CYP2C9 Promoter Variable Number Tandem Repeat Polymorphism in a Dominican Population: Exploring Differences with Genetic Ancestry
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. CYP2C9 Genotyping and Genetic Ancestry Analysis
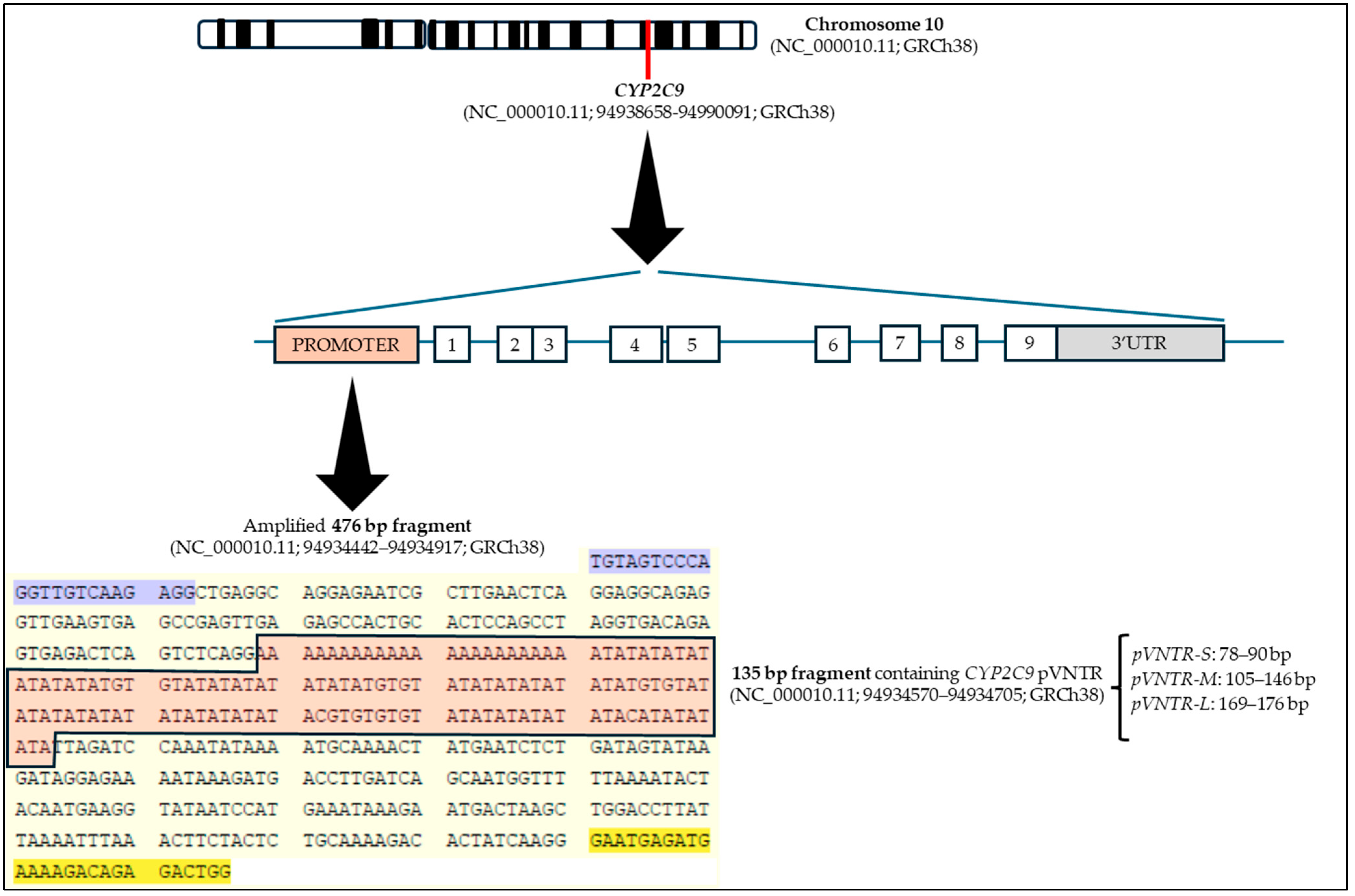
2.3. Determination of CYP2C9 pVNTR
2.4. Data Analysis
3. Results
3.1. Frequency of CYP2C9 pVNTR in the Dominican Population
3.2. Linkage Disequilibrium Analysis Between CYP2C9 pVNTR-S and CYP2C9*3 in the Dominican Population
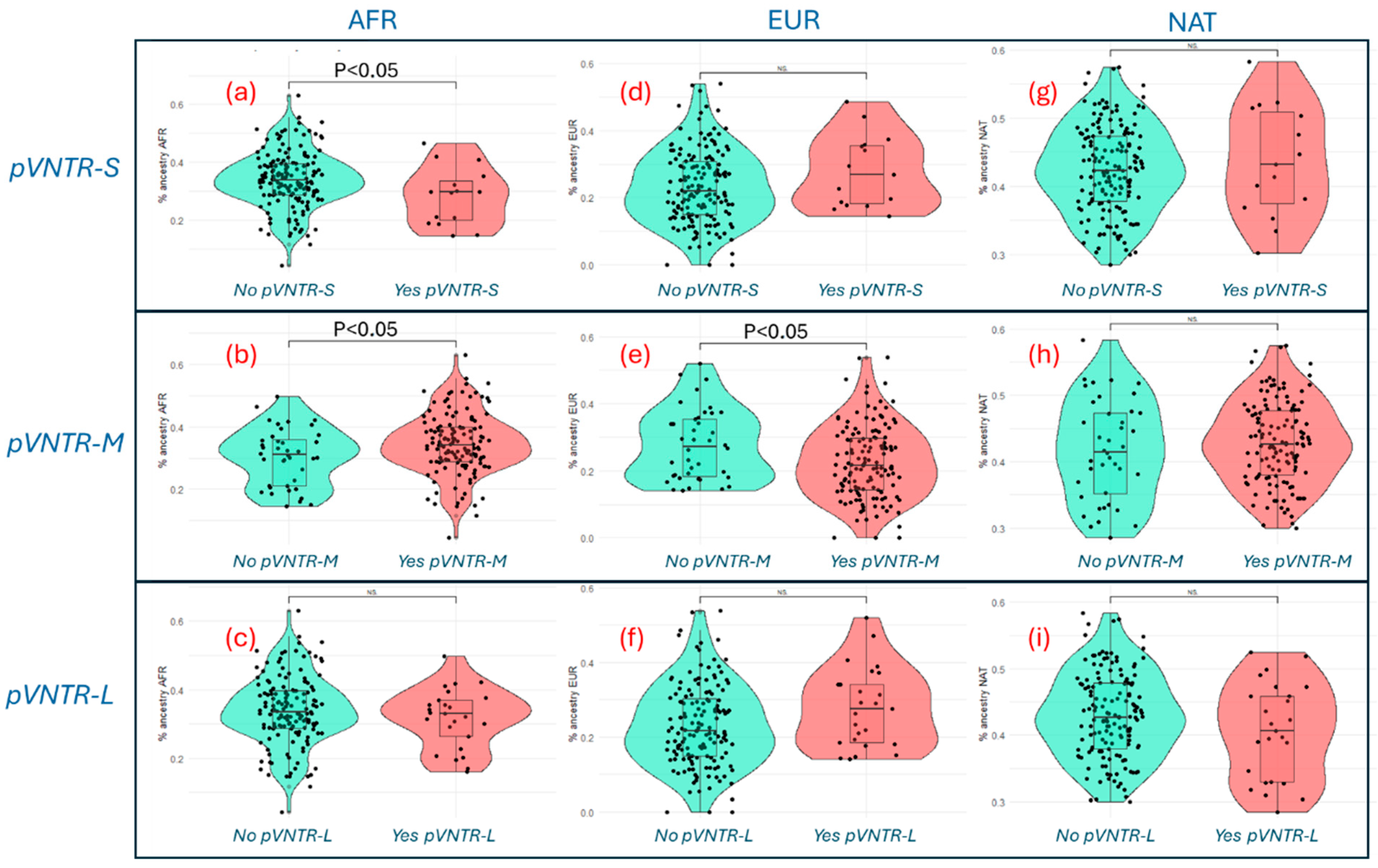
3.3. CYP2C9 pVNTR Ancestry Analysis in the Dominican Population
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Theken, K.N.; Lee, C.R.; Gong, L.; Caudle, K.E.; Formea, C.M.; Gaedigk, A.; Klein, T.E.; Agúndez, J.A.G.; Grosser, T. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2C9 and Nonsteroidal Anti-Inflammatory Drugs. Clin. Pharmacol. Ther. 2020, 108, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.A.; Caudle, K.E.; Gong, L.; Whirl-Carrillo, M.; Stein, C.M.; Scott, S.A.; Lee, M.T.; Gage, B.F.; Kimmel, S.E.; Perera, M.A.; et al. CPIC Warfarin Dosing, 2C9, 4F2, VKOR. Clin. Pharmacol. Ther. 2017, 102, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Karnes, J.H.; Rettie, A.E.; Somogyi, A.A.; Huddart, R.; Fohner, A.E.; Formea, C.M.; Ta Michael Lee, M.; Llerena, A.; Whirl-Carrillo, M.; Klein, T.E.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C9 and HLA-B Genotypes and Phenytoin Dosing: 2020 Update. Clin. Pharmacol. Ther. 2021, 109, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Cooper-DeHoff, R.M.; Niemi, M.; Ramsey, L.B.; Luzum, J.A.; Tarkiainen, E.K.; Straka, R.J.; Gong, L.; Tuteja, S.; Wilke, R.A.; Wadelius, M.; et al. The Clinical Pharmacogenetics Implementation Consortium Guideline for SLCO1B1, ABCG2, and CYP2C9 genotypes and Statin-Associated Musculoskeletal Symptoms. Clin. Pharmacol. Ther. 2022, 111, 1007–1021. [Google Scholar] [CrossRef] [PubMed]
- Sangkuhl, K.; Claudio-Campos, K.; Cavallari, L.H.; Agundez, J.A.G.; Whirl-Carrillo, M.; Duconge, J.; Del Tredici, A.L.; Wadelius, M.; Rodrigues Botton, M.; Woodahl, E.L.; et al. PharmVar GeneFocus: CYP2C9. Clin. Pharmacol. Ther. 2021, 110, 662–676. [Google Scholar] [CrossRef] [PubMed]
- Soars, M.G.; Gelboin, H.V.; Krausz, K.W.; Riley, R.J. A comparison of relative abundance, activity factor and inhibitory monoclonal antibody approaches in the characterization of human CYP enzymology. Br. J. Clin. Pharmacol. 2003, 55, 175–181. [Google Scholar] [CrossRef] [PubMed]
- PharmVar n.d. Available online: https://www.pharmvar.org/gene/CYP2C9 (accessed on 21 March 2025).
- CYP2C9 n.d. Available online: https://www.pharmgkb.org/gene/PA126/haplotype (accessed on 21 March 2025).
- Kirchheiner, J.; Brockmöller, J. Clinical consequences of cytochrome P450 2C9 polymorphisms. Clin. Pharmacol. Ther. 2005, 77, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wang, J.; Huang, S.-Q.; Su, H.-H.; Zhou, S.-F. Genetic polymorphism of the human cytochrome P450 2C9 gene and its clinical significance. Curr. Drug Metab. 2009, 10, 781–834. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.F.; Zhou, Z.W.; Huang, M. Polymorphisms of human cytochrome P450 2C9 and the functional relevance. Toxicology 2010, 278, 165–188. [Google Scholar] [CrossRef] [PubMed]
- Preissner, S.C.; Hoffmann, M.F.; Preissner, R.C.; Dunkel, M.; Gewiess, A. Polymorphic Cytochrome P450 Enzymes (CYPs) and Their Role in Personalized Therapy. PLoS ONE 2013, 8, 82562. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Sun, X.; Gong, Y.; Gawronski, B.E.; Langaee, T.Y.; Shahin, M.H.A.; Khalifa, S.I.; Johnson, J.A. CYP2C9 promoter variable number tandem repeat polymorphism regulates mRNA expression in human livers. Drug Metab. Dispos. 2012, 40, 884–891. [Google Scholar] [CrossRef] [PubMed]
- Montalvo, A.D.; Gong, Y.; Collins, J.M.; Wang, D. The Association Between Promoter Tandem Repeat Polymorphism (pVNTR) and CYP2C9 Gene Expression in Human Liver Samples. Genes 2025, 16, 213. [Google Scholar] [CrossRef] [PubMed]
- Dorado, P.; Santos-Díaz, G.; Gutiérrez-Martín, Y.; Suárez-Santisteban, M.Á. Frequency of CYP2C9 Promoter Variable Number Tandem Repeat Polymorphism in a Spanish Population: Linkage Disequilibrium with CYP2C9*3 Allele. J. Pers. Med. 2022, 12, 782. [Google Scholar] [CrossRef] [PubMed]
- Al-Eitan, L.N.; Almasri, A.Y.; Al-Habahbeh, S.O. Impact of a variable number tandem repeat in the CYP2C9 promoter on warfarin sensitivity and responsiveness in Jordanians with cardiovascular disease. Pharmgenomics. Pers. Med. 2019, 12, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Soares, F.; Peñas-Lledó, E.M.; Tarazona-Santos, E.; Sosa-Macías, M.; Terán, E.; López-López, M.; Rodeiro, I.; Moya, G.E.; Calzadilla, L.R.; Ramírez-Roa, R.; et al. Genomic Ancestry, CYP2D6, CYP2C9, and CYP2C19 Among Latin Americans. Clin. Pharmacol. Ther. 2020, 107, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Guevara, M.; Rodrigues-soares, F.; Gonz, C.; Cruz, D.; Andr, F.D. Afro-Latin American Pharmacogenetics of CYP2D6, CYP2C9, and CYP2C19 in Dominicans: A Study from the RIBEF-CEIBA Consortium. Pharmaceutics 2024, 16, 1399. [Google Scholar] [CrossRef] [PubMed]
- Pratt, V.M.; Cavallari, L.H.; Del Tredici, A.L.; Hachad, H.; Ji, Y.; Moyer, A.M.; Scott, S.A.; Whirl-Carrillo, M.; Weck, K.E. Recommendations for Clinical CYP2C9 Genotyping Allele Selection: A Joint Recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 2019, 21, 746–755. [Google Scholar] [CrossRef] [PubMed]
| CYP2C9 pVNTR | ||||||||
|---|---|---|---|---|---|---|---|---|
| L/M | M/M | M/S | ||||||
| CYP2C9 | n | % | n | % | n | % | n | % |
| *1/*1 | 141 | 73.1 | 21 | 10.9 | 115 | 59.6 | 5 | 2.6 |
| *1/*3 | 9 | 4.6 | - | - | 2 | 1.0 | 7 | 3.6 |
| *1/*5 | 1 | 0.5 | - | - | 1 | 0.5 | - | - |
| *1/*2 | 36 | 18.7 | 4 | 2.1 | 32 | 16.6 | - | - |
| *2/*2 | 3 | 1.6 | - | - | 3 | 1.6 | - | - |
| *2/*3 | 2 | 1.0 | - | - | - | - | 2 | 1.0 |
| *3/*3 | 1 | 0.5 | - | - | - | - | 1 | 0.5 |
| 25 | 13.0 | 153 | 79.3 | 15 | 7.7 | |||
| Population | n | CYP2C9*3 | pVNTR-S | pVNTR-M | pVNTR-L | #D’ (r2) | Ref. |
|---|---|---|---|---|---|---|---|
| African American | 134 | 0.010 | 0.213 *** | 0.742 *** | 0.045 | 0.99 (0.05) | [14] |
| African American | 120 | 0.025 | 0.051 | 0.883 | 0.065 | n.p. (0.53) | [13] |
| Egyptian | 207 | 0.092 | 0.115 *** | 0.785 *** | 0.100 | n.p. (0.59) | [13] |
| Jordanian | 205 | n.e. | 0.295 *** | 0.627 *** | 0.078 | n.e. | [16] |
| Spanish | 209 | 0.074 | 0.081 * | 0.816 ** | 0.103 | 0.93 (0.88) | [15] |
| White American | 804 | 0.050 | 0.058 | 0.789 *** | 0.152 *** | n.p. (0.75) | [13] |
| White American | 113 | 0.040 | 0.049 | 0.814 ** | 0.137 ** | 0.99 (0.79) | [14] |
| Dominican | 193 | 0.034 | 0.039 | 0.896 | 0.065 | 0.76 (0.70) | Present study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de la Cruz, C.G.; Guevara, M.; Rodrigues-Soares, F.; Rodríguez, E.; Peñas-Lledó, E.; LLerena, A.; Dorado, P. CYP2C9 Promoter Variable Number Tandem Repeat Polymorphism in a Dominican Population: Exploring Differences with Genetic Ancestry. Genes 2025, 16, 540. https://doi.org/10.3390/genes16050540
de la Cruz CG, Guevara M, Rodrigues-Soares F, Rodríguez E, Peñas-Lledó E, LLerena A, Dorado P. CYP2C9 Promoter Variable Number Tandem Repeat Polymorphism in a Dominican Population: Exploring Differences with Genetic Ancestry. Genes. 2025; 16(5):540. https://doi.org/10.3390/genes16050540
Chicago/Turabian Stylede la Cruz, Carla González, Mariela Guevara, Fernanda Rodrigues-Soares, Ernesto Rodríguez, Eva Peñas-Lledó, Adrián LLerena, and Pedro Dorado. 2025. "CYP2C9 Promoter Variable Number Tandem Repeat Polymorphism in a Dominican Population: Exploring Differences with Genetic Ancestry" Genes 16, no. 5: 540. https://doi.org/10.3390/genes16050540
APA Stylede la Cruz, C. G., Guevara, M., Rodrigues-Soares, F., Rodríguez, E., Peñas-Lledó, E., LLerena, A., & Dorado, P. (2025). CYP2C9 Promoter Variable Number Tandem Repeat Polymorphism in a Dominican Population: Exploring Differences with Genetic Ancestry. Genes, 16(5), 540. https://doi.org/10.3390/genes16050540









