Olig1/2 Drive Astrocytic Glioblastoma Proliferation Through Transcriptional Co-Regulation of Various Cyclins
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
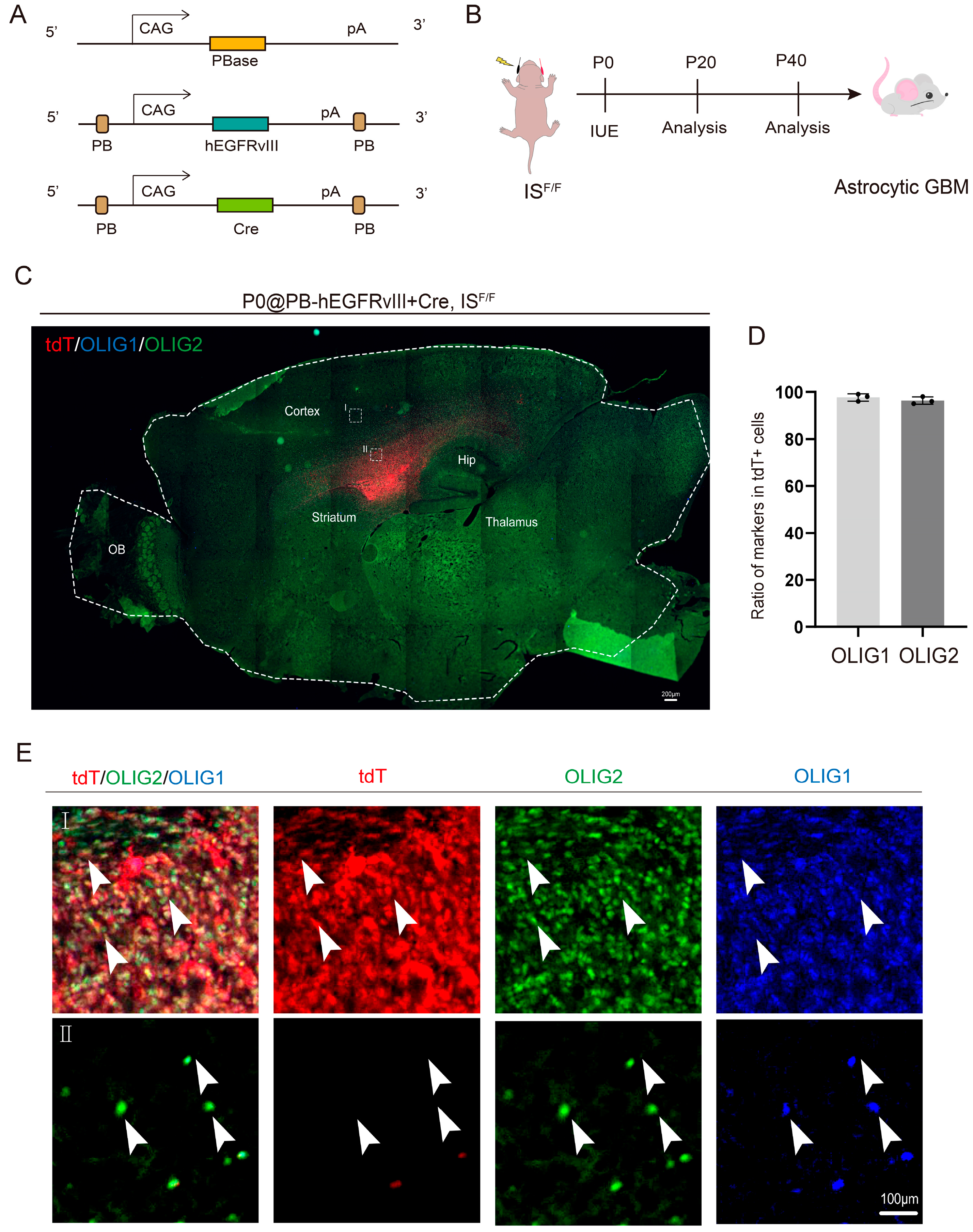
2.2. GBM Mouse Model Generation
2.3. Plasmid Construction
2.4. In Utero Electroporation
2.5. Immunofluorescence Staining
2.6. GEPIA
2.7. UALCAN
2.8. TIMER
2.9. CUT&Tag-Seq
2.10. Image Acquisition and Quantitative Analysis
3. Results
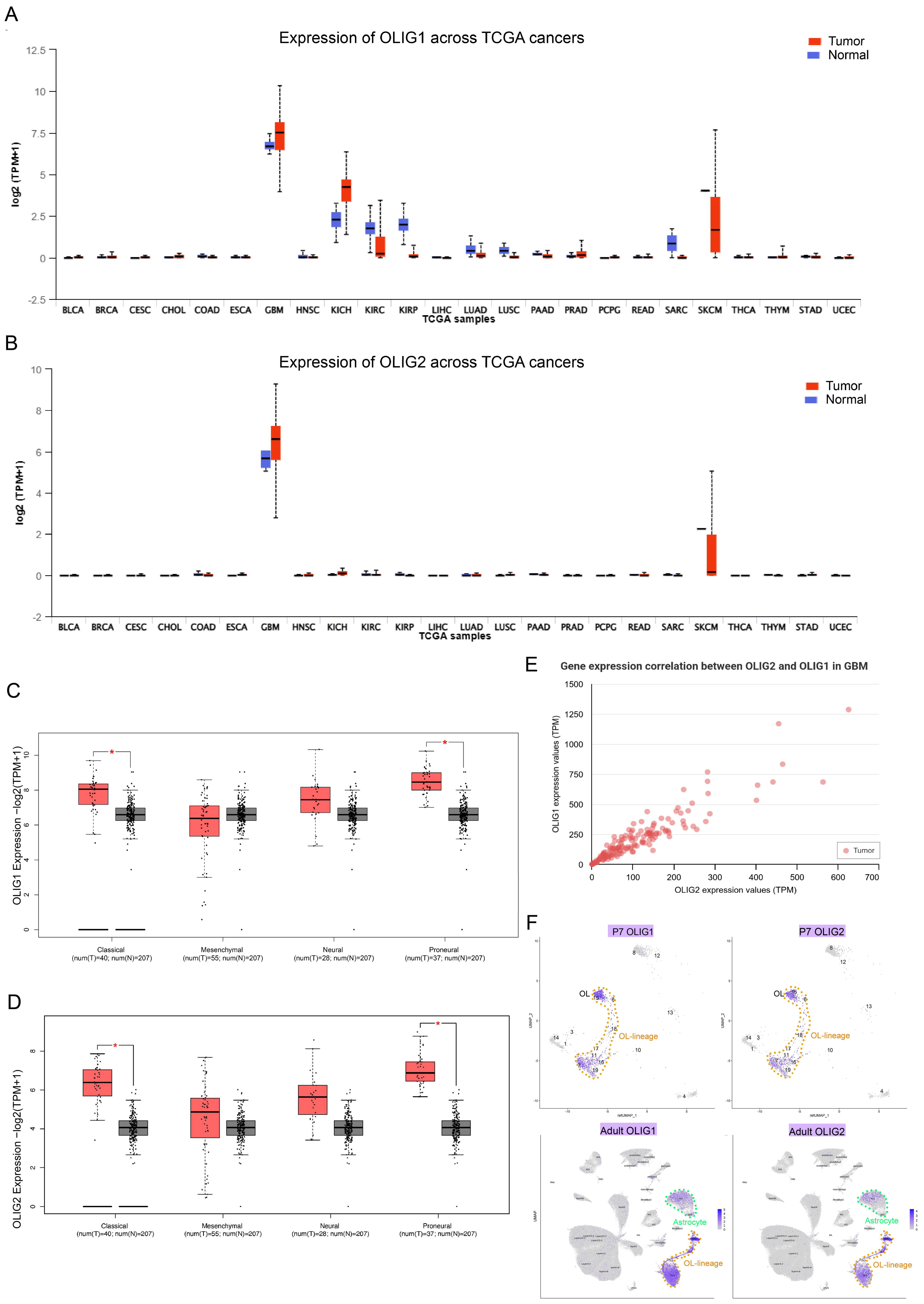
3.1. OLIG1 and OLIG2 Exhibit High Co-Expression in Glioblastoma
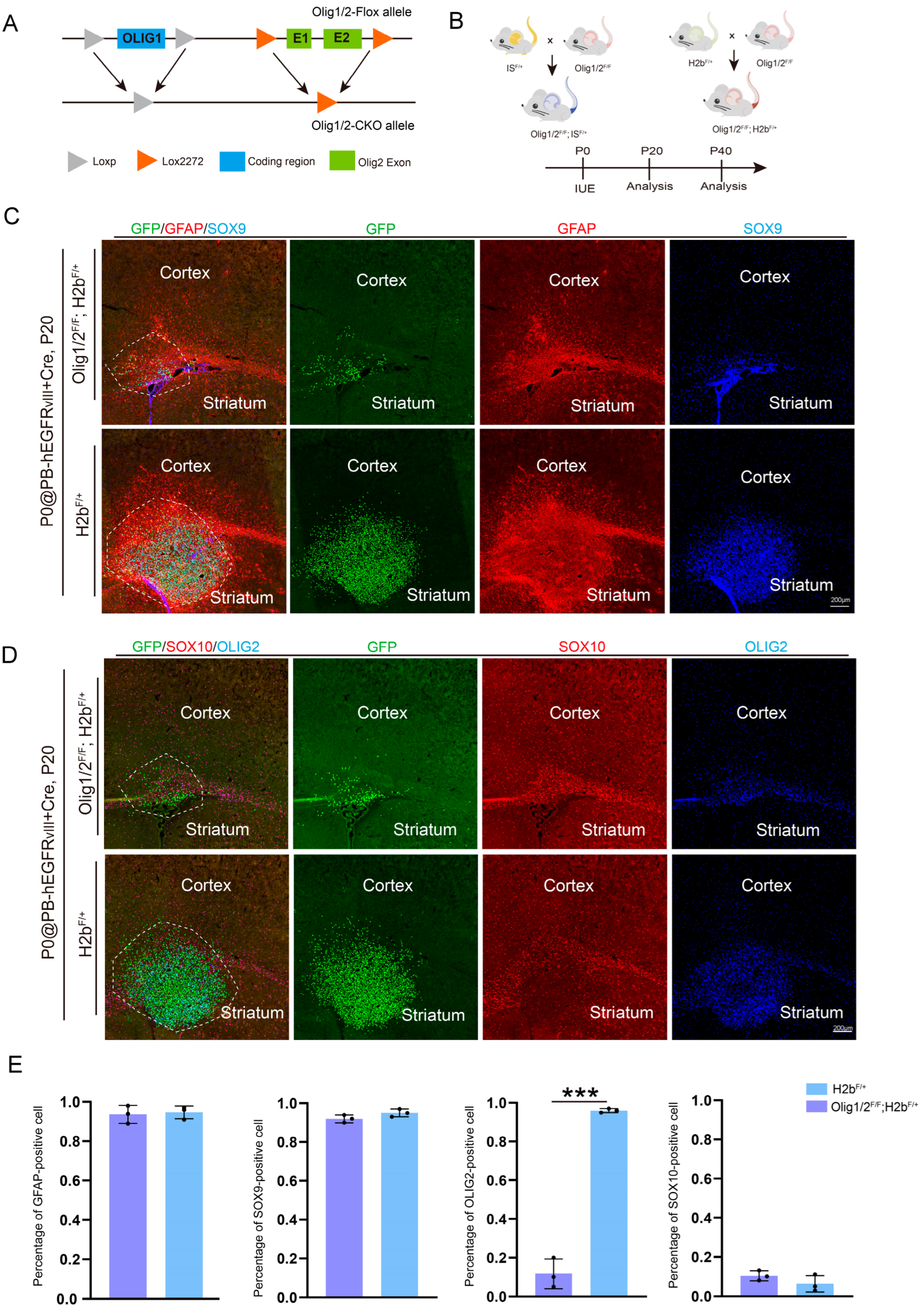
3.2. The Knockout of Olig1 and Olig2 in Astrocytic GBM Does Not Affect Astrocytic Characteristics
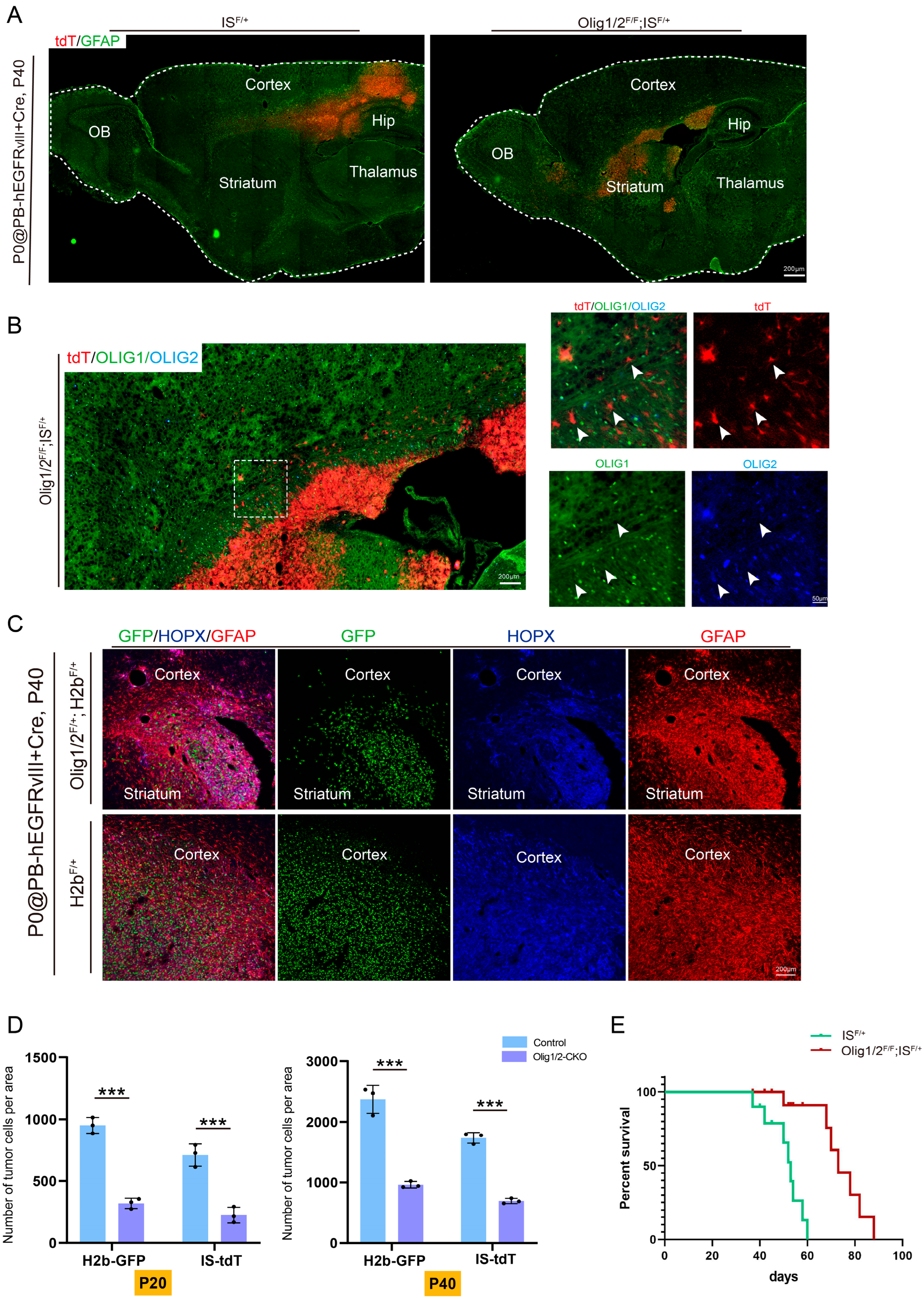
3.3. Olig1/2 Knockout Reduced Tumor Growth Without Affecting Invasion
3.4. Olig1/2 Reduced Tumor Growth by Inhibiting Glioma Cell Proliferation
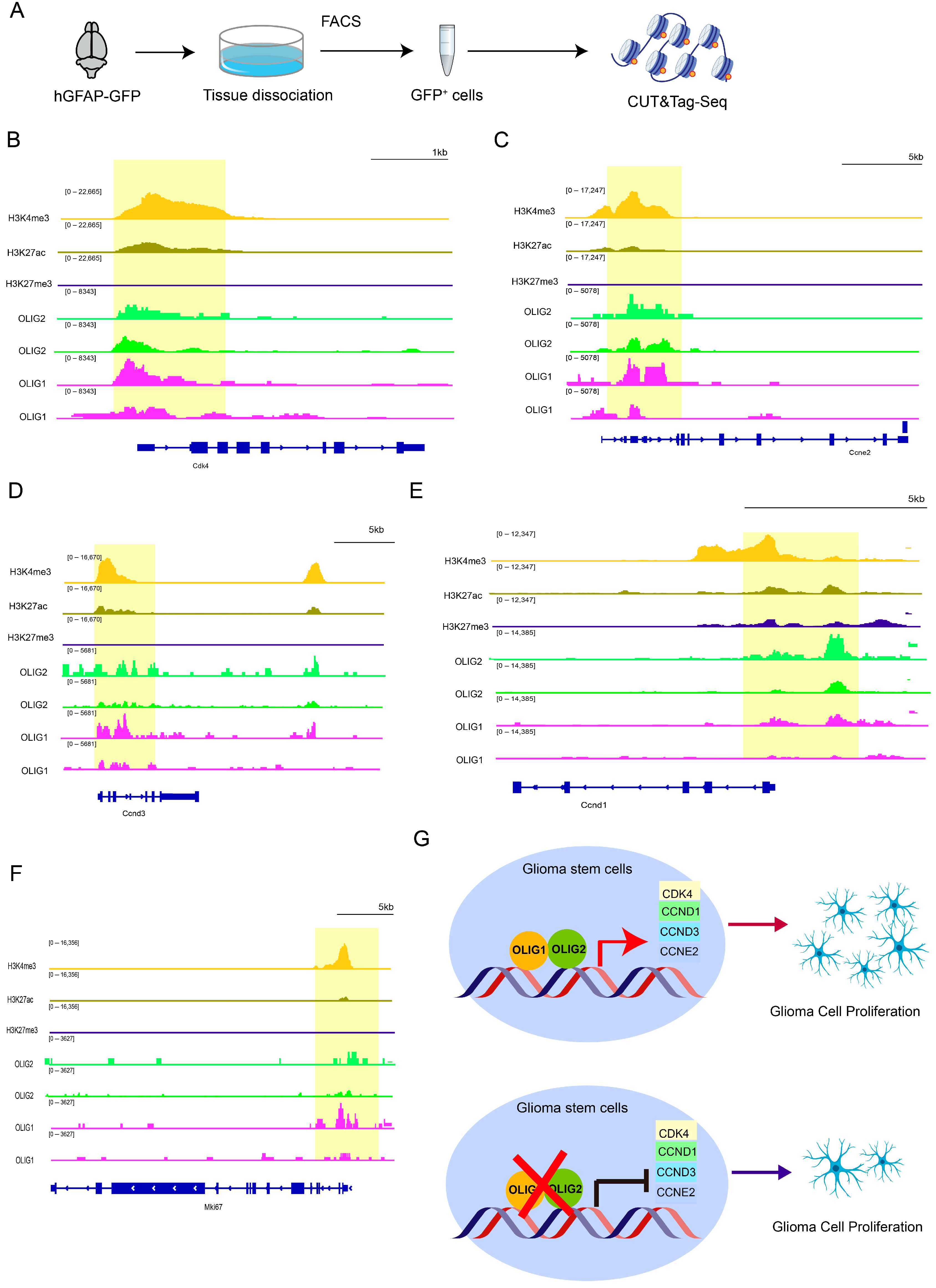
3.5. OLIG1/2 Promote Cell Proliferation by Binding to Various Cyclins
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M.; et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 2019, 178, 835–849.e21. [Google Scholar] [CrossRef] [PubMed]
- Paes de Faria, J.; Kessaris, N.; Andrew, P.; Richardson, W.D.; Li, H. New Olig1 null mice confirm a non-essential role for Olig1 in oligodendrocyte development. BMC Neurosci. 2014, 15, 12. [Google Scholar] [CrossRef]
- Schwartzentruber, J.; Korshunov, A.; Liu, X.Y.; Jones, D.T.; Pfaff, E.; Jacob, K.; Sturm, D.; Fontebasso, A.M.; Quang, D.A.; Tonjes, M.; et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012, 482, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, L.; Jiang, M.; Zhao, C.; Liu, X.; Berry, K.; Waisman, A.; Langseth, A.J.; Novitch, B.G.; Bergles, D.E.; et al. Olig2 Ablation in Immature Oligodendrocytes Does Not Enhance CNS Myelination and Remyelination. J. Neurosci. 2022, 42, 8542–8555. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, Y.; Kim, B.; Wang, H.; Zhao, C.; He, X.; Liu, L.; Liu, W.; Wu, L.M.; Mao, M.; et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 2013, 152, 248–261. [Google Scholar] [CrossRef]
- Zhang, K.; Chen, S.; Yang, Q.; Guo, S.; Chen, Q.; Liu, Z.; Li, L.; Jiang, M.; Li, H.; Hu, J.; et al. The Oligodendrocyte Transcription Factor 2 OLIG2 regulates transcriptional repression during myelinogenesis in rodents. Nat. Commun. 2022, 13, 1423. [Google Scholar] [CrossRef]
- Lu, Q.R.; Sun, T.; Zhu, Z.; Ma, N.; Garcia, M.; Stiles, C.D.; Rowitch, D.H. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 2002, 109, 75–86. [Google Scholar] [CrossRef]
- Meijer, D.H.; Kane, M.F.; Mehta, S.; Liu, H.; Harrington, E.; Taylor, C.M.; Stiles, C.D.; Rowitch, D.H. Separated at birth? The functional and molecular divergence of OLIG1 and OLIG2. Nat. Rev. Neurosci. 2012, 13, 819–831. [Google Scholar] [CrossRef]
- Wegener, A.; Deboux, C.; Bachelin, C.; Frah, M.; Kerninon, C.; Seilhean, D.; Weider, M.; Wegner, M.; Nait-Oumesmar, B. Gain of Olig2 function in oligodendrocyte progenitors promotes remyelination. Brain 2015, 138, 120–135. [Google Scholar] [CrossRef]
- Zhou, Q.; Anderson, D.J. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 2002, 109, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Breunig, J.J.; Levy, R.; Antonuk, C.D.; Molina, J.; Dutra-Clarke, M.; Park, H.; Akhtar, A.A.; Kim, G.B.; Hu, X.; Bannykh, S.I.; et al. Ets Factors Regulate Neural Stem Cell Depletion and Gliogenesis in Ras Pathway Glioma. Cell Rep. 2015, 12, 258–271. [Google Scholar] [CrossRef]
- Kupp, R.; Shtayer, L.; Tien, A.C.; Szeto, E.; Sanai, N.; Rowitch, D.H.; Mehta, S. Lineage-Restricted OLIG2-RTK Signaling Governs the Molecular Subtype of Glioma Stem-like Cells. Cell Rep. 2016, 16, 2838–2845. [Google Scholar] [CrossRef]
- Lu, Q.R.; Park, J.K.; Noll, E.; Chan, J.A.; Alberta, J.; Yuk, D.; Alzamora, M.G.; Louis, D.N.; Stiles, C.D.; Rowitch, D.H.; et al. Oligodendrocyte lineage genes (OLIG) as molecular markers for human glial brain tumors. Proc. Natl. Acad. Sci. USA 2001, 98, 10851–10856. [Google Scholar] [CrossRef] [PubMed]
- Ligon, K.L.; Alberta, J.A.; Kho, A.T.; Weiss, J.; Kwaan, M.R.; Nutt, C.L.; Louis, D.N.; Stiles, C.D.; Rowitch, D.H. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J. Neuropathol. Exp. Neurol. 2004, 63, 499–509. [Google Scholar] [CrossRef]
- Suvà, M.L.; Rheinbay, E.; Gillespie, S.M.; Patel, A.P.; Wakimoto, H.; Rabkin, S.D.; Riggi, N.; Chi, A.S.; Cahill, D.P.; Nahed, B.V.; et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 2014, 157, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Kosty, J.; Lu, F.; Kupp, R.; Mehta, S.; Lu, Q.R. Harnessing OLIG2 function in tumorigenicity and plasticity to target malignant gliomas. Cell Cycle 2017, 16, 1654–1660. [Google Scholar] [CrossRef]
- Liao, Y.; Luo, Z.; Deng, Y.; Zhang, F.; Rao, R.; Wang, J.; Xu, L.; Kumar, S.S.; Sengupta, S.; DeWire-Schottmiller, M.; et al. OLIG2 maintenance is not essential for diffuse intrinsic pontine glioma cell line growth but regulates tumor phenotypes. Neuro-Oncology 2021, 23, 1183–1196. [Google Scholar] [CrossRef]
- Ligon, K.L.; Huillard, E.; Mehta, S.; Kesari, S.; Liu, H.; Alberta, J.A.; Bachoo, R.M.; Kane, M.; Louis, D.N.; Depinho, R.A.; et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron 2007, 53, 503–517. [Google Scholar] [CrossRef]
- Lu, F.; Chen, Y.; Zhao, C.; Wang, H.; He, D.; Xu, L.; Wang, J.; He, X.; Deng, Y.; Lu, E.E.; et al. Olig2-Dependent Reciprocal Shift in PDGF and EGF Receptor Signaling Regulates Tumor Phenotype and Mitotic Growth in Malignant Glioma. Cancer Cell 2016, 29, 669–683. [Google Scholar] [CrossRef]
- Wang, Z.; Tian, Y.; Fu, T.; Yang, F.; Li, J.; Yang, L.; Zhang, W.; Zheng, W.; Jiang, X.; Xu, Z.; et al. Coordinated regulation of cortical astrocyte maturation by OLIG1 and OLIG2 through BMP7 signaling modulation. J. Genet. Genom. 2025, in press. [CrossRef]
- Tian, Y.; Wang, Z.; Yang, F.; Zhang, W.; Li, J.; Yang, L.; Fu, T.; Zheng, W.; Xu, Z.; Ma, T.; et al. Orchestrating the Acquisition of Oligodendrocyte Precursor Cell versus Olfactory Bulb Interneuron Fates through Olig1/2 during Mammalian Cortical Gliogenesis and Gliomagenesis. bioRxiv 2024. [Google Scholar] [CrossRef]
- Chen, J.; Li, Y.; Yu, T.S.; McKay, R.M.; Burns, D.K.; Kernie, S.G.; Parada, L.F. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 2012, 488, 522–526. [Google Scholar] [CrossRef]
- Verma, R.; Chen, X.; Xin, D.; Luo, Z.; Ogurek, S.; Xin, M.; Rao, R.; Berry, K.; Lu, Q.R. Olig1/2-Expressing Intermediate Lineage Progenitors Are Predisposed to PTEN/p53-Loss-Induced Gliomagenesis and Harbor Specific Therapeutic Vulnerabilities. Cancer Res. 2023, 83, 890–905. [Google Scholar] [CrossRef]
- Ligon, K.L.; Fancy, S.P.; Franklin, R.J.; Rowitch, D.H. Olig gene function in CNS development and disease. Glia 2006, 54, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Szu, J.I.; Tsigelny, I.F.; Wojcinski, A.; Kesari, S. Biological functions of the Olig gene family in brain cancer and therapeutic targeting. Front. Neurosci. 2023, 17, 1129434. [Google Scholar] [CrossRef]
- Cloughesy, T.F.; Cavenee, W.K.; Mischel, P.S. Glioblastoma: From molecular pathology to targeted treatment. Annu. Rev. Pathol. 2014, 9, 1–25. [Google Scholar] [CrossRef]
- Li, J.L.; Yang, F.H.; Tian, Y.; Wang, Z.W.; Qi, D.S.; Yang, Z.G.; Song, J.G.; Ding, J.; Wang, X.; Zhang, Z.Z. Lateral/caudal ganglionic eminence makes limited contribution to cortical oligodendrocytes. eLife 2024, 13, RP94317. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Han, D.; Song, X.; Gao, Y.; Li, Z.; Li, X.; Yang, Z.; Xu, Z. Context-dependent regulation of Notch signaling in glial development and tumorigenesis. Sci. Adv. 2023, 9, eadi2167. [Google Scholar] [CrossRef]
- Li, Z.; Shang, Z.; Sun, M.; Jiang, X.; Tian, Y.; Yang, L.; Wang, Z.; Su, Z.; Liu, G.; Li, X.; et al. Transcription factor Sp9 is a negative regulator of D1-type MSN development. Cell Death Discov. 2022, 8, 301. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Wang, Z.; Lindtner, S.; Yang, L.; Shang, Z.; Tian, Y.; Guo, R.; You, Y.; Zhou, W.; Rubenstein, J.L.; et al. Dlx1/2-dependent expression of Meis2 promotes neuronal fate determination in the mammalian striatum. Development 2022, 149, dev200035. [Google Scholar] [CrossRef]
- An, Z.; Aksoy, O.; Zheng, T.; Fan, Q.W.; Weiss, W.A. Epidermal growth factor receptor and EGFRvIII in glioblastoma: Signaling pathways and targeted therapies. Oncogene 2018, 37, 1561–1575. [Google Scholar] [CrossRef]
- Cai, Y.; Zhao, Z.; Shi, M.; Zheng, M.; Gong, L.; He, M. Embryonic origins of forebrain oligodendrocytes revisited by combinatorial genetic fate mapping. eLife 2024, 13, RP95406. [Google Scholar] [CrossRef]
- Yakovlev, A.G.; Faden, A.I. Caspase-dependent apoptotic pathways in CNS injury. Mol. Neurobiol. 2001, 24, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, G.; Yang, L.; Sun, M.; Zhang, Z.; Xu, Z.; Gao, Y.; Jiang, X.; Su, Z.; Li, X.; et al. BMP7 expression in mammalian cortical radial glial cells increases the length of the neurogenic period. Protein Cell 2024, 15, 21–35. [Google Scholar] [CrossRef]
- Shang, Z.; Yang, L.; Wang, Z.; Tian, Y.; Gao, Y.; Su, Z.; Guo, R.; Li, W.; Liu, G.; Li, X.; et al. The transcription factor Zfp503 promotes the D1 MSN identity and represses the D2 MSN identity. Front. Cell Dev. Biol. 2022, 10, 948331. [Google Scholar] [CrossRef]
- Wen, Y.; Su, Z.; Wang, Z.; Yang, L.; Liu, G.; Shang, Z.; Duan, Y.; Du, H.; Li, Z.; You, Y.; et al. Transcription Factor VAX1 Regulates the Regional Specification of the Subpallium Through Repressing Gsx2. Mol. Neurobiol. 2021, 58, 3729–3744. [Google Scholar] [CrossRef]
- Lee, S.K.; Lee, B.; Ruiz, E.C.; Pfaff, S.L. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005, 19, 282–294. [Google Scholar] [CrossRef]
- Li, H.; de Faria, J.P.; Andrew, P.; Nitarska, J.; Richardson, W.D. Phosphorylation regulates OLIG2 cofactor choice and the motor neuron-oligodendrocyte fate switch. Neuron 2011, 69, 918–929. [Google Scholar] [CrossRef] [PubMed]
- Furusho, M.; Ono, K.; Takebayashi, H.; Masahira, N.; Kagawa, T.; Ikeda, K.; Ikenaka, K. Involvement of the Olig2 transcription factor in cholinergic neuron development of the basal forebrain. Dev. Biol. 2006, 293, 348–357. [Google Scholar] [CrossRef]
- Lu, Q.R.; Yuk, D.; Alberta, J.A.; Zhu, Z.; Pawlitzky, I.; Chan, J.; McMahon, A.P.; Stiles, C.D.; Rowitch, D.H. Sonic hedgehog–regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron 2000, 25, 317–329. [Google Scholar] [CrossRef]
- Tatsumi, K.; Isonishi, A.; Yamasaki, M.; Kawabe, Y.; Morita-Takemura, S.; Nakahara, K.; Terada, Y.; Shinjo, T.; Okuda, H.; Tanaka, T.; et al. Olig2-Lineage Astrocytes: A Distinct Subtype of Astrocytes That Differs from GFAP Astrocytes. Front. Neuroanat. 2018, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Tatsumi, K.; Kinugawa, K.; Isonishi, A.; Kitabatake, M.; Okuda, H.; Takemura, S.; Tanaka, T.; Mori, E.; Wanaka, A. Olig2-astrocytes express neutral amino acid transporter SLC7A10 (Asc-1) in the adult brain. Mol. Brain 2021, 14, 163. [Google Scholar] [CrossRef]
- Wang, H.; Xu, L.; Lai, C.; Hou, K.; Chen, J.; Guo, Y.; Sambangi, A.; Swaminathan, S.; Xie, C.; Wu, Z.; et al. Region-specific distribution of Olig2-expressing astrocytes in adult mouse brain and spinal cord. Mol. Brain 2021, 14, 36. [Google Scholar] [CrossRef]
- Marshall, C.A.; Novitch, B.G.; Goldman, J.E. Olig2 directs astrocyte and oligodendrocyte formation in postnatal subventricular zone cells. J. Neurosci. 2005, 25, 7289–7298. [Google Scholar] [CrossRef] [PubMed]
- Myers, B.L.; Brayer, K.J.; Paez-Beltran, L.E.; Villicana, E.; Keith, M.S.; Suzuki, H.; Newville, J.; Anderson, R.H.; Lo, Y.; Mertz, C.M.; et al. Transcription factors ASCL1 and OLIG2 drive glioblastoma initiation and co-regulate tumor cell types and migration. Nat. Commun. 2024, 15, 10363. [Google Scholar] [CrossRef]
- Zhang, L.; He, X.; Liu, X.; Zhang, F.; Huang, L.F.; Potter, A.S.; Xu, L.; Zhou, W.; Zheng, T.; Luo, Z.; et al. Single-Cell Transcriptomics in Medulloblastoma Reveals Tumor-Initiating Progenitors and Oncogenic Cascades during Tumorigenesis and Relapse. Cancer Cell 2019, 36, 302–318.e7. [Google Scholar] [CrossRef]
- Zou, F.; Xie, G.; Ma, J.A.; Zhou, D.A.; Jiang, Y.I.; Zheng, J.Y. Epidermal growth factor receptor mutation heterogeneity analysis of pulmonary sarcomatoid carcinoma successfully treated with erlotinib: A case report. Oncol. Lett. 2015, 9, 2239–2243. [Google Scholar] [CrossRef][Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Y.; Wang, Z.; Sun, M.; Li, J.; Zheng, W.; Yang, F.; Zhang, Z. Olig1/2 Drive Astrocytic Glioblastoma Proliferation Through Transcriptional Co-Regulation of Various Cyclins. Genes 2025, 16, 573. https://doi.org/10.3390/genes16050573
Tian Y, Wang Z, Sun M, Li J, Zheng W, Yang F, Zhang Z. Olig1/2 Drive Astrocytic Glioblastoma Proliferation Through Transcriptional Co-Regulation of Various Cyclins. Genes. 2025; 16(5):573. https://doi.org/10.3390/genes16050573
Chicago/Turabian StyleTian, Yu, Ziwu Wang, Mengge Sun, Jialin Li, Wenhui Zheng, Feihong Yang, and Zhuangzhi Zhang. 2025. "Olig1/2 Drive Astrocytic Glioblastoma Proliferation Through Transcriptional Co-Regulation of Various Cyclins" Genes 16, no. 5: 573. https://doi.org/10.3390/genes16050573
APA StyleTian, Y., Wang, Z., Sun, M., Li, J., Zheng, W., Yang, F., & Zhang, Z. (2025). Olig1/2 Drive Astrocytic Glioblastoma Proliferation Through Transcriptional Co-Regulation of Various Cyclins. Genes, 16(5), 573. https://doi.org/10.3390/genes16050573





