Association Analysis of ENPP1 Tissue Expression, Polymorphism, and Growth Traits in Xiangsu Pigs
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Whole-Blood DNA, Tissue RNA Extraction, and cDNA Synthesis
2.3. Primer Design
2.4. PCR Sequence Amplification and Real-Time Fluorescence Quantitative Analysis
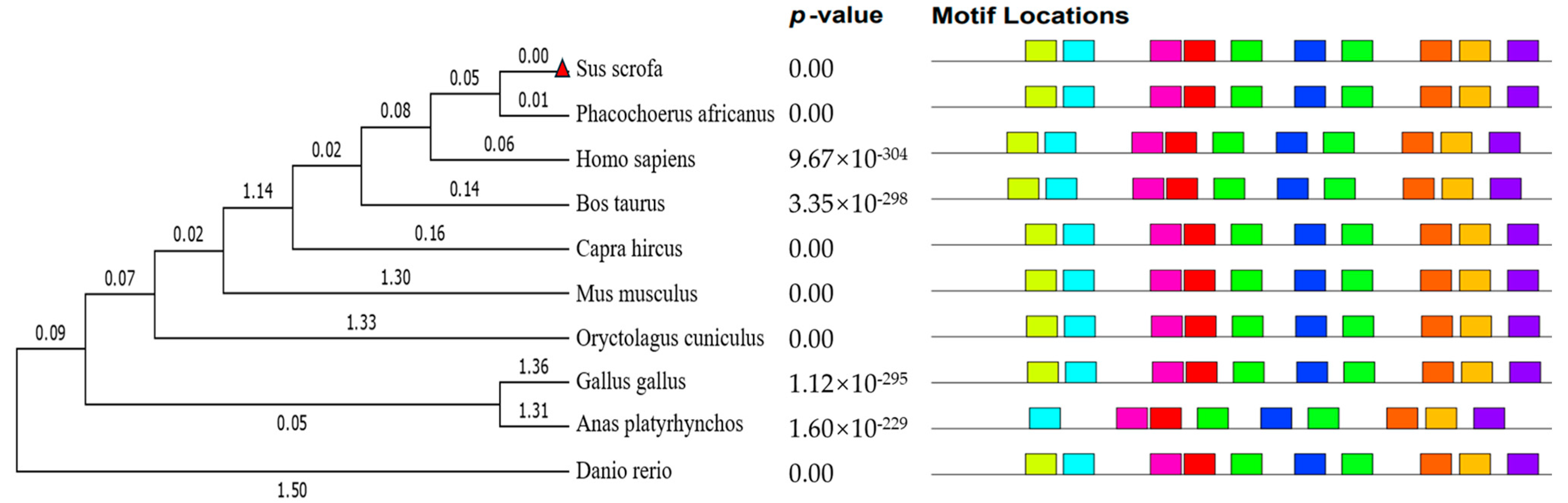
2.5. Bioinformatics Analysis
2.6. Statistical Analysis
3. Results
3.1. Expression Levels of ENPP1 in Different Tissues of Xiangsu Pigs
3.2. Analysis of ENPP1 Gene Polymorphism
3.3. Biodiversity Analysis
3.4. Analysis of Growth Data of Xiangsu Hybrid Pig Population
3.5. Analysis of Population Genetic Diversity
3.6. Correlation Analysis Between Genotype and Growth Traits
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yuan, H.; Wei, W.; Zhang, Y.; Li, C.; Zhao, S.; Chao, Z.; Xia, C.; Quan, J.; Gao, C. Unveiling the Influence of Copy Number Variations on Genetic Diversity and Adaptive Evolution in China’s Native Pig Breeds via Whole-Genome Resequencing. Int. J. Mol. Sci. 2024, 25, 5843. [Google Scholar] [CrossRef]
- Xu, J.; Ruan, Y.; Sun, J.; Shi, P.; Huang, J.; Dai, L.; Xiao, M.; Xu, H. Association Analysis of PRKAA2 and MSMB Polymorphisms and Growth Traits of Xiangsu Hybrid Pigs. Genes 2022, 14, 113. [Google Scholar] [CrossRef]
- Huang, J.; Ruan, Y.; Xiao, M.; Dai, L.; Jiang, C.; Li, J.; Xu, J.; Chen, X.; Xu, H. Association between polymorphisms in NOBOX and litter size traits in Xiangsu pigs. Front. Vet. Sci. 2024, 11, 1359312. [Google Scholar] [CrossRef]
- Borza, R.; Salgado-Polo, F.; Moolenaar, W.H.; Perrakis, A. Structure and function of the ecto-nucleotide pyrophosphatase/phosphodiesterase (ENPP) family: Tidying up diversity. J. Biol. Chem. 2022, 298, 101526. [Google Scholar] [CrossRef]
- Onyedibe, K.I.; Wang, M.; Sintim, H.O. ENPP1, an old enzyme with new functions, and small molecule inhibitors-A sting in the tale of ENPP. Molecules 2019, 24, 4192. [Google Scholar] [CrossRef]
- Bollen, M.; Gijsbers, R.; Ceulemans, H.; Stalmans, W.; Stefan, C. Nucleotide pyrophosphatases/phosphodiesterases on the move. Crit. Rev. Biochnol. 2000, 35, 393–432. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Fang, L.; Feng, M.; Guo, S.; Xie, L.; Chen, C.; Zhou, J.; Zhang, H. Ecto-nucleotide pyrophosphatase/phosphodiesterase 1 inhibitors: Research progress and prospects. Eur. J. Med. Chem. 2024, 267, 116211. [Google Scholar] [CrossRef]
- Dong, H.; Maddux, B.A.; Altomonte, J.; Meseck, M.; Accili, D.; Youngren, J.F.; Goldfine, I.D. Increased Hepatic Levels of the Insulin Receptor Inhibitor, PC-1/NPP1, Induce Insulin Resistance and Glucose Intolerance. Diabetes 2025, 54, 367–372. [Google Scholar] [CrossRef][Green Version]
- Arnold, A.; Dennison, E.; Kovacs, C.S.; Mannstadt, M.; Rizzoli, R.; Brandi, M.L.; Clarke, B.; Thakker, R.V. 2021. Hormonal regulation of biomineralization. Nat. Rev. Endocrinol. 2021, 17, 261–275. [Google Scholar] [CrossRef]
- Cheng, Z.; O’Brien, K.; Howe, J.; Sullivan, C.; Schrier, D.; Lynch, A.; Jungles, S.; Sabbagh, Y.; Thompson, D. INZ-701 prevents ectopic tissue calcification and restores bone architecture and growth in ENPP1-deficient mice. J. Bone Miner. Res. 2020, 3, 1594–1604. [Google Scholar] [CrossRef]
- Brachet, C.; Mansbach, A.L.; Clerckx, A.; Deltenre, P.; Heinrichs, C. Hearing loss is part of the clinical picture of ENPP1 loss of function mutation. Horm. Res. Paediat. 2014, 81, 63–66. [Google Scholar] [CrossRef]
- Roberts, F.L.; Rashdan, N.A.; Phadwal, K.; Markby, G.R.; Zoll, J.; Farquharson, C.; MacRae, V.E. Osteoblast-specific deficiency of ectonucleotide pyrophosphatase or phosphodiesterase-1 engenders insulin resistance in high-fat diet fed mice. J. Cell. Physiol. 2021, 236, 4614–4624. [Google Scholar] [CrossRef]
- Neamati, N.; Hosseini, S.R.; Hajiahmadi, M.; Halalkhor, S.; Nooreddini, H.; Niaki, H.A.; Korani, B.; Parsian, H. The ENPP1 K121Q polymorphism modulates developing of bone disorders in type 2 diabetes: A cross sectional study. Gene 2017, 637, 100–107. [Google Scholar] [CrossRef]
- Pan, W.; Ciociola, E.; Saraf, M.; Tumurbaatar, B.; Tuvdendorj, D.; Prasad, S.; Chandalia, M.; Abate, N. Metabolic consequences of ENPP1 overexpression in adipose tissue. Am. J. Physiol. Endocrinol. Metab. 2011, 301, E901–E911. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, K.; Liu, X.; Von, K.S.; Stabach, P.; Lester, E.R.; Chu, E.Y.; Srivastava, S.; Somerman, M.J.; Tommasini, S.M.; Busse, B.; et al. Catalysis-Independent ENPP1 Protein Signaling Regulates Mammalian Bone Mass. J. Bone Miner. Res. 2022, 37, 1733–1749. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Guo, W.; Liao, Y.; Xu, D.; Sun, B.; Song, H.; Wang, T.; Kuang, Y.; Jing, B.; Li, K.; et al. Dysregulated ENPP1 increases the malignancy of human lung cancer by inducing epithelial-mesenchymal transition phenotypes and stem cell features. Am. J. Cancer Res. 2019, 9, 134–144. [Google Scholar]
- Wu, G.; Yu, M.; Liu, T.; Zhang, D.; Chang, Y.; Liu, Z.; Liu, D.; Xu, C. Integration of Multiomics Data Reveals Selection Characteristics of ITGB1 That Are Associated with Size Differentiation in Pigs. Int. J. Mol. Sci. 2025, 26, 1569. [Google Scholar] [CrossRef] [PubMed]
- Goldfine, I.D.; Maddux, B.A.; Youngren, J.F.; Reaven, G.; Accili, D.; Vigneri, R.; Frittitta, L. The role of membrane glycoprotein plasma cell antigen 1/ectonucleotide pyrophosphatase phosphodiesterase 1 in the pathogenesis of insulin resistance and related abnormalities. Endocr. Rev. 2008, 29, 62–75. [Google Scholar] [CrossRef]
- Zhu, M.; Wang, L.; Ma, Z.; Mao, Y.; Wang, G.; Zheng, R.; Zuo, B.; Wang, Y. Analysis of the Genetic Diversity and Genetic Structure of Jiangshan Black Pigs Using Single Nucleotide Polymorphism (SNP) Chips. Animals 2024, 14, 2660. [Google Scholar] [CrossRef]
- Zwierzchowski, L.; Ostrowska, M.; Żelazowska, B.; Bagnicka, E. Single nucleotide polymorphisms in the bovine SLC2A12 and SLC5A1 glucose transporter genes—The effect on gene expression and milk traits of Holstein Friesian cows. Anim. Biotechnol. 2023, 34, 225–235. [Google Scholar] [CrossRef]
- Liu, C.; Ran, X.; Wang, J.; Li, S.; Liu, J. Detection of genomic structural variations in Guizhou indigenous pigs and the comparison with other breeds. PLoS ONE 2018, 13, e0194282. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.T.; Ran, X.Q.; Mao, N.; Zhang, F.P.; Niu, X.; Ruan, Y.Q.; Yi, F.L.; Li, S.; Wang, J.F. Analysis of alternative splicing events by RNA sequencing in the ovaries of Xiang pig at estrous and diestrous. Theriogenology 2018, 119, 60–68. [Google Scholar] [CrossRef]
- Luo, Z.Y.; Dai, X.L.; Ran, X.Q.; Cen, Y.X.; Niu, X.; Li, S.; Huang, S.H.; Wang, J.F. Identification and profile of microRNAs in Xiang pig testes in four different ages detected by Solexa sequencing. Theriogenology 2018, 117, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Bao, L.; Gong, S.; Dou, G.; Li, Z.; Wang, Z.; Yu, L.; Ding, F.; Yang, X.; Liu, S. T Cell-Derived Apoptotic Extracellular Vesicles Hydrolyze cGAMP to Alleviate Radiation Enteritis via Surface Enzyme ENPP1. Adv. Sci. 2024, 11, e2401634. [Google Scholar] [CrossRef]
- Zhou, X.; Pan, Y.; Wang, Y.; Wang, B.; Yan, Y.; Qu, Y.; Ke, X. Tanshinones induce tumor cell apoptosis via directly targeting FHIT. Sci. Rep. 2021, 11, 12217. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, Y.; Tian, Y.; Ding, X.; Lin, R.; Xiao, L.; Peng, F.; Zhang, K.; Yang, Z. Role of ENPP1 in cancer pathogenesis: Mechanisms and clinical implications (Review). Oncol. Lett. 2024, 28, 590. [Google Scholar] [CrossRef]
- Zhu, Z.; He, Z.; Tang, T.; Wang, F.; Chen, H.; Zhou, J.; Lin, C.; Chen, G.; Wang, J.; Li, J.; et al. Effect of mechanical stimulation on tissue heterotopic ossification: An in vivo experimental study. Front. Physiol. 2023, 14, 1225898. [Google Scholar] [CrossRef]
- Furukawa, H.; Oka, S.; Kondo, N.; Nakagawa, Y.; Shiota, N.; Kumagai, K.; Ando, K.; Takeshita, T.; Oda, T.; Yoshikawa, N.; et al. The Contribution of Deleterious Rare Alleles in ENPP1 and Osteomalacia Causative Genes to Atypical Femoral Fracture. J. Clin. Endocrinol. Metab. 2022, 107, e1890–e1898. [Google Scholar] [CrossRef]
- Li, Q.; van de Wetering, K.; Uitto, J. Pseudoxanthoma Elasticum as a Paradigm of Heritable Ectopic Mineralization Disorders: Pathomechanisms and Treatment Development. Am. J. Pathol. 2018, 189, 216–225. [Google Scholar] [CrossRef]
- Ferreira, C.R.; Kavanagh, D.; Oheim, R.; Zimmerman, K.; Stürznickel, J.; Li, X.; Stabach, P.; Rettig, R.L.; Levine, M.A.; Horowitz, M.C.; et al. Response of the ENPP1-Deficient Skeletal Phenotype to Oral Phosphate Supplementation and/or Enzyme Replacement Therapy: Comparative Studies in Humans and Mice. J. Bone Miner. Res. 2021, 36, 942–955. [Google Scholar] [CrossRef]
- Zhu, L.; Zhang, L.; Cha, J.; Li, C.; Mao, B. Loss of ZC4H2, an Arthrogryposis Multiplex Congenita Associated Gene, Promotes Osteoclastogenesis in Mice. Genes 2024, 15, 1134. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Li, J.; Tao, L. Purine metabolism in the development of osteoporosis. Biomed. Pharmacother. 2022, 155, 113784. [Google Scholar] [CrossRef]
- Chang, C.C.; Lee, K.L.; Chan, T.S.; Chung, C.C.; Liang, Y.C. Histone Deacetylase Inhibitors Downregulate Calcium Pyrophosphate Crystal Formation in Human Articular Chondrocytes. Int. J. Mol. Sci. 2022, 23, 2604. [Google Scholar] [CrossRef] [PubMed]
- Mäki-Tanila, A.; Webster, L. Heritability, SNP, inbreeding, dairy cattle, genomic selection—And other keywords. J. Anim. Genet. 2019, 136, 1–2. [Google Scholar] [CrossRef]
- Azimu, W.; Manatbay, B.; Li, Y.; Kaimaerdan, D.; Wang, H.E.; Reheman, A.; Muhatai, G. Genetic diversity and population structure analysis of eight local chicken breeds of Southern Xinjiang. Br. Poult. Sci. 2018, 59, 629–635. [Google Scholar] [CrossRef]
- Wang, M.; Shu, H.; Xie, J.; Huang, Y.; Wang, K.; Feng, R.; Yu, X.; Guan, J.; Feng, W.; Liu, M. An intron mutation of HNF1A causes abnormal splicing and impairs its activity as a transcription facto. Mol. Cellular. Endocrinol. 2022, 545, 111575. [Google Scholar] [CrossRef]
- Abou Tayoun, A.N.; Pesaran, T.; DiStefano, M.T.; Oza, A.; Rehm, H.L.; Biesecker, L.G.; Harrison, S.M. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Human Mutat. 2018, 39, 1517–1524. [Google Scholar] [CrossRef]
- Chiang, H.L.; Chen, Y.T.; Su, J.Y.; Lin, H.N.; Yu, C.A.; Hung, Y.J.; Lin, C.L. Mechanism and modeling of human disease-associated near-exon intronic variants that perturb RNA splicing. Nat. Struct. Mol. Biol. 2022, 29, 1043–1055. [Google Scholar] [CrossRef]
- Mauro, V.P.; Chappell, S.A. A critical analysis of codon optimization in human therapeutics. Trends Mol. Med. 2014, 20, 604–613. [Google Scholar] [CrossRef]
- Safdar, M.; Khan, M.S.; Karim, A.Y.; Omar, S.A.; Smail, S.W.; Saeed, M.; Zaheer, S.; Ali, M.; Ahmad, B.; Tasleem, M.; et al. SNPs at 3′UTR of APOL1 and miR-6741-3p target sites associated with kidney diseases more susceptible to SARS-CoV-2 infection: In silco and in vitro. Mamm. Genome 2021, 32, 389–400. [Google Scholar] [CrossRef]
- Clark, D.L.; Bohrer, B.M.; Tavárez, M.A.; Boler, D.D.; Beever, J.E.; Dilger, A.C. Effects of the porcine IGF2 intron 3-G3072A mutation on carcass cutability, meat quality, and bacon processing. J. Anim. Sci. 2014, 92, 5778–5788. [Google Scholar] [CrossRef] [PubMed]
- Frittitta, L.; Ercolino, T.; Bozzali, M. A cluster of three single nucleotide polymorphisms in the 3′-untranslated region of human glycoprotein PC-1 gene stabilizes PC-1 mRN and is associated with increased PC-1 protein content and insulin resistance-related abnormalities. Diabetes 2021, 50, 1952–1955. [Google Scholar] [CrossRef]




| Genes | Product Size (bp) | Primer Sequence (5′-3′) | Tm (°C) |
|---|---|---|---|
| ENPP1-Exon1 | 808 | F: AGTGCCCGAAATCAGACAGGAAG R: CCTTGTCGGCTTTTCCTAAGACC | 56 |
| ENPP1-Exon2 | 241 | F: GCACACATTGTATGACCAGCACTA R: CGAATCTCAGTCACATTCTGAAGG | 57 |
| ENPP1-Exon3 | 992 | F: CAGCAATGCCAGATCCTTAACCC R: CCAGTCACTGTTGGAGAACTCAGAA | 57 |
| ENPP1-Exon4 | 875 | F: ACTCGGACTTAGGAAATAGCCCA R: TACCTGTGCTAAACCCAGCTAAGAC | 63 |
| ENPP1-Exon5 | 644 | F: GTGGATAGAAGGAGGGTTGTCAGTT R: GAGTTCTCTTCTTGGCTCAGCAGTT | 65 |
| ENPP1-Exon6 | 712 | F: CTTTAGACGACATGCCGACATCAG R: GGCTACAGCTCTGATTAGACCCCTA | 62 |
| ENPP1-Exon7 | 645 | F: CTTTTCTTAGCACTGGTGCCAAAG R: GAACACTGTAAACCAGCCGAAATG | 62 |
| ENPP1-Exon8 | 665 | F: ACCAAAGCAAAGAGGAGTTTGCTC R: ACCCAGCTCTGGAATAAACTCCAC | 63 |
| ENPP1-Exon9 | 325 | F: CTTAAGAGGAAAGCACTGGTGTTG R: ACAGGTCTCATCGTTCAGACAACA | 61 |
| ENPP1-Exon10 | 486 | F: GCGTCCACTTCCCAGTTTTACTA R: TCAGAGCCAAGAGCAAAGTCTTC | 57 |
| ENPP1-Exon11 | 408 | F: AGGAATGATACCCCAGTGTTGTG R: CCACTGGTTCAACTTAGCACTGTAG | 59 |
| ENPP1-Exon12 | 475 | F: ATGTGCCATTCTGTGCCCTTGT R: CGGAGCCACAACAGGAACTCTTAT | 59 |
| ENPP1-Exon13 | 896 | F: CTCTGGATATATGTCCAGGAATGGG R: TCATCTCTTCCAGCATTCCTCCTT | 57 |
| ENPP1-Exon14 | 598 | F: GGCTGAGTGGTTCCGAAAAATCT R: AACCAGAGCCACAGTGGTGAGAAT | 59 |
| ENPP1-Exon15 | 543 | F: AACTAGGGACTGAATCTAGGCATCC R: GCCAAGGTTAAGAGACCCTGAGATA | 59 |
| ENPP1-Exon16 | 843 | F: TAGCATGGTCAGGGTTCAAAGTC R: TCTCACTCTGCCCAACATACACA | 60 |
| ENPP1-Exon17 | 363 | F:CATGGTAAATGTACACCAGGGGTT R:CCTGACCTATGACCTCCTTTTGAT | 62 |
| ENPP1-Exon18 | 702 | F: CAAGGTCTGTCTGTATTCAGCTACC R: AGGTCTGCCTGTGTGACTAGAATTG | 56 |
| ENPP1-Exon19 | 792 | F: GGGAGAGATCTAGCTTAGTCCTGGA R: TCTGTTTCATCAGGTGGTCCTCC | 56 |
| ENPP1-Exon20 | 977 | F: CGAATCCGACTAGGAACCATGAG R: CGGGGTTTCAAAGTCACGTAGATAG | 57 |
| ENPP1-Exon21 | 791 | F: GCATATGGAGGTCACCAGCTTATG R: TAGCTCCAATTAGACCCCTAGCCT | 59 |
| ENPP1-Exon22 | 917 | F: TAGTTGAAATGAGGCAGAGGAGG R: GTTATCCGGGCTGATGGTCAATA | 57 |
| ENPP1-Exon23 | 677 | F: GGGATGCTTGAAGTACAGCCGAT R: GACAAGATCGTAACAGCAGCCACA | 57 |
| ENPP1-Exon24 | 557 | F: CAGTCTGGCAGGAAATATTGAGG R: GCCATACGTAGCCAGTGTAAGGTTT | 57 |
| ENPP1-Exon25 | 617 | F: TGATGAGAACGGAGGAGATGGA R: AAGGGTTTGCCAGCATCAGAGA | 61 |
| ENPP1-Exon26 | 1006 | F: GCAGAAGTGAATGTTGTCTGGTCAC R: CATCAGAATGGATTAGGCAGAGGA | 59 |
| ENPP1-Exon27 | 1478 | F: CCATACCCTTTAGTACCAACCTGTG R: GATGCTCTACATAGAGGCAACTGTG | 61 |
| ENPP1-qPCR | 109 | F: GAGAACATTTGGGAACTGTCGC R: GCTGCAAGTCCATATCCGTTCT | 63 |
| GAPDH-qPCR | 169 | F: TTGTGATGGGCGTGAACC R: GTCTTCTGGGTGGCAGTGAT | 58 |
| Index | W/kg | B S/cm | B H/cm | C C/cm | A C/cm | T C/cm | C D/cm | C W/cm | L H C/cm |
|---|---|---|---|---|---|---|---|---|---|
| Mean | 91.08 | 110.18 | 75.39 | 110.75 | 120.12 | 20.22 | 40.09 | 30.12 | 71.53 |
| Max | 99.92 | 119 | 83 | 119 | 130 | 23 | 46 | 34 | 75 |
| Min | 82.47 | 100 | 68 | 103 | 109 | 17 | 34 | 26 | 65 |
| SD | 4.55 | 4.70 | 3.97 | 3.65 | 3.99 | 1.25 | 2.88 | 1.96 | 2.01 |
| SNPs | Genotypic Frequencies | Allelic Frequency | χ2 | |||
|---|---|---|---|---|---|---|
| g.64275T→C | TT | TC | g.64275T→C | T | C | 0.953 |
| 0.073(12) | 0.339(56) | 0.588(97) | 0.242 | 0.758 | ||
| g.64429G→A | GG | GA | g.64429G→A | G | A | 3.802 |
| 0.436(72) | 0.394(65) | 0.170(28) | 0.633 | 0.367 | ||
| g.64527T→C | TT | TC | g.64527T→C | T | C | 0.014 |
| 0.424(70) | 0.4(66) | 0.176(29) | 0.624 | 0.376 | ||
| g.64850T→C | TT | TC | g.64850T→C | T | C | 0.213 |
| 0.333(55) | 0.503(83) | 0.164(27) | 0.585 | 0.415 | ||
| g.64911G→A | GG | GA | g.64911G→A | G | A | 0.103 |
| 0.358(59) | 0.491(81) | 0.152(25) | 0.603 | 0.397 | ||
| SNPs | He | Ho | Ne | PIC |
|---|---|---|---|---|
| g.64275T→C | 0.367 | 0.633 | 1.581 | 0.300 |
| g.64429G→A | 0.464 | 0.536 | 1.867 | 0.357 |
| g.64527T→C | 0.499 | 0.502 | 1.994 | 0.374 |
| g.64850T→C | 0.456 | 0.514 | 1.994 | 0.368 |
| g.64911G→A | 0.479 | 0.521 | 1.919 | 0.364 |
| SNPs | Genotype | W/kg | B S/cm | B H/cm | C C/cm | A C/cm | T C/cm | C D/cm | C W/cm | L H C/cm |
|---|---|---|---|---|---|---|---|---|---|---|
| g.64275T→C | TT(12) | 89.91 ± 4.41 | 111.25 ± 4.99 | 74.67 ± 3.94 | 109.33 ± 1.92 | 118.58 ± 2.15 | 20.42 ± 0.67 | 38.92 ± 2.47 | 29.92 ± 2.31 | 71.58 ± 2.07 |
| TC(56) | 90.13 ± 4.51 a | 109.65 ± 4.80 | 74.58 ± 3.95 a | 109.85 ± 3.31 A | 119.49 ± 3.82 | 20.00 ± 1.17 | 39.45 ± 2.87 a | 30.07 ± 1.94 | 71.38 ± 1.88 | |
| CC(97) | 91.76 ± 4.50 b | 110.35 ± 4.62 | 75.93 ± 3.94 b | 111.43 ± 3.85 B | 120.65 ± 4.17 | 20.33 ± 1.33 | 40.60 ± 2.85 b | 30.17 ± 1.95 | 71.61 ± 2.09 | |
| g.64429G→A | GG(72) | 90.38 ± 4.54 | 109.94 ± 5.05 | 74.68 ± 3.89 | 110.28 ± 3.60 | 119.78 ± 4.14 | 20.17 ± 1.20 | 39.50 ± 2.88 A | 30.01 ± 2.00 | 71.65 ± 1.82 |
| GA(64) | 91.73 ± 4.72 | 110.14 ± 4.48 | 75.89 ± 4.19 | 110.89 ± 3.61 | 120.18 ± 3.75 | 20.28 ± 1.32 | 40.26 ± 2.87 | 30.12 ± 1.91 | 71.48 ± 2.34 | |
| AA(29) | 91.39 ± 4.02 | 110.89 ± 4.37 | 76.04 ± 3.46 | 111.64 ± 3.79 | 120.82 ± 4.16 | 20.25 ± 1.24 | 41.25 ± 2.58 B | 30.39 ± 2.04 | 71.36 ± 1.68 | |
| g.64527T→C | TT(70) | 90.50 ± 4.51 | 109.97 ± 5.06 | 74.94 ± 3.86 | 110.36 ± 3.60 | 119.87 ± 4.16 | 20.17 ± 1.20 | 39.54 ± 2.91 a | 29.99 ± 2.00 | 71.74 ± 1.78 |
| TC(66) | 91.50 ± 4.80 | 110.06 ± 4.50 | 75.67 ± 4.25 | 110.79 ± 3.65 | 120.11 ± 3.77 | 20.27 ± 1.32 | 40.24 ± 2.85 | 30.20 ± 1.88 | 71.38 ± 2.36 | |
| CC(29) | 91.53 ± 4.02 | 110.97 ± 4.31 | 75.83 ± 3.58 | 111.62 ± 3.73 | 120.72 ± 4.12 | 20.24 ± 1.21 | 41.10 ± 2.65 b | 30.28 ± 2.10 | 71.38 ± 1.66 | |
| g.64850T→C | TT(55) | 90.81 ± 4.39 | 111.04 ± 4.53 | 75.36 ± 3.70 | 110.67 ± 3.39 | 120.04 ± 3.57 | 20.18 ± 1.11 | 40.31 ± 2.77 | 30.20 ± 2.12 | 71.27 ± 1.84 |
| TC(83) | 91.18 ± 4.55 | 109.63 ± 4.50 | 75.34 ± 4.23 | 110.45 ± 3.71 | 119.82 ± 4.17 | 20.19 ± 1.30 | 39.76 ± 2.95 | 30.08 ± 1.88 | 71.61 ± 2.24 | |
| CC(27) | 91.33 ± 4.98 | 110.15 ± 5.53 | 75.59 ± 3.84 | 111.85 ± 3.88 | 121.19 ± 4.18 | 20.41 ± 1.37 | 40.70 ± 2.84 | 30.07 ± 1.96 | 71.81 ± 1.57 | |
| g.64911G→A | GG(59) | 90.98 ± 4.40 | 110.85 ± 4.56 | 75.27 ± 3.73 | 110.63 ± 3.41 | 119.93 ± 3.54 | 20.20 ± 1.11 | 40.36 ± 2.70 | 30.27 ± 2.07 | 71.31 ± 1.79 |
| GA(81) | 91.11 ± 4.51 | 109.65 ± 4.46 | 75.44 ± 4.21 | 110.53 ± 3.69 | 119.89 ± 4.17 | 20.21 ± 1.31 | 39.75 ± 2.99 | 30.04 ± 1.87 | 71.62 ± 2.27 | |
| AA(25) | 91.22 ± 5.15 | 110.32 ± 5.69 | 75.48 ± 3.87 | 111.76 ± 4.02 | 121.28 ± 4.34 | 20.32 ± 1.38 | 40.60 ± 2.92 | 30.04 ± 2.03 | 71.80 ± 1.61 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Huang, J.; Xu, H. Association Analysis of ENPP1 Tissue Expression, Polymorphism, and Growth Traits in Xiangsu Pigs. Genes 2025, 16, 395. https://doi.org/10.3390/genes16040395
Chen J, Huang J, Xu H. Association Analysis of ENPP1 Tissue Expression, Polymorphism, and Growth Traits in Xiangsu Pigs. Genes. 2025; 16(4):395. https://doi.org/10.3390/genes16040395
Chicago/Turabian StyleChen, Jiaqi, Jiajin Huang, and Houqiang Xu. 2025. "Association Analysis of ENPP1 Tissue Expression, Polymorphism, and Growth Traits in Xiangsu Pigs" Genes 16, no. 4: 395. https://doi.org/10.3390/genes16040395
APA StyleChen, J., Huang, J., & Xu, H. (2025). Association Analysis of ENPP1 Tissue Expression, Polymorphism, and Growth Traits in Xiangsu Pigs. Genes, 16(4), 395. https://doi.org/10.3390/genes16040395





