A Patient with a Small Deletion Affecting Only Exon 1-Intron 1 of the NXF5 Gene: Potential Evidence Supporting Its Role in Neurodevelopmental Disorders
Abstract
1. Introduction
2. Materials and Methods
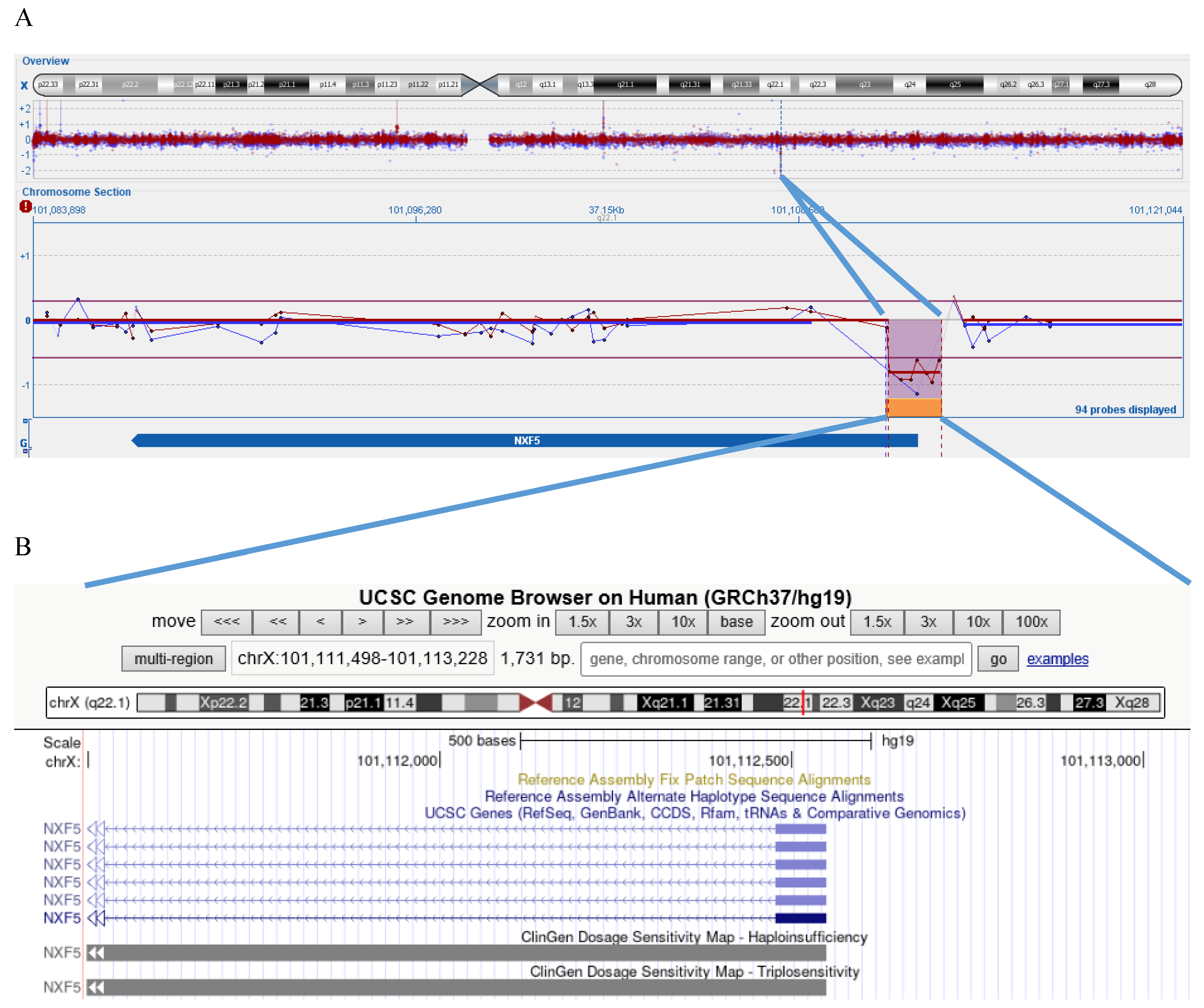
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ID | Intellectual Disability |
| ASD | Autism Spectrum Disorders |
| MRI | Brain Magnetic Resonance Imaging |
| EEG | Electroencephalogram |
| aCGH | Array Comparative Genomic Hybridization |
| NXF5 | Nuclear RNA Export Factor 5 |
| NXF2 | Nuclear RNA Export Factor 2 |
| NXF2B | Nuclear RNA Export Factor 2B |
| NXF4 | Nuclear RNA Export Factor 4 |
| NXF3 | Nuclear RNA Export Factor 3 |
| CNVs | Copy Number Variants |
| M | Male |
| F | Female |
| FISH | Fluorescent In Situ Hybridization |
| ADHD | Attention Deficit Hyperactivity Disorder |
| mat | Maternal |
| dn | De Novo |
| DD | Developmental Delay |
| ADI-R | Autism Diagnostic Interview-Revised |
| XLID | X-Linked Intellectual Disabilities |
References
- Maulik, P.K.; Mascarenhas, M.N.; Mathers, C.D.; Dua, T.; Saxena, S. Prevalence of intellectual disability: A meta-analysis of population-based studies. Res. Dev. Disabil. 2011, 32, 419–436. [Google Scholar] [CrossRef] [PubMed]
- Vissers, L.E.; Gilissen, C.; Veltman, J.A. Genetic studies in intellectual disability and related disorders. Nat. Rev. Genet. 2016, 17, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Leonard, H.; Wen, X. The epidemiology of mental retardation: Challenges and opportunities in the new millennium. Ment. Retard. Dev. Disabil. Res. Rev. 2002, 8, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Jun, L.; Frints, S.; Duhamel, H.; Herold, A.; Abad-Rodrigues, J.; Dotti, C.; Izaurralde, E.; Marynen, P.; Froyen, G. NXF5, a novel member of the nuclear RNA export factor family, is lost in a male patient with a syndromic form of mental retardation. Curr. Biol. 2001, 11, 1381–1391. [Google Scholar] [CrossRef] [PubMed]
- Frints, S.G.M.; Jun, L.; Fryns, J.-P.; Devriendt, K.; Teulingkx, R.; Van den Berghe, L.; De Vos, B.; Borghgraef, M.; Chelly, J.; Des Portes, V.; et al. Inv(X)(p21.1; q22.1) in a man with mental retardation, short stature, general muscle wasting, and facial dysmorphism: Clinical study and mutation analysis of the NXF5 gene. Am. J. Med. Genet. Part A 2003, 119A, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Froyen, G.; Van Esch, H.; Bauters, M.; Hollanders, K.; Frints, S.G.M.; Vermeesch, J.R.; Devriendt, K.; Fryns, J.-P.; Marynen, P. Detection of genomic copy number changes in patients with idiopathic mental retardation by high-resolution X-array-CGH: Important role for increased gene dosage of XLMR genes. Hum. Mutat. 2007, 28, 1034–1042. [Google Scholar] [CrossRef] [PubMed]
- Grillo, L.; Reitano, S.; Belfiore, G.; Spalletta, A.; Amata, S.; Bottitta, M.; Barone, C.; Falco, M.; Fichera, M.; Romano, C. Familial 1.1 Mb deletion in chromosome Xq22.1 associated with mental retardation and behavioural disorders in female patients. Eur. J. Med. Genet. 2010, 53, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-P.; Su, Y.-N.; Lin, H.-H.; Chern, S.-R.; Tsai, F.-J.; Wu, P.-C.; Lee, C.-C.; Chen, Y.-T.; Wang, W. De novo duplication of Xq22.1→q24 with a disruption of the NXF gene cluster in a mentally retarded woman with short stature and premature ovarian failure. Taiwan. J. Obstet. Gynecol. 2011, 50, 334–339. [Google Scholar] [CrossRef][Green Version]
- Kearney, H.M.; Thorland, E.C.; Brown, K.K.; Quintero-Rivera, F.; South, S.T.; Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee. American College of Medical Genetics standard and Guidelines for interpretation and reporting of postnatal constitutional of copy number variants. Genet. Med. 2011, 13, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Hastings, R.; Howell, R.; Dagna Bricarelli, F.; Kristoffersson, U.; Cavani, S. Specific Constitutional Cytogenetic Guidelines of European Cytogeneticist Association E.C.A.2012. Available online: https://www.e-c-a.eu (accessed on 4 April 2025).
- Martin, K.C.; Barad, M.; Kandel, E.R. Local protein synthesis and its role in synapse-specific plasticity. Curr. Opin. Neurobiol. 2000, 10, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Corbin, F.; Bouillon, M.; Fortin, A.; Morin, S.; Rousseau, F.; Khandjian, E.W. The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes. Hum. Molec. Genet. 1997, 6, 1465–1472. [Google Scholar] [CrossRef]
- Piton, A.; Redin, C.; Mandel, J.-L. XLID-causing mutations and associated genes challenged in light of data from large-scale human exome sequencing. Am. J. Hum. Genet. 2013, 93, 368–383. [Google Scholar] [CrossRef] [PubMed]
- Mendes, M.; Chen, D.Z.; Engchuan, W.; Leal, T.P.; Thiruvahindrapuram, B.; Trost, B.; Howe, J.L.; Pellecchia, G.; Nalpathamkalam, T.; Alexandrova, R.; et al. Chromosome X-wide common variant association study in autism spectrum disorder. Am. J. Hum. Genet. 2025, 112, 135–153. [Google Scholar] [CrossRef] [PubMed]
- Esposito, T.; Lea, R.A.; Maher, B.H.; Moses, D.; Cox, H.C.; Magliocca, S.; Angius, A.; Nyholt, D.R.; Titus, T.; Kay, T.; et al. Unique X-linked familial FSGS with co-segregating heart block disorder is associated with a mutation in the NXF5 gene. Hum. Mol. Genet. 2013, 22, 3654–3666. [Google Scholar] [CrossRef] [PubMed]
- Vanmarsenille, L.; Verbeeck, J.; Belet, S.; Roebroek, A.J.; Van de Putte, T.; Nevelsteen, J.; Callaerts-Vegh, Z.; D’Hooge, R.; Marynen, P.; Froyen, G. Generation and characterization of an Nxf7 knockout mouse to study NXF5 deficiency in a patient with intellectual disability. PLoS ONE 2013, 8, e64144. [Google Scholar] [CrossRef] [PubMed]
- Tisserant, E.; Vitobello, A.; Callegarin, D.; Verdez, S.; Bruel, A.L.; Aho Glele, L.S.; Sorlin, A.; Viora-Dupont, E.; Konyukh, M.; Marle, N.; et al. Copy number variants calling from WES data through eXome hidden Markov model (XHMM) identifies additional 2.5% pathogenic genomic imbalances smaller than 30 kb undetected by array-CGH. Ann. Hum. Genet. 2022, 86, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, K.D.; Shaffer, L.G.; Aradhya, S.; Gastier-Foster, J.M.; Patel, A.; Rudd, M.K.; Biggerstaff, J.S.; Sanger, W.G.; Schwartz, S.; Tepperberg, J.H.; et al. Variability in interpreting and reporting copy number changes detected by array-based technology in clinical laboratories. Genet. Med. 2009, 11, 866–873. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Saw, P.E. Integration in Biomedical Science 2024: Emerging Trends in the Post-Pandemic Era. BIO Integr. 2024, 5, 998. [Google Scholar] [CrossRef]
| Patients Published in | Sex | ID | Peculiar Characteristics | Body Parameters | Language | ASD | Other Features | Unbalance and Method |
|---|---|---|---|---|---|---|---|---|
| [5] | M | Moderate–severe | Present | Short stature; | Absent | Present | Muscle waste | Inversion |
| low weight; | FISH | |||||||
| microcephaly | Breakpoint in NXF5 | |||||||
| [6] | M | Moderate | Present | Normal | Dealy | Present | ADHD | Microduplication involving 15 genes |
| hypotonia | aCGH | |||||||
| [7] | F | Mild | Present | Short stature | Normal | No | Borderline personality disorder; | Microdeletion, de novo, involving 15 genes; |
| epilepsy | X-inactivation skewed 90:10 | |||||||
| [7] | F | Severe | Present | Short stature; | Absent | Present | Hypotonia; | Microdeletion, mat, involving 15 genes; |
| microcephaly | scoliosis | X-inactivation random |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tapia, Y.Y.; Ciaccio, C.; Bacınoğlu, M.B.; D’Arrigo, S.; Sciacca, F.L. A Patient with a Small Deletion Affecting Only Exon 1-Intron 1 of the NXF5 Gene: Potential Evidence Supporting Its Role in Neurodevelopmental Disorders. Genes 2025, 16, 571. https://doi.org/10.3390/genes16050571
Tapia YY, Ciaccio C, Bacınoğlu MB, D’Arrigo S, Sciacca FL. A Patient with a Small Deletion Affecting Only Exon 1-Intron 1 of the NXF5 Gene: Potential Evidence Supporting Its Role in Neurodevelopmental Disorders. Genes. 2025; 16(5):571. https://doi.org/10.3390/genes16050571
Chicago/Turabian StyleTapia, Yessica Yesenia, Claudia Ciaccio, Merve Begüm Bacınoğlu, Stefano D’Arrigo, and Francesca Luisa Sciacca. 2025. "A Patient with a Small Deletion Affecting Only Exon 1-Intron 1 of the NXF5 Gene: Potential Evidence Supporting Its Role in Neurodevelopmental Disorders" Genes 16, no. 5: 571. https://doi.org/10.3390/genes16050571
APA StyleTapia, Y. Y., Ciaccio, C., Bacınoğlu, M. B., D’Arrigo, S., & Sciacca, F. L. (2025). A Patient with a Small Deletion Affecting Only Exon 1-Intron 1 of the NXF5 Gene: Potential Evidence Supporting Its Role in Neurodevelopmental Disorders. Genes, 16(5), 571. https://doi.org/10.3390/genes16050571







