3, with modulation by Six1/4 factors, is exemplified by the -111 kb-Myf5 enhancer. Dev. Biol. 2013, 376, 236–244. [Google Scholar] [CrossRef]
- Sato, T.; Rocancourt, D.; Marques, L.; Thorsteinsdottir, S.; Buckingham, M. A Pax3/Dmrt2/Myf5 regulatory cascade functions at the onset of myogenesis. PLoS Genet. 2010, 6, e1000897. [Google Scholar] [CrossRef]



| Antibody | Supplier | Catalog Number |
|---|---|---|
| PAX3 | Invitrogen | 38-1801 |
| IgG Control | EpiCypher | 13-0042k |
| H3K4me3 | EpiCypher | 13-0041 |
| K3K4me1 | EpiCypher | 13-0057 |
| H3K27me3 | EpiCypher | 13-0055 |
| H3K27ac | Millipore | MABE647 |
| Classification | H3K4me1 | H3K4me3 | H3k27me3 | H3K27ac | ATAC Peak |
|---|---|---|---|---|---|
| Active | + | + | − | + | + |
| Primed | + | + | − | − | + |
| Poised | + | + | + | − | + |
| Repressed | − | − | + | − | − |
| Closed | +/− | +/− | − | +/− | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moore, S.P.G.; Ganesh Krishnan, S.; Jaswanth Kothari, R.; Prince, N.B.; Kenny, C.; Zhang, C.; Lang, D. PAX3 Regulatory Signatures and Gene Targets in Melanoma Cells. Genes 2025, 16, 577. https://doi.org/10.3390/genes16050577
Moore SPG, Ganesh Krishnan S, Jaswanth Kothari R, Prince NB, Kenny C, Zhang C, Lang D. PAX3 Regulatory Signatures and Gene Targets in Melanoma Cells. Genes. 2025; 16(5):577. https://doi.org/10.3390/genes16050577
Chicago/Turabian StyleMoore, Stephen P. G., Shripushkar Ganesh Krishnan, Rutu Jaswanth Kothari, Noah B. Prince, Colin Kenny, Chao Zhang, and Deborah Lang. 2025. "PAX3 Regulatory Signatures and Gene Targets in Melanoma Cells" Genes 16, no. 5: 577. https://doi.org/10.3390/genes16050577
APA StyleMoore, S. P. G., Ganesh Krishnan, S., Jaswanth Kothari, R., Prince, N. B., Kenny, C., Zhang, C., & Lang, D. (2025). PAX3 Regulatory Signatures and Gene Targets in Melanoma Cells. Genes, 16(5), 577. https://doi.org/10.3390/genes16050577






