Optimizing Rhamnolipid Performance by Modulating the Expression of Fatty Acid Synthesis Genes fabA and fabZ in Pseudomonas aeruginosa PAO1
Abstract
1. Introduction
2. Materials and Methods
2.1. Oligonucleotides, Plasmids, and Bacterial Strains
2.2. Plasmid Construction
2.3. Construction of Plasmid-Based Ts-Mutant fabZ(Ts) and ΔrhlC Strains
2.4. Isolation of Suppressors
2.5. Spot-Plating Assay
2.6. Rhamnolipid Extraction and Purification
2.7. Quantification of Rhamnolipids via the Orcinol–Sulfuric Acid Method
2.8. Thin-Layer Chromatography (TLC) Analysis
2.9. Emulsification Capacity Assay
2.10. Determination of the Critical Micelle Concentration (CMC)
2.11. LC–MS/MS Analysis
2.12. Statistical Analysis
2.13. Data Availability
3. Results
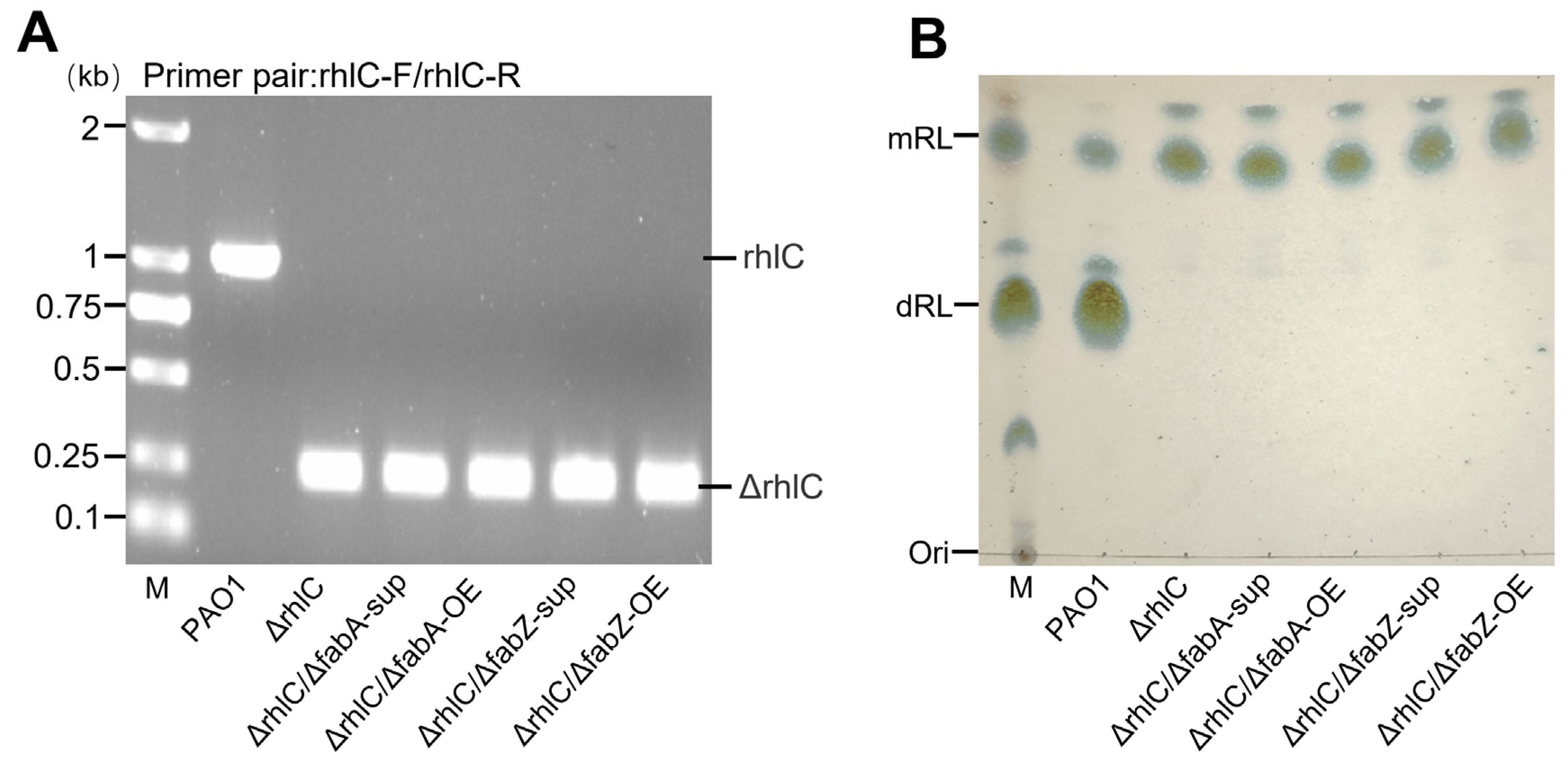
3.1. Construction of rhlC-Deleted Strains to Investigate fabA and fabZ Effects on Mono-Rhamnolipid Fatty Acyl Composition
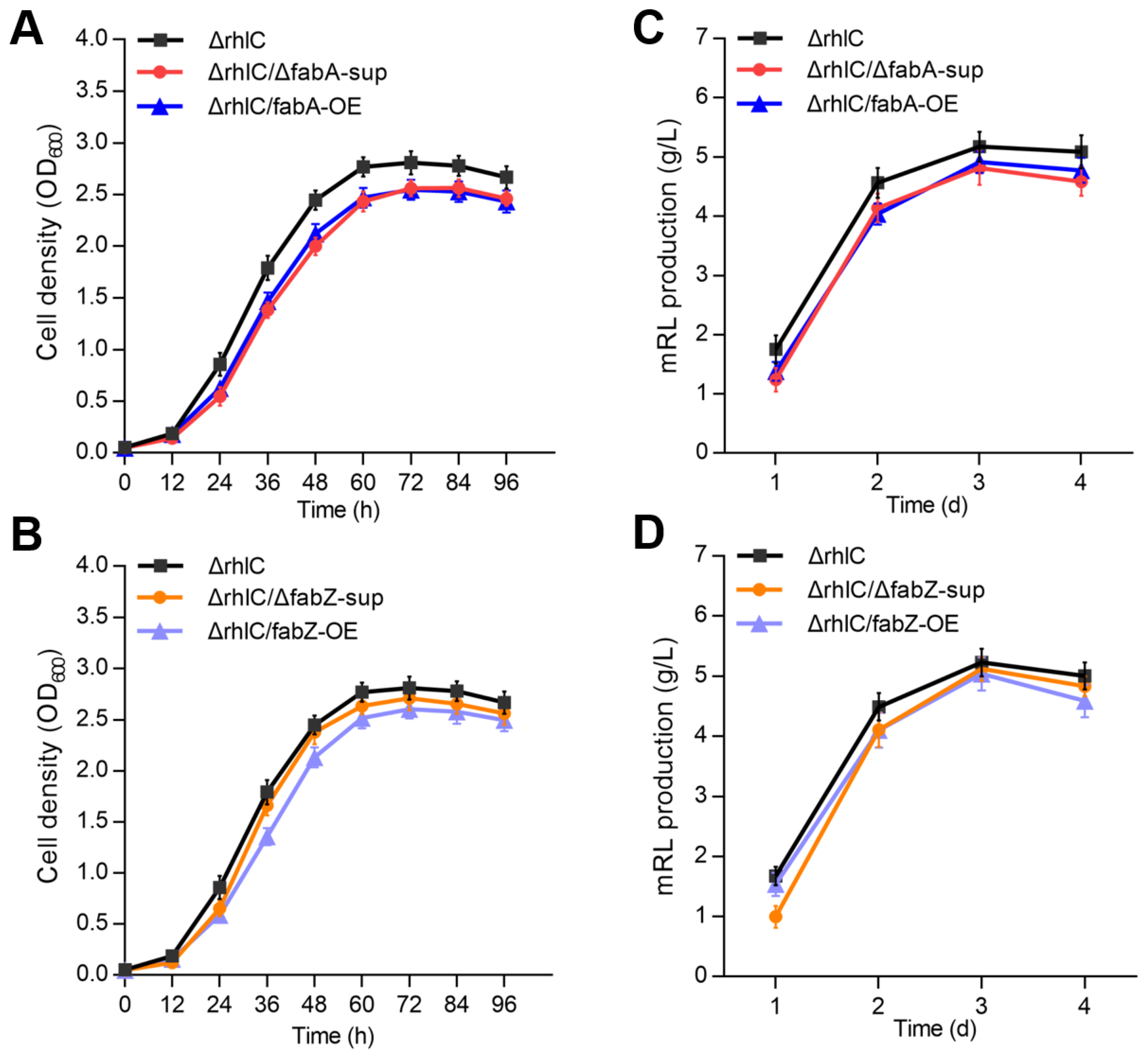
3.2. Effects of fabA and fabZ Deletion and Overexpression on Bacterial Growth and RL Production
3.3. Effects of fabA and fabZ Deletion and Overexpression on the Fatty Acyl Composition of mRLs
3.4. Emulsification Activity of mRLs with Modified Acyl Chain Composition Due to fabA and fabZ Perturbations
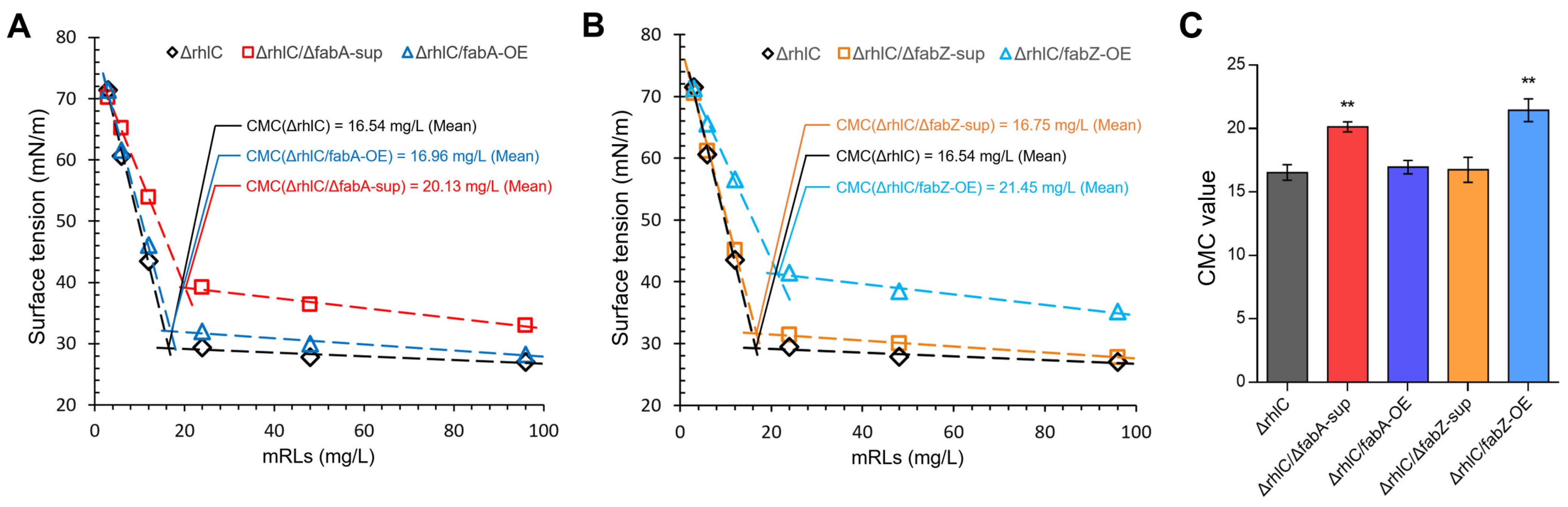
3.5. Critical Micelle Concentration (CMC) Analysis of mRLs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Irfan-Maqsood, M.; Seddiq-Shams, M. Rhamnolipids: Well-characterized glycolipids with potential broad applicability as biosurfactants. Ind. Biotechnol. 2014, 10, 285–291. [Google Scholar] [CrossRef]
- Thakur, P.; Saini, N.K.; Thakur, V.K.; Gupta, V.K.; Saini, R.V.; Saini, A.K. Rhamnolipid the Glycolipid Biosurfactant: Emerging trends and promising strategies in the field of biotechnology and biomedicine. Microb. Cell Factories 2021, 20, 1. [Google Scholar] [CrossRef]
- Soberón-Chávez, G.; Lépine, F.; Déziel, E. Production of rhamnolipids by Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2005, 68, 718–725. [Google Scholar] [CrossRef] [PubMed]
- Rahman, K.; Rahman, T.J.; McClean, S.; Marchant, R.; Banat, I.M. Rhamnolipid biosurfactant production by strains of Pseudomonas aeruginosa using low-cost raw materials. Biotechnol. Prog. 2002, 18, 1277–1281. [Google Scholar] [CrossRef]
- White, S.W.; Zheng, J.; Zhang, Y.-M.; Rock, C.O. The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 2005, 74, 791–831. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Rock, C.O. RhlA converts β-hydroxyacyl-acyl carrier protein intermediates in fatty acid synthesis to the β-hydroxydecanoyl-β-hydroxydecanoate component of rhamnolipids in Pseudomonas aeruginosa. J. Bacteriol. 2008, 190, 3147–3154. [Google Scholar] [CrossRef]
- Abdel-Mawgoud, A.M.; Lépine, F.; Déziel, E. Rhamnolipids: Diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 2010, 86, 1323–1336. [Google Scholar] [CrossRef]
- Costa, S.G.; Nitschke, M.; Lépine, F.; Déziel, E.; Contiero, J. Structure, properties and applications of rhamnolipids produced by Pseudomonas aeruginosa L2-1 from cassava wastewater. Process Biochem. 2010, 45, 1511–1516. [Google Scholar] [CrossRef]
- Gong, Z.; He, Q.; Che, C.; Liu, J.; Yang, G. Optimization and scale-up of the production of rhamnolipid by Pseudomonas aeruginosa in solid-state fermentation using high-density polyurethane foam as an inert support. Bioprocess Biosyst. Eng. 2020, 43, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Blunt, W.; Blanchard, C.; Morley, K. Effects of environmental parameters on microbial rhamnolipid biosynthesis and bioreactor strategies for enhanced productivity. Biochem. Eng. J. 2022, 182, 108436. [Google Scholar] [CrossRef]
- Zhao, F.; Cui, Q.; Han, S.; Dong, H.; Zhang, J.; Ma, F.; Zhang, Y. Enhanced rhamnolipid production of Pseudomonas aeruginosa SG by increasing copy number of rhlAB genes with modified promoter. RSC Adv. 2015, 5, 70546–70552. [Google Scholar] [CrossRef]
- Heath, R.J.; Rock, C.O. Roles of the FabA and FabZ β-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J. Biol. Chem. 1996, 271, 27795–27801. [Google Scholar] [CrossRef]
- Kimber, M.S.; Martin, F.; Lu, Y.; Houston, S.; Vedadi, M.; Dharamsi, A.; Fiebig, K.M.; Schmid, M.; Rock, C.O. The structure of (3R)-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Pseudomonas aeruginosa. J. Biol. Chem. 2004, 279, 52593–52602. [Google Scholar] [CrossRef]
- Ruppe, S.; Mains, K.; Fox, J.M. A kinetic rationale for functional redundancy in fatty acid biosynthesis. Proc. Natl. Acad. Sci. USA 2020, 117, 23557–23564. [Google Scholar] [CrossRef]
- Lee, S.A.; Gallagher, L.A.; Thongdee, M.; Staudinger, B.J.; Lippman, S.; Singh, P.K.; Manoil, C. General and condition-specific essential functions of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2015, 112, 5189–5194. [Google Scholar] [CrossRef]
- Goodall, E.C.; Robinson, A.; Johnston, I.G.; Jabbari, S.; Turner, K.A.; Cunningham, A.F.; Lund, P.A.; Cole, J.A.; Henderson, I.R. The essential genome of Escherichia coli K-12. mBio 2018, 9, e02096-17. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhang, Z.; Zhu, J.; Ma, Y.; Wang, J.; Liu, J. Analysis of the plasmid-based ts allele of PA0006 reveals its function in regulation of cell morphology and biosynthesis of core lipopolysaccharide in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2022, 88, e00480-22. [Google Scholar] [CrossRef]
- Tian, L.; Yang, Z.; Wang, J.; Liu, J. Analysis of the Plasmid-Based ts-Mutant Δ fabA/pTS-fabA Reveals Its Lethality under Aerobic Growth Conditions That Is Suppressed by Mild Overexpression of desA at a Restrictive Temperature in Pseudomonas aeruginosa. Microbiol. Spectr. 2023, 11, e01338-23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, Z.; Liu, J. Genetic Analysis of the Plasmid-Based Temperature-Lethal Mutant pa1792| lpxH (Ts) in Pseudomonas aeruginosa. Genes 2024, 15, 784. [Google Scholar] [CrossRef]
- Zhu, J.; Zhao, H.; Yang, Z. Genetic Analysis of the ts-Lethal Mutant Δpa0665/pTS-pa0665 Reveals Its Role in Cell Morphology and Oxidative Phosphorylation in Pseudomonas aeruginosa. Genes 2024, 15, 590. [Google Scholar] [CrossRef]
- Müller, M.M.; Hörmann, B.; Syldatk, C.; Hausmann, R. Pseudomonas aeruginosa PAO1 as a model for rhamnolipid production in bioreactor systems. Appl. Microbiol. Biotechnol. 2010, 87, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Rahim, R.; Ochsner, U.A.; Olvera, C.; Graninger, M.; Messner, P.; Lam, J.S.; Soberón-Chávez, G. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol. Microbiol. 2001, 40, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Guzman, L.-M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop II, R.M.; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef]
- Huang, W.; Wilks, A. A rapid seamless method for gene knockout in Pseudomonas aeruginosa. BMC Microbiol. 2017, 17, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Laabei, M.; Jamieson, W.D.; Lewis, S.E.; Diggle, S.P.; Jenkins, A.T.A. A new assay for rhamnolipid detection—Important virulence factors of Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2014, 98, 7199–7209. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.G.; Goldenberg, B.G. Surface-active agents from two Bacillus species. Appl. Environ. Microbiol. 1987, 53, 224–229. [Google Scholar] [CrossRef]
- Brückner, J. Estimation of monosaccharides by the orcinol–sulphuric acid reaction. Biochem. J. 1955, 60, 200. [Google Scholar] [CrossRef]
- Abalos, A.; Pinazo, A.; Infante, M.R.; Casals, M.; Garcia, F.; Manresa, A. Physicochemical and antimicrobial properties of new rhamnolipids produced by Pseudomonas a eruginosa AT10 from soybean oil refinery wastes. Langmuir 2001, 17, 1367–1371. [Google Scholar] [CrossRef]
- Déziel, É.; Lépine, F.; Dennie, D.; Boismenu, D.; Mamer, O.A.; Villemur, R. Liquid chromatography/mass spectrometry analysis of mixtures of rhamnolipids produced by Pseudomonas aeruginosa strain 57RP grown on mannitol or naphthalene. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 1999, 1440, 244–252. [Google Scholar] [CrossRef]
- Monteiro, S.A.; Sassaki, G.L.; de Souza, L.M.; Meira, J.A.; de Araújo, J.M.; Mitchell, D.A.; Ramos, L.P.; Krieger, N. Molecular and structural characterization of the biosurfactant produced by Pseudomonas aeruginosa DAUPE 614. Chem. Phys. Lipids 2007, 147, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Li, Y.; Tian, Y.; Hao, Z.; Chen, F.; Ma, Y. Enhanced rhamnolipid production of Pseudomonas aeruginosa DN1 by metabolic engineering under diverse nutritional factors. J. Pet. Environ. Biotechnol. 2018, 9, 384. [Google Scholar]
- Sinumvayo, J.P.; Ishimwe, N. Agriculture and food applications of rhamnolipids and its production by Pseudomonas aeruginosa. J. Chem. Eng. Process Technol. 2015, 6, 1. [Google Scholar] [CrossRef]
- Chong, H.; Li, Q. Microbial production of rhamnolipids: Opportunities, challenges and strategies. Microb. Cell Factories 2017, 16, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pang, A.-P.; Wang, Y.; Zhang, T.; Gao, F.; Shen, J.-D.; Huang, L.; Zhou, J.; Zhang, B.; Liu, Z.-Q.; Zheng, Y.-G. Highly efficient production of rhamnolipid in P. putida using a novel sacB-based system and mixed carbon source. Bioresour. Technol. 2024, 394, 130220. [Google Scholar] [CrossRef] [PubMed]
- Sakthipriya, N.; Doble, M.; Sangwai, J.S. Biosurfactant from Pseudomonas species with waxes as carbon source–Their production, modeling and properties. J. Ind. Eng. Chem. 2015, 31, 100–111. [Google Scholar] [CrossRef]
- Wittgens, A.; Santiago-Schuebel, B.; Henkel, M.; Tiso, T.; Blank, L.M.; Hausmann, R.; Hofmann, D.; Wilhelm, S.; Jaeger, K.-E.; Rosenau, F. Heterologous production of long-chain rhamnolipids from Burkholderia glumae in Pseudomonas putida—A step forward to tailor-made rhamnolipids. Appl. Microbiol. Biotechnol. 2018, 102, 1229–1239. [Google Scholar] [CrossRef]
- Tiso, T.; Zauter, R.; Tulke, H.; Leuchtle, B.; Li, W.-J.; Behrens, B.; Wittgens, A.; Rosenau, F.; Hayen, H.; Blank, L.M. Designer rhamnolipids by reduction of congener diversity: Production and characterization. Microb. Cell Factories 2017, 16, 1–14. [Google Scholar] [CrossRef]
- Xiao, X.; Yu, X.; Khosla, C. Metabolic flux between unsaturated and saturated fatty acids is controlled by the FabA: FabB ratio in the fully reconstituted fatty acid biosynthetic pathway of Escherichia coli. Biochemistry 2013, 52, 8304–8312. [Google Scholar] [CrossRef][Green Version]
- Dodge, G.J.; Patel, A.; Jaremko, K.L.; McCammon, J.A.; Smith, J.L.; Burkart, M.D. Structural and dynamical rationale for fatty acid unsaturation in Escherichia coli. Proc. Natl. Acad. Sci. USA 2019, 116, 6775–6783. [Google Scholar] [CrossRef]
- Guzmán, E.; Ortega, F.; Rubio, R.G. Exploring the world of rhamnolipids: A critical review of their production, interfacial properties, and potential application. Curr. Opin. Colloid Interface Sci. 2024, 69, 101780. [Google Scholar] [CrossRef]
- Palos Pacheco, R.; Kegel, L.L.; Pemberton, J.E. Interfacial and solution aggregation behavior of a series of bioinspired rhamnolipid congeners rha-C14-c x (x= 6, 8, 10, 12, 14). J. Phys. Chem. B 2021, 125, 13585–13596. [Google Scholar] [CrossRef] [PubMed]
- Mitrinova, Z.; Tcholakova, S.; Popova, Z.; Denkov, N.; Dasgupta, B.R.; Ananthapadmanabhan, K. Efficient control of the rheological and surface properties of surfactant solutions containing C8–C18 fatty acids as cosurfactants. Langmuir 2013, 29, 8255–8265. [Google Scholar] [CrossRef]
- Youssef, N.H.; Nguyen, T.; Sabatini, D.A.; McInerney, M.J. Basis for formulating biosurfactant mixtures to achieve ultra low interfacial tension values against hydrocarbons. J. Ind. Microbiol. Biotechnol. 2007, 34, 497–507. [Google Scholar] [CrossRef] [PubMed]


| (A) Oligonucleotides | ||
|---|---|---|
| Name | Sequence (5′-3′) | Usage |
| rhlC-F | AAGAACGATCATGGACCGGATA | Assay rhlC alleles in chr |
| rhlC-R | GAATGCGTTTCGCCGACTAG | Ditto |
| fabZ-F | CACCACGTGCGGACCGATCA | Assay fabZ alleles in chr and ts-plasmid |
| fabZ-R | CCTGGCTGCCGTGACCTCAA | Ditto |
| (B) Plasmids | ||
| Name | Relevant Genotype | Reference |
| pDel | pUC-Gmr-sacB | [17,18,19,20] |
| pTS or pRES | pUC-Tcr-orits | [17,18,19,20] |
| pOE | pBBRMCS-5-araC-PBAD-Gmr | [17,18,19,20] |
| pDel-fabZ | fabZ Deletion cassette in pDel | This study |
| pTS-fabZ | fabZ rescue cassette in pTS | This study |
| pDel-rhlC | rhlC Deletion cassette in pDel | This study |
| pOE-fabZ | araC-PBAD-fabZ in pOE | [18] |
| pOE-fabA | araC-PBAD-fabA in pOE | [18] |
| (C) Strains | ||
| Name | Relevant Genotype | Reference |
| PAO1 | Wild type P. aeruginosa | [17,18,19,20] |
| ΔrhlC | ΔrhlC in P. aeruginosa | This study |
| ΔfabA-sup | suppressor of ΔfabA | [18] |
| ΔrhlC/ΔfabA-sup | ΔrhlC in ΔfabA-sup | This study |
| fabA-OE | pOE-fabA in PAO1 | [18] |
| ΔrhlC/fabA-OE | ΔrhlC in fabA-OE | This study |
| ΔfabZ-sup | suppressor of ΔfabZ | This study |
| ΔrhlC/ΔfabZ-sup | ΔrhlC in ΔfabZ-sup | This study |
| fabZ-OE | pOE-fabZ in PAO1 | [18] |
| ΔrhlC/fabZ-OE | ΔrhlC in fabZ-OE | This study |
| Formula | M/Z | ΔrhlC (% Level, Mean) | ΔrhlC/ΔfabA-Sup (% Level, Mean) | ΔrhlC/fabA-OE (% Level, Mean) | ΔrhlC/ΔfabZ-Sup (% Level, Mean) | ΔrhlC/fabZ-OE (% Level, Mean) | Structure | |
|---|---|---|---|---|---|---|---|---|
| 1 | C22H40O9 | 448.27 | 3.70% | 0.48% | 27.54% | 0.20% | 20.29% | Rha-C8-C8 or Rha-C10-C6 |
| 2 | C24H44O9 | 476.3 | 7.23% | 3.34% | 9.30% | 1.81% | 4.57% | Rha-C10-C8 or Rha-C8-C10 |
| 3 | C26H48O9 | 504.33 | 27.61% | 12.94% | 10.10% | 43.44% | 16.32% | Rha-C10-C10 |
| 4 | C28H50O9 | 530.35 | 30.31% | 59.30% | 19.41% | 45.82% | 30.90% | Rha-C10-C12:1 or Rha-C12:1-C10 |
| 5 | C28H52O9 | 532.36 | 17.35% | 6.48% | 7.10% | 1.62% | 5.61% | Rha-C10-C12 or Rha-C12-C10 |
| 6 | C30H54O9 | 557.37 | 8.46% | 14.07% | 23.40% | 3.97% | 18.33% | Rha-C10-C14:1 or Rha-C14:1-C10 |
| 7 | C30H56O9 | 559.38 | 5.34% | 3.39% | 3.16% | 3.14% | 3.97% | Rha-C10-C14 or Rha-C14-C10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, J.; Chen, Z.; Zhu, H.; Tang, Q.; Yang, Z. Optimizing Rhamnolipid Performance by Modulating the Expression of Fatty Acid Synthesis Genes fabA and fabZ in Pseudomonas aeruginosa PAO1. Genes 2025, 16, 515. https://doi.org/10.3390/genes16050515
Lu J, Chen Z, Zhu H, Tang Q, Yang Z. Optimizing Rhamnolipid Performance by Modulating the Expression of Fatty Acid Synthesis Genes fabA and fabZ in Pseudomonas aeruginosa PAO1. Genes. 2025; 16(5):515. https://doi.org/10.3390/genes16050515
Chicago/Turabian StyleLu, Junpeng, Zhenhua Chen, Huiming Zhu, Qinghai Tang, and Zhili Yang. 2025. "Optimizing Rhamnolipid Performance by Modulating the Expression of Fatty Acid Synthesis Genes fabA and fabZ in Pseudomonas aeruginosa PAO1" Genes 16, no. 5: 515. https://doi.org/10.3390/genes16050515
APA StyleLu, J., Chen, Z., Zhu, H., Tang, Q., & Yang, Z. (2025). Optimizing Rhamnolipid Performance by Modulating the Expression of Fatty Acid Synthesis Genes fabA and fabZ in Pseudomonas aeruginosa PAO1. Genes, 16(5), 515. https://doi.org/10.3390/genes16050515






