Abstract
Background: Onychostoma virgulatum is an endemic freshwater fish in South China, first described as a new species in 2009. However, little is known about this species and no complete mitochondrial genomes of O. virgulatum has been reported to date. This study seeks to elucidate the characteristics of the mitochondrial genome of O. virgulatum and investigate the phylogenetic relationships within the Acrossocheilinae subfamily, particularly among the genera Onychostoma, Acrossocheilus, and Folifer. Methods: The mitochondrial genome of O. virgulatum was sequenced and assembled. We analyzed its sequence length, nucleotide composition, and evolutionary relationships within the Acrossocheilinae by incorporate data from 58 previously published mitochondrial genomes. Results: The complete circular sequence is 16,606 bp in length and contains 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, and a typical control region (D-loop), all arranged in a typical order. The genomic base composition is biased toward A+T content (56.5%), with 31.4% A, 25.1% T, 27.4% C, and 16.1% G. Among about 30 Acrossocheilina species, the nonsynonymous (Ka) to synonymous substitutions (Ks) for all 13 protein-coding genes (PCGs) are significantly less than 1, suggesting strong negative or purifying selection in these species. The phylogenetic trees inferred from the mitogenome and 13 PCGs of 58 Acrossocheilinae sequences consistently indicate that: (1) O. virgulatum shares the closest genetic relationship with Onychostoma barbatulum; (2) Acrossocheilinae species are clustered into three major clades, with neither Acrossocheilus nor Onychostoma forming monophyletic groups. Conclusions: This study provides new insights into the taxonomy and phylogenetic relationships of Acrossocheilinae, particularly O. virgulatum, contributing to a better understanding of the systematics, origin, and evolution of this subfamily.
1. Introduction
The endemic Chinese and Southeast Asian genus Onychostoma Günther, 1896, is a speciose group of the subfamily Acrossocheilinae (Teleostei: Cyprinidae) [1,2,3], which was long classified under the subfamily Barbinae. It comprises small and medium-sized benthic freshwater fish with omnivorous habits and is often found in cooler, fast-flowing mountain streams and rivers [4,5,6,7,8,9,10,11,12,13] (Fishbase: https://www.fishbase.org, accessed on 10 April 2025). The species distribution within the genus Onychostoma is relatively complex, ranging from widely distributed species and those with restricted ranges. Most species possess significant economic value. Currently, the genus comprises 23 valid species, recorded across China, Vietnam, Thailand, Laos, and Cambodia. China is the center of diversity, hosting 19 species—12 of which are endemic [4,5,6,7,8,9,10,11,12,13] (Fishbase: https://www.fishbase.org, accessed on 10 April 2025). The taxonomic validity of this genus has long been debated. Traditionally, species identification within Onychostoma has relied primarily on morphological traits, including the length, size, and structure of the last unbranched (simple) dorsal fin ray as well as the shape and width of the mouth opening. However, these morphological features exhibit adaptive evolution to flowing-water habitats. Additionally, some species display sexual dimorphism in mouth morphology and ontogenetic variations in body coloration. Undoubtedly, these differences complicate species delineation and the taxonomy of certain fish within this genus remains contentious. Previous studies have confirmed the nonmonophyletic nature of Onychostoma and its closer relationship with the other two genera (Acrossocheilus and Folifer) in Acrossocheilinae [14,15,16,17,18,19,20]. However, a definitive classification at the generic level has yet to be established.
Xin, Zhang, and Cao, 2009, described Onychostoma virgulatum as a new species in 2009 [10] (Figure 1). This endemic freshwater fish has a narrow distribution range, occurring only in the upper reaches of the Qiupu River (Anhui Province) and the Suichuan River (Jiangxi Province)—tributaries of the southern bank of the lower Yangtze River in South China [10,21]. However, according to The Fishes of Jiangxi (2024) [22], specimens originally identified as Varicorhinus (Onychostoma) lini (Wu) from the Ganjiang, Xinjiang, and Xunwu Rivers [23,24] should be reclassified as O. virgulatum. Notably, the Xunwu River belongs to the upper reaches of the Dongjiang River (Pear River system), whereas the Ganjiang and Xinjiang Rivers are tributaries of the lower Yangtze River.

Figure 1.
The appearance of Onychostoma virgulatum.
The species appears to exhibit a patchy distribution limited to a few areas, a pattern consistent with many Acrossocheilinae fishes [4,5,6,7,8,9,10,11,12,13,16]. However, current knowledge about the species remains limited and no complete mitochondrial genomes of O. virgulatum have been reported to date. This study aims to supplement the mitochondrial DNA (mtDNA) sequence data of O. virgulatum and, by integrating the existing relevant data, investigate the phylogenetic relationships within the Acrossocheilinae subfamily—Onychostoma, Acrossocheilus, and Folifer.
2. Materials and Methods
2.1. Sampling, DNA Extraction, PCR Amplification, and Sequencing
In August 2024, ten individuals of O. virgulatum were collected using a net from the Qiupu River (30°11′25″ N, 117°40′47″ E) in Shitai County, Anhui Province, China—the type locality of the species. All individuals were brought back to the laboratory in oxygen bags for aquaculture and molecular biology experiments. From these, one individual was randomly selected as the sample, deeply anesthetized with 100 mg/L MS-222 (Sigma, St. Louis, MO, USA), then fixed and stored in 100% ethanol and stored in the Fishery Institute of Anhui Academy of Agricultural Sciences. The genomic DNA was extracted from the muscle tissue of the sample using a DNeasy Blood and Tissue Kit (Qiagen, Dusseldorf, Germany) and then kept at −20 °C for PCR amplification.
According to references [25,26,27], a total of 18 primer pairs (Table S1) were selected to amplify the mitogenome sequences. The amplifications were performed in 25 μL reaction mixtures containing 2.5 μL 10 × buffer (Mg2+), 2 μL dNTPs(10 mmol/L), 1 µL of each primer (10 µmol/L), 1 U Taq polymerase (TaKaRa, Dalian, China), and approximately 100 ng template genomic DNA. PCR conditions were as follows: initial denaturation at 95 °C for 5 min, then 35 cycles of denaturation (95 °C for 35 s, 51–55 °C for 45 s, and 72 °C for 1 min), and final extension at 72 °C for 10 min. Subsequently, the targeted fragments were purified and directly sequenced in both directions by the Sangon Biotechnology Company (Sangon, Shanghai, China) after 1% agarose gel electrophoresis.
2.2. Genome Assembly, Annotation, and Selection Pressure Analysis
The DNA sequence fragments were manually assembled by the software Chromas 2.6.6 (http://technelysium.com.au/, accessed on 12 December 2024), Clustal X (http://www.clustal.org/, accessed on 12 December 2024), and Seaview 4.4.2 [28]. The protein-coding gene, tRNA gene, rRNA genes, and D-loop region were identified by comparing the mitogenome sequences with other Onychostoma sequences (downloaded from Genbank) and through homology searches. A circular map of the mitochondrial genome was generated using the Proksee server (https://proksee.ca/, accessed on 21 February 2025). The AT-skew and GC-skew were calculated using the following general formulae: AT-skew = (A% − T%)/(A% + T%) and GC-skew = (G% − C%)/(G% + C%), respectively [29]. The base composition, nucleotide substitution, and relative synonymous codon usage (RSCU) of each protein-coding gene (PCG) were determined using MEGA 12.0.9 [30], and the column diagram of RSCU was generated by the software suite PhyloSuite 1.2.3 [31].
The mean nonsynonymous (Ka), synonymous substitutions (Ks), and ratio of Ka/Ks in each PCG of 29 reference sequences within the Acrossocheilinae and mitogenome of O. virgulatum were calculated by DnaSP 6.0 [32], then the obtained data were used to make a column chart in Microsoft Excel.
2.3. Phylogenetic Analysis
We retrieved and downloaded all currently available complete mitochondrial genomes of all Acrossocheilinae species and Spinibarbus sinensis from the GenBank database. By PhyloSuite 1.2.3 [31], 59 sequences were reserved after deleting redundant sequences and were aligned using MAFFT 7.505 [33] with default settings. We eventually successfully extracted the nucleotide sequences and 13 PCGs of 58 mitochondrial genomes covering about 29 species and 3 unidentified sequences, including 13 Onychostomas species, 14 Acrossocheilus species, Folifer brevifilis, and S. sinensis (Table 1). A phylogenetic tree of these Acrossocheilinae species was constructed based on the mitogenome sequences and concatenated sequences of 13 PCGs using the maximum likelihood (ML) and Bayesian (BI) methods with S. sinensis as the outgroup.

Table 1.
Information on 59 mitogenome sequences of Acrossocheilinae used in this study.
The best substitution model GTR+F+R3 (mitogenome) or partition model with a gene + codon partitioning strategy (allowing merging of similar partitions) (PCGs) were chosen based on the Bayesian information criterion (BIC) using the smart model selection algorithm [34,35], and the ML trees were constructed using IQ-TREE 2.4.0 software with a standard bootstrap test inferred from 1000 replicates [36,37]. The GTR model (mitogenome) or partition model (PCGs) were chosen based on the Bayesian information criterion (BIC) using the smart model selection algorithm [34,35], and the BI trees were conducted using MrBayes 3.2.8 software and two independent Markov chain Monte Carlo (MCMC) chains were run for 10 million generations and beginning with a random tree in each BI analysis [35,38]. Also, all values were sampled every 1000 generations; the first 25% of samples were used as burn-in, and the remaining trees were used to construct the data. The phylogenetic trees were edited by the online tool Interactive Tree of Life (iTOL) (https://itol.embl.de/, accessed on 26 March 2025).
3. Results
3.1. Mitochondrial Genomic Structure and Composition
The complete mitochondrial genome of O. virgulatum is a typical closed circular double-stranded DNA molecule with 16,606 bp in size and the sequence was deposited in GenBase (Accession Number: CAA104068) (Figure 2). The mitogenome typically contains 37 genes with 13 protein-coding genes, 2 ribosomal RNA (rRNA) genes, 22 tRNAs, and a control region (D-Loop) (Figure 2, Table 2). Most of the 37 genes are encoded on the H-strand but the ND6 and 8 tRNA genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser(UCN), tRNA-Glu, and tRNA-Pro) are encoded on the L-strand. In total, 12 of the 13 protein-coding genes of the mitogenome start with a typical ATG codon, except for the COX1 gene (GTG codon). A total of 6 protein-coding genes end with the termination codon TAA (ND1, COX1, ATP6, ND4L, ND5, and ND6), while 6 genes terminate with a single base T, and the COX3 gene terminates with TA. The 22 tRNA genes range in size from 67 to 76 bp, and the tRNA-Cys gene (67 bp) is the shortest in size, whereas the longest are the tRNA-Leu(UUR) and tRNA-Lys genes (76 bp). Among the 2 rRNA genes, 12S rRNA is located between tRNA-Phe and tRNA-Val with a length of 956 bp, and 16S rRNA is located between tRNA-Val and tRNA-Leu(UUR) with a length of 1684 bp. The noncoding control region (D-loop) is located between tRNA-Pro and tRNA-Phe with a length of 945 bp, in which an extended terminal associated sequence (ETAS), central conserved domain (CD) including three conserved sequence blocks (CSB-F, CSB-E, and CSB-D), and conserved sequence block (CSB) consisting of three conserved sequence blocks (CSB-1, CSB-2, and CSB-3) are identified (Supplementary Figure S1). In addition, there are 11 small gene spacers (1–33 bp in size) in the mitogenome, the longest gap of which is 33 bp, located between tRNA-Asn and tRNA-Cys and 7 overlapping regions (1–7 bp in size) with the longest overlapping regions (7 bp) existing between ATP8/ATP6 and ND4L/ND4. A sequence capable of initiating L-chain replication (OL) is identified in the longest gap region, which consists of 10 bases forming a conserved stem-ring structure (Figure S2).
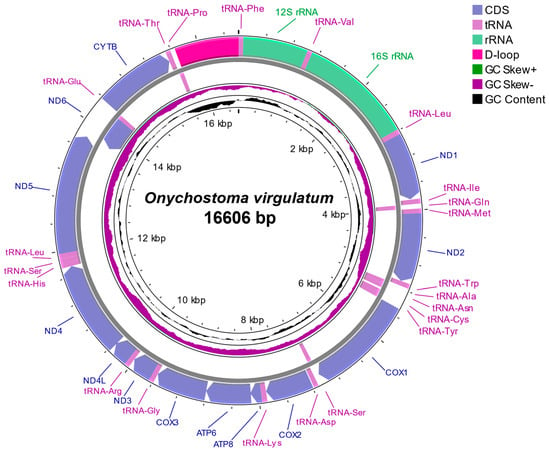
Figure 2.
Complete mitogenome organization and gene arrangement of O. virgulatum.

Table 2.
Composition and structure of O. virgulatum mitochondrial genome.
The overall base composition of the mitogenome is 31.4% A, 25.1% T, 27.4% C, and 16.1% G, exhibiting a slight bias towards A+T content (56.5%) (Table 3). However, the D-loop exhibits a significant bias toward A+T content (66.1%), which is subject to less evolutionary selection pressure and has a fast evolutionary rate. In addition, the A+T content of the first (48.0%), second (59.0%), and third (62.1%) codon position of the PCGs are significantly different, and all PCGs except ND6 exhibit strong anti-G bias (11.5–17.5%), especially in the second (13.7%) and third (7.3%) positions of codon.

Table 3.
Nucleotide compositions of different regions of O. virgulatum.
The AT-skew of the mitogenome, rRNAs, tRNAs, D-loop, concatenated PCGs, and three PCGs (COXI, ND4L, and ND6) are all positive, while ND3 is zero and nine PCGs are negative. The AT-skew of PCGs-2nd are positive but the AT-skew of PCGs-1st and PCGs-3rd is negative. Conversely, the GC-skew of mitogenome, rRNAs, D-loop, concatenated PCGs, three codon positions of PCGs, and 12 PCGs are all negative, while the tRNAs and ND6 are positive.
3.2. Characteristics of Codon Usage and Selection Pressure
The 13 PCGs encode a total of 3794 amino acids (except for stop codons), with the highest content being 630 Leucine (16.6%) and the lowest content being 26 Cysteine (0.69%) (Figure 3). The relative synonymous codon usage (RSCU) statistics show that there are 27 preferred codons (RSCU > 1) in total, with the highest being 2.65 (Figure 3). There are 4 codons with RSCU values greater than 2: CUA, UCA, CCA, and CGA, all of which end in A.
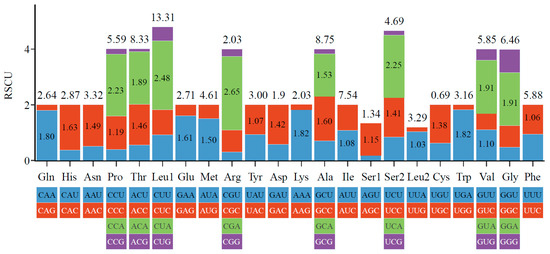
Figure 3.
Amino acid content (top number, %) and RSCU (internal numbers) of O. virgulatum mitochondrial PCGs. Only RSCU values greater than 1 are displayed.
Obviously, the Ka values (0.005–0.075) are much smaller than the Ks values (0.405–0.643) in all PCGs, and so the Ka/Ks ratio of each PCG is far less than 1, with the highest Ka/Ks ratio (0.139) in ND6, whereas the lowest ratio (0.009) is in COX1 (Figure 4).
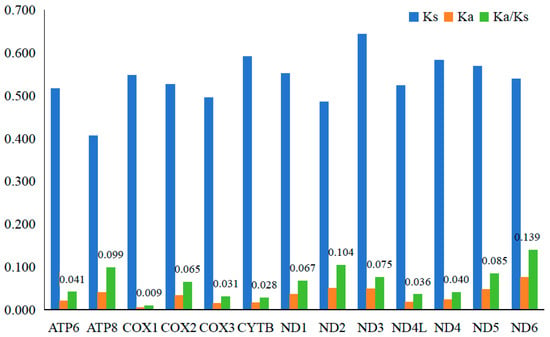
Figure 4.
The mean Ka, Ks, and Ka/Ks values of PCGs among 30 Acrossocheilinae species.
3.3. Phylogenetic Relationship
The four phylogenetic trees obtained by ML and BI analyses are analogous with the same topologies, in which all Acrossocheilinae species are clustered into three major clades with a high node support rate (>98%) in each clade (Figure 5 and Figure 6). Clade I is considered as an ancestral group, which is a mixed clade consisting of 6 species of Acrossocheilina: (F. brevifilis + Onychostoma alticorpus) sister to (Acrossocheilus yunnanensis + Acrossocheilus monticola + sister pair (Onychostoma ovale + Onychostoma rarum)); Clade II represents the Onychostoma groups, including all other Onychostoma sequences. Clade III represents Acrossocheilus groups, including all other Acrossocheilus sequences. Clade I is sister to sister pair (Clade II + Clade III).
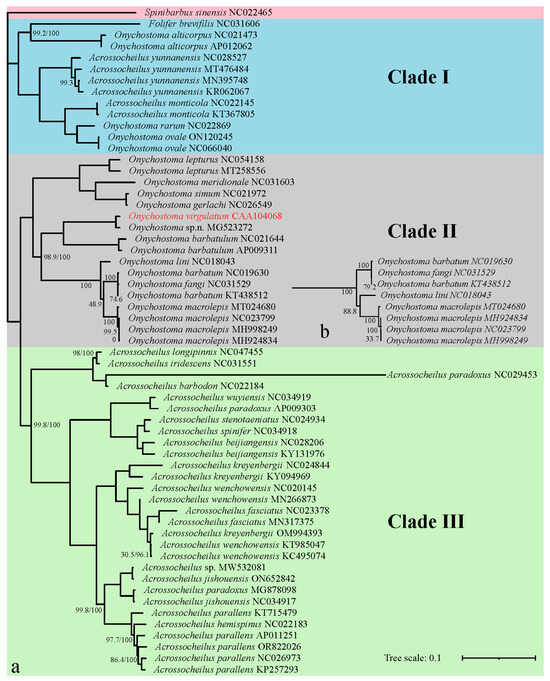
Figure 5.
Phylogenetic trees of Acrossocheilinae based on 13 PCGs using maximum-likelihood (ML) (a) and Bayesian (BI) (b) analysis. GenBank accession number follows the species name. The numbers on nodes indicate NJ (left) and BI (right) support values, with no number indicating 100% support.
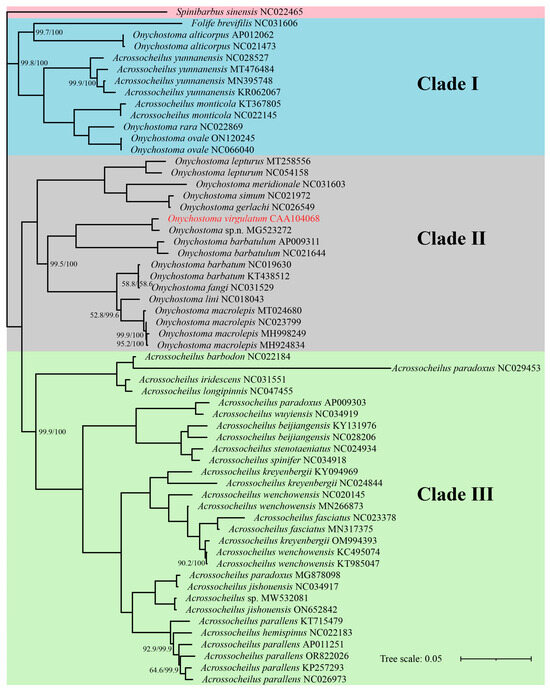
Figure 6.
Phylogenetic trees of Acrossocheilinae based on mitogenome using maximum-likelihood (ML) and Bayesian (BI) analysis. GenBank accession number follows the species name. The two numbers on some nodes indicate ML (left) and BI (right) support values, with no number indicating 100% support.
Additionally, these trees also consistently show that (Figure 5 and Figure 6): (1) the three sequence-group A. yunnanensis sequences include the unverified A. yunnanensis sequence (KR062067), Onychostoma sp. n. (MG523272) and O. virgulatum in the study, Acrossocheilus sp. (MW532081) and Acrossocheilus jishouensis (ON652842), are clustered into a single branch, respectively; (2) Onychostoma simum (NC021972) and Onychostoma gerlachi (NC026549) are clustered into a single branch with an abnormally close relationship; (3) the sequence identified as Onychostoma fangi (NC031529) is unreasonably clustered together with two Onychostoma barbatum sequences; (4) three sequences identified as Acrossocheilus paradoxus cannot be clustered in one cluster, but they are clustered with three different species, respectively; (5) Acrossocheilus longipinnis, a senior synonym of Acrossocheilus stenotaeniatus (Fishbase: https://www.fishbase.org, accessed on 10 April 2025), is clustered together with Acrossocheilus iridescens, while A. stenotaeniatus is clustered together with Acrossocheilus spinifer; (6) those individuals belonging to each of Acrossocheilus fasciatus, Acrossocheilus kreyenbergii, and Acrossocheilus wenchowensis failed to cluster according to their taxonomic circumscription; (7) the only Acrossocheilus hemispinus sequence (NC022183) was clustered together with the sequences of Acrossocheilus parallens. All the above results have a high node support value (>85%). Slight differences in the topologies among the four phylogenetic trees are observed in the positions of O. lini in the ML-tree based on 13 PCGs.
4. Discussion
We first identified the mitogenome of O. virgulatum, which is a typical closed circular DNA molecule with a length of 16,606 bp and which comprises 13 PCGs genes, 22 tRNA genes, 2 rRNA genes, and a typical control region (D-loop). The gene arrangements and composition exhibit similarities to those of other Onychostoma as well as various previously analyzed Acrossocheilinae mitogenomes [39,40,41,42]. O. virgulatum has long been favored by local residents, but because of habitat destruction and overfishing, the population size of the species has drastically declined in the past 10 years. It has been 15 years since the species was identified as a new species from Qiupu River in Anhui province [10], whereas the species was rarely studied because of its narrow distribution range, small population size, and semimigratory habits (wintering at the bottom of the river or in caves) [21,22]. Therefore, the determination of the mitochondrial genomes has great value, which is helpful in strengthening the conservation and management of the species.
The Ka/Ks ratio of each PCG of the Acrossocheilina mitogenome were shown to be significantly less than 1, which generally indicates strong negative or purifying selection in these species and further revealed the strong conservation of all PCGs in these species. The primary function of mitochondrial PCGs is energy production, which involves oxygen and food, so the purifying selection might be related to the common characteristics of Acrossocheilina: high dissolved oxygen demand in water and omnivorous (Fishbase: https://www.fishbase.org, accessed on 10 April 2025). The different Ka/Ks ratios among 13 genes (0.009–0.139) suggest different degrees of conservation among these genes, reflecting different degrees of functional constraints in energy metabolism. For instance, the extremely low Ka/Ks ratio in COX1 (0.009) is significantly lower than that of ND6 (0.139), indicating that COX1 has undergone stronger purifying selection during evolution, meaning its amino acid sequence is highly conserved and functional mutations are strictly limited. In contrast, the relatively high Ka/Ks ratio of ND6 suggests that it has weaker evolutionary constraints and may allow for the accumulation of more neutral or slightly deleterious mutations. Because the mitochondrial genes (such as COX1 and ND6) of vertebrate are highly conserved in function, we believe that this difference mainly stems from the following evolutionary pressures [43,44,45]: COX1/CYTB are highly conserved: as they are involved in the core structure of the electron transport chain (complexes IV/III), their functions need to be strictly maintained; ND6/ATP8 are prone to variation: ND6 is the only hydrophobic protein encoded by a light chain in complex I and is susceptible to the accumulation of mutations; ATP8 has fewer functional constraints. This difference has been found in other groups such as mammals.
Molecular phylogenies are not only used to support the placement of the newly collected specimens within the genus Onychostoma, even Acrossocheilina, but also to resolve the phylogenetic relationships within the group. In this study, Onychostoma sp.n. (MG523272) was identified as O. virgulatum, and they have the closest relationship with Onychostoma barbatulum, with a 100% support value. The sample collection location of the sequence (MG52327) was not recorded, so it is not yet possible to confirm whether there is a new distribution. The sequence of O. barbatulum (NC021644) was sampled from the type locality—Taiwan island. Furthermore, O. barbatulum is one of only two species of Onychostoma in Taiwan; the other is O. alticorpus, found only in Taiwan [46]. So, we considered the identification of both species to be credible. In addition, the unverified A. yunnanensis sequence (KR062067) should be A. yunnanensis, and Acrossocheilus sp. (MW532081) has an intraspecific relationship with A. jishouensis (ON652842) [42].
All phylogenetic trees obtained by ML and BI analyses are analogous with the same topologies, which consistently revealed the nonmonophyletic relationship between Onychostoma and Acrossocheilus, while a few species of both genera are closely related to F. brevifilis. This conclusion has been confirmed by many previous studies [14,15,16,17,18,19,20,25,40,41,42], such as Wang et al. (2007) [14] and Li et al. (2008) [15], who, based on RAG2 and 16S rRNA sequences, respectively, found that Onychostoma and Acrossocheilus were a nonmonophyletic group; Xin (2008) [16] carried out the taxonomic reassignment of species within Onychostoma based on morphological and skeletal characteristics, with a sample size more adequate than previous studies, and also found the nonmonophyly of the two genera. In recent years, several phylogenetic analyses of Acrossocheilina based on mitogenome have also come to similar conclusion [20,40,41,42]. The discovery of the nonmonophyly of the genera Onychostoma and Acrossocheilus has far-reaching implications for fish taxonomy, phylogenetic research, and biodiversity conservation, specifically manifested as: (1) The reassessment of taxonomy at the genus level in Acrossocheilus. The nonmonophyletic relationship indicates that the current definitions of genera may be based on convergent morphological features rather than true evolutionary relationships; the boundaries of genera need to be redefined in combination with molecular systematics (such as multigene or genomic data), and some species (such as O. alticorpus) may need to be established in new genera. (2) Adaptive radiation and convergent evolution: Nonmonophyly may reflect convergent evolution under similar ecological niches (such as similar jaw morphology resulting from benthic feeding). Studying the association between functional morphology and ecological adaptability can reveal the driving mechanisms of the diversification of Onychostoma and Acrossocheilus and even East Asian stream fishes. (3) Reassessment of species’ endangered status and precision in habitat management: Taxonomic changes may alter the uniqueness of certain species (for instance, A. yunnanensis is actually an independently evolved branch), necessitating a re-evaluation of their conservation priorities. After clarifying the true distribution range of monophyletic groups, a network of protected areas can be designed for key evolutionary units, preventing conservation gaps due to taxonomic errors.
In addition, species identification errors were found in many mitogenome sequences based on phylogenetic trees in the study. For instance, at least one sequence has been incorrectly identified between O. fangi (NC031529) and O. barbatum (NC019630, KT438512). Among the three sequences annotated as A. paradoxus, at least two demonstrate incongruent phylogenetic positioning, suggesting potential misidentification at the species level. There is confusion in species identification of three closely-related species: A. fasciatus, A. kreyenbergii. and A. wenchowensis, which have high morphological similarity and adjacent distribution. In common sense, A. hemispinus (NC022183) also should be attributed to the wrong species. After consulting the author who found A. spinifer as a new species [47] and uploaded the sequence of A. spinifer (NC034918), species identification of this sequence (NC034918) should be correct and A. stenotaeniatus (NC024934) is likely to be problematic. Given the limited species information, however, we cannot yet be sure whether A. longipinnis (NC047455) and A. iridescens (NC031551) are correct. As mentioned in the introduction, adaptive evolution to running water (convergence), sexual dimorphism, and ontogenetic variations in some morphological characters bring confusion in the delineation of these species. These results highlight the limitations of classification based solely on morphology. It should be noted that there are species identification errors in many taxon in the mitochondrial public data. Therefore, to correctly identify species and further ensure the accuracy of mitochondrial data in public databases (such as GenBank and GenBase), we propose a checklist for quality control of public mitochondrial data as follows: (1) Improve basic information: clearly define the submitted information of samples (required fields: species, specimen photos, geographical location, collection time, and contact information of the submitter) and record the DNA extraction and sequencing methods. (2) Taxonomic validation: combined with integrative taxonomy [48]: provide morphological evidence, additional nuclear gene markers that support the consistency of mitochondrial data and ecological niche modeling that check whether the geographical distribution of the samples matches the ecological niches of the known species. (3) Data quality assessment: check sequence integrity and consistency, establish a public platform, or utilize a database feedback system to flag suspicious data. (4) Long-term maintenance: regular review: database managers collaborate with taxonomists to re-evaluate disputed data and encourage users to supplement missing information. It is fortunate that these errors are only found in species within the genus from clade II or clade III. It suggests, at the very least, that the species identification between genera is clear, but that the definition at the genus level is problematic, as mentioned above, which needs more new genus in Acrossocheilina.
Limited information is currently available regarding the mitochondrial genome of some species of Acrossocheilina. Therefore, further investigations incorporating extensive taxon sampling are imperative to accurately validate the phylogenetic connections within the genera Onychostoma, Acrossocheilus. Our findings significantly contribute to the study of the genetic diversity and taxonomic status of O. virgulatum, also offering valuable insights to better understand the evolution of Acrossocheilina.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes16050541/s1, Figure S1: the structure and sequence of the control region of the O. virgulatum mitochondrial genome; Figure S2: stem-loop structure of L-strand replication initiation region of O. virgulatum; Table S1: sequences of primers used in amplification of the complete mitochondrial genome in O. virgulatum.
Author Contributions
Conceptualization, Y.H.; Data curation, H.W. and A.L.; Funding acquisition, G.D.; Investigation, G.D. and H.Z.; Methodology, Y.H.; Resources, G.D.; Software, Y.H.; Validation, Y.H.; Writing—original draft, Y.H.; Writing—review and editing, H.W. All authors will be updated at each stage of manuscript processing, including submission, revision, and revision reminder, via emails from our system or the assigned Assistant Editor. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Anhui Aquatic Industry Technology System (approved by the Department of Agriculture and Rural Affairs of Anhui province [2021] No. 711).
Institutional Review Board Statement
The experimental protocol was established according to the ethical guidelines of the Basel Declaration and was approved by the Experimental Animal Welfare and Ethical of Anhui Academy of Agricultural Sciences (NO. AAAS 2024-16).
Informed Consent Statement
Not applicable.
Data Availability Statement
The mitogenome sequence of O. virgulatum supporting the findings of this study has been deposited in the GenBase database (https://ngdc.cncb.ac.cn/genbase/) under the accession number CAA104068. The sequence will be made openly available starting 29 May 2025.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bǎnǎrescu, P. A review of the species of the subgenus Onychostoma s. str. with description of a new species (Pisces, Cyprinidae). Rev. Roum. Biol. Ser. Zool. 1971, 16, 241–248. [Google Scholar]
- Bǎnǎrescu, P. Revision of the Onychostoma subgenus Scaphesthes (Pisces, Cyprinidae). Rev. Roum. Biol. Ser. Zool. 1971, 16, 357–364. [Google Scholar]
- Tan, M.; Armbruster, J.W. Phylogenetic classification of extant genera of fishes of the order Cypriniformes (Teleostei: Ostariophysi). Zootaxa 2018, 4476, 6–39. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.H.; Lin, R.R.; Yue, P.Q.; Chu, X.L. Barbinae. In Fauna Sinica Osteichthyes: Cypriniformes (III); Yue, P.Q., Ed.; Science Press: Beijing, China, 2000; pp. 126–130. [Google Scholar]
- Rainboth, W.J. Fishes of the Cambodian Mekong; Food and Agriculture Organization of the United Nations: Rome, Italy, 1996; p. 265. [Google Scholar]
- Kottelat, M. Fishes of the Nam Theun and Xe Bangfai basin, Loas, with diagnosis of twenty-two new species (Teleostei: Cypribidae, Baltoridae, Cobitidae, Coiidae and Odontobutidae). Ichthyol. Explor. Freshw. 1998, 9, 1–128. [Google Scholar]
- Kottelat, M. Fishes of Laos; Wildlife Heritage Trust Publications: Colombo, Sri Lanka, 2001; p. 198. [Google Scholar]
- Kottelat, M. Freshwater Fishes of Northern Vietnam; World Bank: Washington, DC, USA, 2001; p. 123. [Google Scholar]
- Nguyen, V.H.; Ngo, S.V. Freshwater Fishes of Vietnam; Volume I: Family Cyprinidae; Agriculture Publish House: Hanoi, Vietnam, 2001. [Google Scholar]
- Xin, Q.; Zhang, E.; Cao, W.X. Onychostoma virgulatum, a new species of cyprinid fish (Pisces: Teleostei) from southern Anhui Province, South China. Ichthyol. Explor. Freshw. 2009, 20, 255–266. [Google Scholar]
- Jang-Liaw, N.H.; Chen, I.S. Onychostoma minnanensis, a new cyprinid species (Teleostei: Cyprinidae) from Fujian, southern mainland China with comments on the mitogenetic differentiation among related species. Ichthyol. Res. 2013, 60, 62–74. [Google Scholar] [CrossRef]
- Hoang, H.D.; Pham, H.M.; Tran, N.T. Two new species of shovel-jaw carp Onychostoma (Teleostei: Cyprinidae) from southern Vietnam. Zootaxa 2015, 3962, 123–138. [Google Scholar] [CrossRef]
- Song, X.L.; Cao, L.; Zhang, E. Onychostoma brevibarba, a new cyprinine fish (Pisces: Teleostei) from the middle Chang Jiang basin in Hunan province, south China. Zootaxa 2018, 4410, 147–163. [Google Scholar] [CrossRef]
- Wang, X.Z.; Li, J.B.; He, S.P. Molecular evidence for the monophyly of East Asian groups of Cyprinidae (Teleostei: Cypriniformes) derived from the nuclear recombination activating gene 2 sequences. Mol. Phylogenet. Evol. 2007, 42, 157–170. [Google Scholar] [CrossRef]
- Li, J.B.; Wang, X.Z.; Kong, X.H.; Zhao, K.; He, S.P.; Richard, L.M. Variation patterns of the mitochondrial 16S rRNA gene with secondary structure constraints and their application to phylogeny of cyprinine fishes (Teleostei: Cypriniformes). Mol. Phylogenet. Evol. 2008, 47, 472–487. [Google Scholar] [CrossRef]
- Xin, Q. Taxonomic Revision of Species and Phylogenetic Analysis of Interspecific Relationships Within the Cyprinid Genus Onychostoma Sensu Lato Günther, 1896. Master’s Thesis, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China, 2008. [Google Scholar]
- Yuan, L.Y. Monophyly, Affinity and Taxonomic Revision of the Cyprinid Genus Acrossocheilus Oshima, 1919. Ph.D. Thesis, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China, 2009. [Google Scholar]
- Yang, L.; Sado, T.; Hirt, M.V.; Pasco-Viel, E.; Arunachalam, M.; Li, J.B.; Wang, X.Z.; Freyhof, J.; Saitoh, K.; Simons, A.M.; et al. Phylogeny and polyploidy: Resolving the classification of cyprinine fishes (Teleostei: Cypriniformes). Mol. Phylogenet. Evol. 2015, 85, 97–116. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.Y.; Liu, X.X.; Zhang, E. Mitochondrial phylogeny of Chinese barred species of the cyprinid genus Acrossocheilus Oshima, 1919 (Teleostei: Cypriniformes) and its taxonomic implications. Zootaxa 2015, 4059, 151–168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Cheng, Q.; Geng, H.; Lin, A.H.; Wang, H.Y. Molecular phylogeny of Onychostoma (Cyprinidae) based on mitochondrial genomes. Acta Hydrobiol. Sin. 2018, 42, 512–516. [Google Scholar] [CrossRef]
- Wang, Z.T.; Zhang, E. An updated species checklist of freshwater fishes from the Ganjiang River. Biodivers. Sci. 2021, 29, 1256–1264. Available online: https://www.biodiversity-science.net/CN/10.17520/biods.2021119 (accessed on 27 April 2025). [CrossRef]
- Cheng, W.J.; Fu, H.Y. The Fishes of Jiangxi; Science Press: Beijing, China, 2024; p. 78. [Google Scholar]
- Guo, Z.Z.; Liu, R.L. Study on fishes from Jiangxi province. J. Nanchang Univ. (Nat. Sci.) 1995, 19, 222–232. [Google Scholar]
- Zou, D.L. Fish resources in Xunwushui, Jiangxi province. Chin. J. Zool. 1988, 23, 15–17. [Google Scholar]
- Cheng, C. Complete Mitochondrial Genome Sequencing and Analysis of Onychostoma gerlachi and Phylogenetic Research of Genus Onychostoma. Master’s Thesis, South-Central University for Nationalities, Wuhan, China, 2013. [Google Scholar]
- Xiao, W.H.; Zhang, Y.P.; Liu, H.Z. Molecular systematics of Xenocyprinae (teleostei: Cyprinidae): Taxonomy, biogeography, and coevolution of a special group restricted in East Asia. Mol. Phylogenet. Evol. 2001, 18, 163–173. [Google Scholar] [CrossRef]
- Liu, H.Z.; Tzeng, C.S.; Teng, H.Y. Sequence variations in the mitochondrial DNA control region and their implications for the phylogeny of the Cypriniformes. Can. J. Zool. 2002, 80, 569–581. [Google Scholar] [CrossRef]
- Galtier, N.; Gouy, M.; Gautier, C. SEAVIEW and PHY-LO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Bioinformatics 1996, 12, 543–548. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, M.; Tamura, K. MEGA 12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.L.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.-I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Vincent, L.; Jean-Emmanuel, L.; Olivier, G. SMS: Smart model selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Xiang, C.Y.; Gao, F.L.; Jakovlić, I.; Lei, H.P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Wang, I.C.; Lin, H.D.; Liang, C.M.; Huang, C.C.; Wang, R.D.; Yang, J.Q.; Wang, W.K. Complete mitochondrial genome of the freshwater fish Onychostoma lepturum (Teleostei, Cyprinidae): Genome characterization and phylogenetic analysis. ZooKeys 2020, 1005, 57–72. [Google Scholar] [CrossRef]
- Chen, S.W.; Tang, Q.Y. The complete mitochondrial genome of a vulnerable cyprinid fish Onychostoma macrolepis (teleostei: Cypriniformes) from Qinling-Bashan mountain area in China. Mitochondrial DNA Part B 2020, 5, 1640–1641. [Google Scholar] [CrossRef]
- Zhang, R.Y.; Zhu, T.T.; Luo, Q. The complete mitochondrial genome of the freshwater fish Onychostoma ovale (Cypriniformes, Cyprinidae): Genome characterization and phylogenetic analysis. Genes 2023, 14, 1227. [Google Scholar] [CrossRef]
- Lan, X.Y.; Wang, J.X.; Zhang, M.Y.; Zhou, Q.; Xiang, H.M.; Jiang, W.S. Molecular identification of Acrossocheilus jishouensis (Teleostei: Cyprinidae) and its complete mitochondrial genome. Biochem. Genet. 2024, 62, 1396–1412. [Google Scholar] [CrossRef] [PubMed]
- Saccone, C.; Gissi, C.; Lanave, C.; Larizza, A.; Pesole, G.; Reyes, A. Evolution of the mitochondrial genetic system: An overview. Gene 2000, 261, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Pesini, E.; Lott, M.T.; Procaccio, V.; Poole, J.C.; Brandon, M.C.; Mishmar, D.; Yi, C.; Kreuziger, J.; Baldi, P.; Wallace, D.C. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007, 35, D823–D828. [Google Scholar] [CrossRef]
- Nabholz, B.; Glémin, S.; Galtier, N. Extreme variation of mtDNA neutral substitution rate across mammalian species—The longevity hypothesis. Mol. Biol. Evol. 2008, 25, 795. [Google Scholar] [CrossRef]
- Shen, S.C. Fishes of Taiwan; Department of Zoology, National Taiwan University: Taipei, China, 1993; p. 960. [Google Scholar]
- Yuan, L.Y.; Wu, Z.Q.; Zhang, E. Acrossocheilus spinifer, a new species of barred cyprinid fish from south China (Pisces: Teleostei). J. Fish. Biol. 2006, 68, 163–173. [Google Scholar] [CrossRef]
- Dayrat, B. Towards integrative taxonomy. Biol. J. Linn. Soc. 2005, 85, 407–417. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).