Interstitial 1q Deletion Syndrome: A New Patient with Congenital Diaphragmatic Hernia and Multiple Midline Anomalies
Abstract
1. Introduction
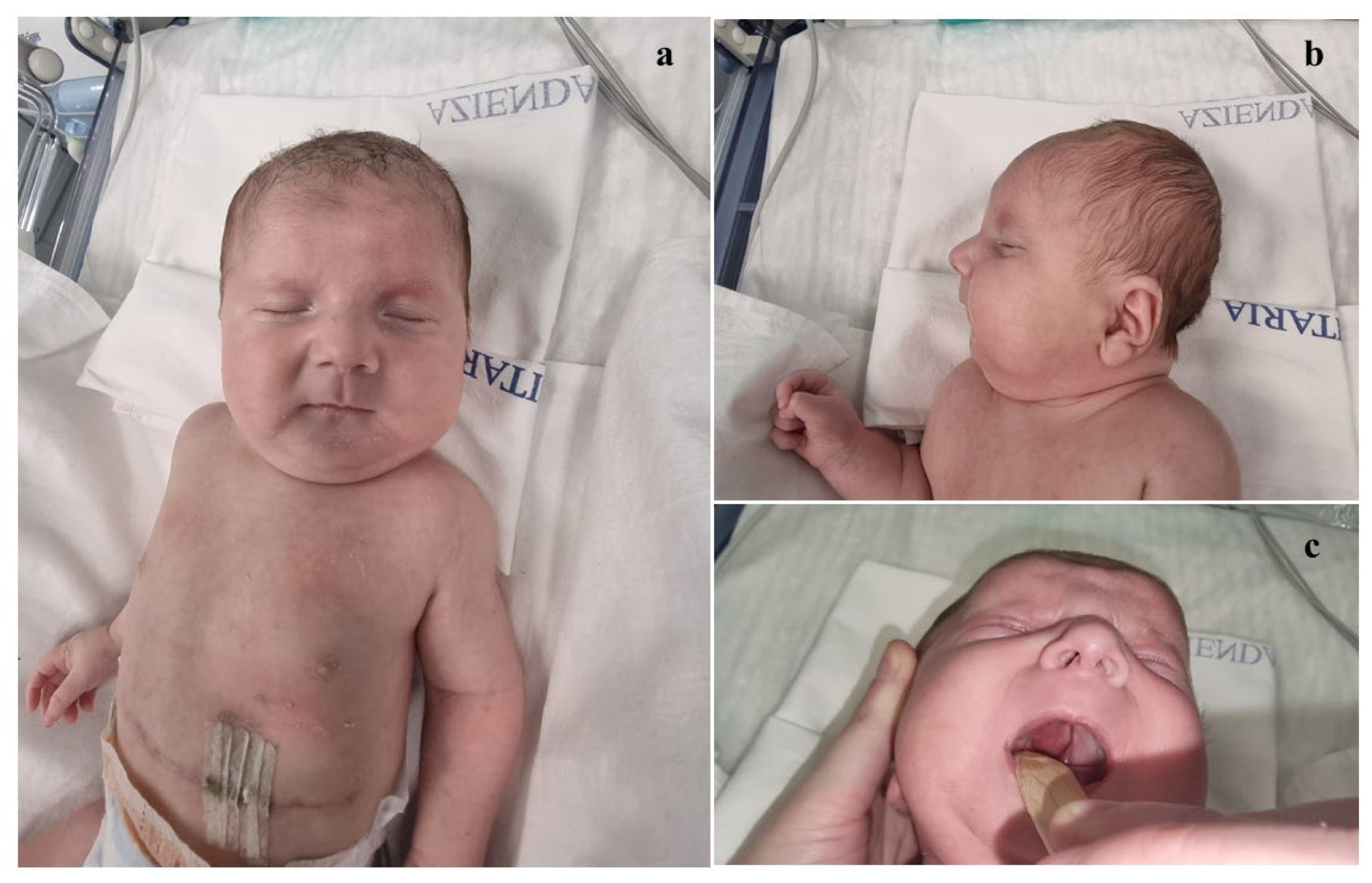
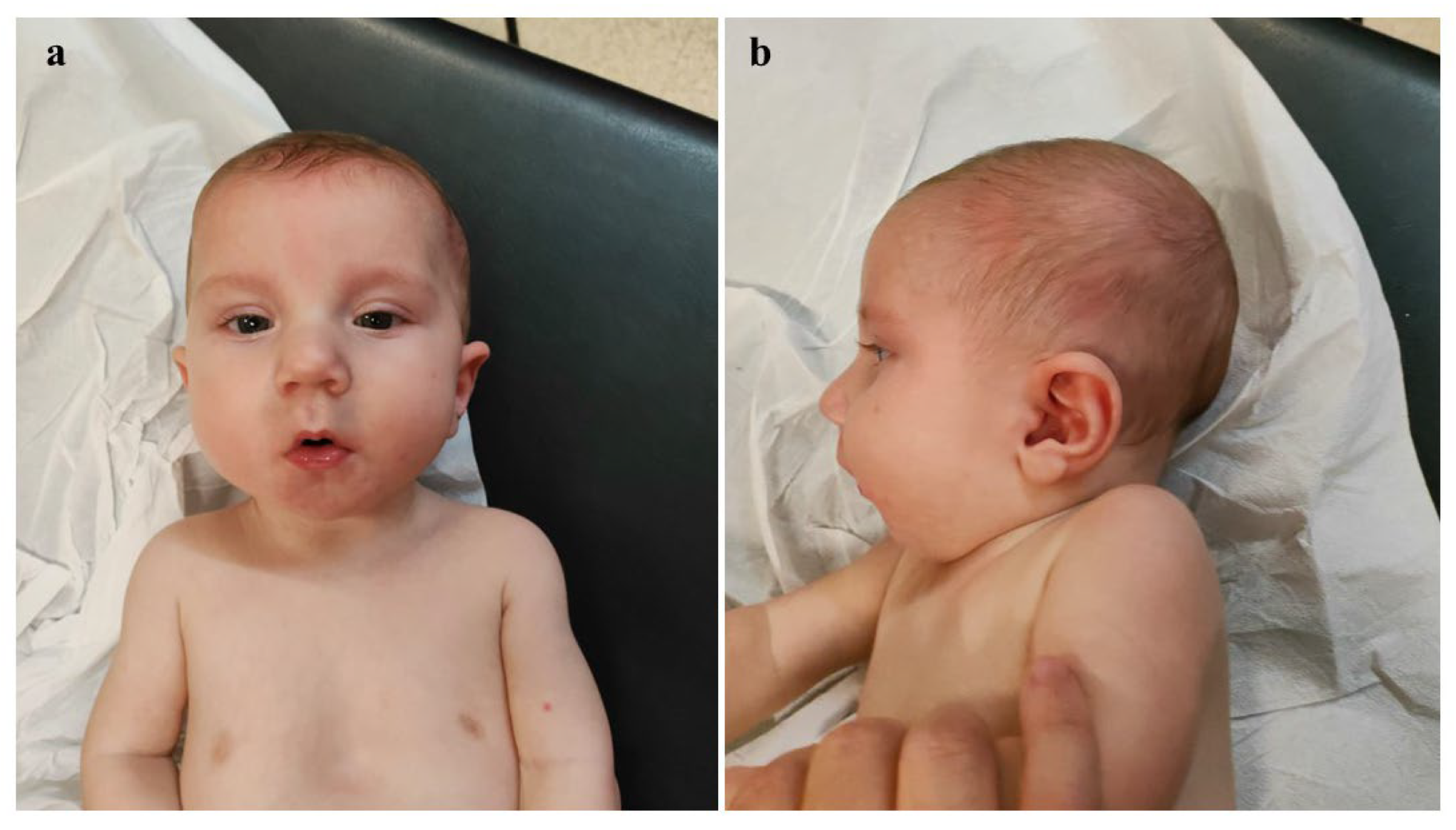
2. Case Presentation
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chatterjee, D.; Ing, R.J.; Gien, J. Update on Congenital Diaphragmatic Hernia. Anesth. Analg. 2020, 131, 808–821. [Google Scholar] [CrossRef] [PubMed]
- Mehollin-Ray, A.R. Congenital diaphragmatic hernia. Pediatr. Radiol. 2020, 50, 1855–1871. [Google Scholar] [CrossRef] [PubMed]
- Serra, G.; Antona, V.; Schierz, M.; Vecchio, D.; Piro, E.; Corsello, G. Esophageal atresia and Beckwith-Wiedemann syndrome in one of the naturally conceived discordant newborn twins: First report. Clin. Case Rep. 2018, 6, 399–401. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, H.L.; Adzick, N.S. Congenital diaphragmatic hernia (CDH) in the neonate: Clinical features and diagnosis. UpToDate 2024. preprint. [Google Scholar]
- Scott, D.A.; Gofin, Y.; Berry, A.M.; Adams, A.D. Underlying genetic etiologies of congenital diaphragmatic hernia. Prenat. Diagn. 2022, 42, 373–386. [Google Scholar] [CrossRef]
- Serra, G.; Memo, L.; Cavicchioli, P.; Cutrone, M.; Giuffrè, M.; La Torre, M.L.; Schierz, I.A.M.; Corsello, G. Novel mutations of the ABCA12, KRT1 and ST14 genes in three unrelated newborns showing congenital ichthyosis. Ital. J. Pediatr. 2022, 48, 145. [Google Scholar] [CrossRef]
- Piccione, M.; Serra, G.; Sanfilippo, C.; Andreucci, E.; Sani, I.; Corsello, G. A new mutation in EDA gene in X-linked hypohidrotic ectodermal dysplasia associated with keratoconus. Minerva Pediatr. 2012, 64, 59–64. [Google Scholar]
- Piro, E.; Schierz, I.A.M.; Antona, V.; Pappalardo, M.P.; Giuffrè, M.; Serra, G.; Corsello, G. Neonatal hyperinsulinemic hypoglycemia: Case report of kabuki syndrome due to a novel KMT2D splicing-site mutation. Ital. J. Pediatr. 2020, 46, 136. [Google Scholar] [CrossRef]
- Valutazione Antropometrica Neonatale. Riferimento Carte INeS. Available online: http://www.inescharts.com (accessed on 6 January 2025).
- Goede, J.; Hack, W.W.; Sijstermans, K.; van der Voort-Doedens, L.M.; Van der Ploeg, T.; Meij-de Vries, A.; Delemarre-van de Waal, H.A. Normative values for testicular volume measured by ultrasonography in a normal population from infancy to adolescence. Horm. Res. Paediatr. 2011, 76, 56–64. [Google Scholar] [CrossRef]
- Kaplan, S.L.; Edgar, J.C.; Ford, E.G.; Adgent, M.A.; Schall, J.I.; Kelly, A.; Umbach, D.M.; Rogan, W.J.; Stallings, V.A.; Darge, K. Size of testes, ovaries, uterus and breast buds by ultrasound in healthy full-term neonates ages 0–3 days. Pediatr. Radiol. 2016, 46, 1837–1847. [Google Scholar] [CrossRef]
- Riggs, E.R.; Andersen, E.F.; Cherry, A.M.; Kantarci, S.; Kearney, H.; Patel, A.; Raca, G.; Ritter, D.I.; South, S.T.; Thorland, E.C.; et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2020, 22, 245–257. [Google Scholar] [CrossRef] [PubMed]
- UK-WHO Growth Charts–Neonatal and Infant Close Monitoring (NICM). Available online: https://www.rcpch.ac.uk/resources/uk-who-growth-charts-neonatal-infant-close-monitoring-nicm (accessed on 6 January 2025).
- Milani, D.; Bedeschi, M.F.; Iascone, M.; Chiarelli, G.; Cerutti, M.; Menni, F. De novo deletion of 1q31.1–q32.1 in a patient with developmental delay and behavioral disorders. Cytogenet. Genome Res. 2012, 136, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Hyder, Z.; Fairclough, A.; Douzgou, S. Chromosome 1q31.2q32.1 deletion in an adult male with intellectual disability, dysmorphic features and obesity. Clin. Dysmorphol. 2019, 28, 131–136. [Google Scholar] [CrossRef]
- Carter, J.; Zombor, M.; Máté, A.; Sztriha, L.; Waters, J.J. De Novo Interstitial Microdeletion at 1q32.1 in a 10-Year-Old Boy with Developmental Delay and Dysmorphism. Case Rep. Genet. 2016, 2016, 2501741. [Google Scholar] [CrossRef][Green Version]
- Piccione, M.; Serra, G.; Consiglio, V.; Di Fiore, A.; Cavani, S.; Grasso, M.; Malacarne, M.; Pierluigi, M.; Viaggi, C.; Corsello, G. 14q13.1-21.1 deletion encompassing the HPE8 locus in an adolescent with intellectual disability and bilateral microphthalmia, but without holoprosencephaly. Am. J. Med. Genet. A 2012, 158A, 1427–1433. [Google Scholar] [CrossRef]
- Serra, G.; Giambrone, C.; Antona, V.; Cardella, F.; Carta, M.; Cimador, M.; Corsello, G.; Giuffrè, M.; Insinga, V.; Maggio, M.C.; et al. Congenital hypopituitarism and multiple midline defects in a newborn with non-familial Cat Eye syndrome. Ital. J. Pediatr. 2022, 48, 170. [Google Scholar] [CrossRef]
- Schierz, I.A.M.; Giuffrè, M.; Cimador, M.; D’Alessandro Serra, G.; Favata, F.; Antona, V.; Piro, E.; Corsello, G. Hypertrophic pyloric stenosis masked by kidney failure in a male infant with a contiguous gene deletion syndrome at Xp22.31 involving the steroid sulfatase gene: Case report. Ital. J. Pediatr. 2022, 48, 19. [Google Scholar] [CrossRef]
- Hamano, S.; Fukushima, Y.; Yamada, T.; Shimizu, H.; Okuyama, M.; Ito, F.; Maekawa, K. A case of interstitial 1q deletion [46,XY,del(q25q32.1)]. Ann. Genet. 1987, 30, 105–108. [Google Scholar]
- Hill, A.D.; Chang, B.S.; Hill, R.S.; Garraway, L.A.; Bodell, A.; Sellers, W.R.; Walsh, C.A. A 2-Mb critical region implicated in the microcephaly associated with terminal 1q deletion syndrome. Am. J. Med. Genet. A 2007, 143A, 1692–1698. [Google Scholar] [CrossRef]
- Hemming, I.A.; Forrest, A.R.; Shipman, P.; Woodward, K.J.; Walsh, P.; Ravine, D.G.; Heng, J.I. Reinforcing the association between distal 1q CNVs and structural brain disorder: A case of a complex 1q43–q44 CNV and a review of the literature. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2016, 171B, 458–467. [Google Scholar] [CrossRef]
- Stoll, C.; Alembick, Y.; Roth, M.P. Associated anomalies in Pierre Robin sequence. Am. J. Med. Genet. A. 2023, 191, 2312–2323. [Google Scholar] [CrossRef] [PubMed]
- Meshram, G.G.; Kaur, N.; Hura, K.S. Pierre Robin Sequence: Diagnostic Difficulties Faced while Differentiating Isolated and Syndromic Forms. Acta Medica 2020, 63, 86–90. [Google Scholar] [CrossRef] [PubMed]
- De Souza, C.D.R.; Padovani, L.F.; Ferreira-Donati, G.C.; Moraes, M.C.A.F.; Corrêa, C.C.; Maximino, L.P. Babies with Pierre Robin Sequence: Neuropsychomotor Development. Pediatr. Neurol. 2023, 141, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Nagamani, S.C.; Erez, A.; Bay, C.; Pettigrew, A.; Lalani, S.R.; Herman, K.; Graham, B.H.; Nowaczyk, M.J.; Proud, M.; Craigen, W.J.; et al. Delineation of a deletion region critical for corpus callosal abnormalities in chromosome 1q43–q44. Eur. J. Hum. Genet. 2012, 20, 176–179. [Google Scholar] [CrossRef][Green Version]
- Brea-Fernández, A.J.; Souto-Trinei, F.A.; Iglesias, E.; Caamaño, P.; Rodríguez Sánchez, B.; Gómez Lado, C.; Eiris, J.; Fernández-Prieto, M.; Barros, F.; Brea, R.J.; et al. Expanding the Clinical and Molecular Spectrum of FOXG1- and ZBTB18-Associated Neurodevelopmental Disorders. Cytogenet. Genome Res. 2023, 163, 301–306. [Google Scholar] [CrossRef]
- Yang, F.; Ding, Y.; Wang, Y.; Zhang, Q.; Li, H.; Yu, T.; Chang, G.; Wang, X. A de novo variant in ZBTB18 gene caused autosomal dominant non-syndromic intellectual disability 22 syndrome: A case report and literature review. Medicine 2024, 103, e35908. [Google Scholar] [CrossRef]
- Shaffer, L.G.; Theisen, A.; Bejjani, B.A.; Ballif, B.C.; Aylsworth, A.S.; Lim, C.; McDonald, M.; Ellison, J.W.; Kostiner, D.; Saitta, S.; et al. The discovery of microdeletion syndromes in the post-genomic era: Review of the methodology and characterization of a new 1q41q42 microdeletion syndrome. Genet. Med. 2007, 9, 607–616. [Google Scholar] [CrossRef]
- Della Monica, M.; Lonardo, F.; Faravelli, F.; Pierluigi, M.; Luquetti, D.V.; De Gregori, M.; Zuffardi, O.; Scarano, G. A case of autism with an interstitial 1q deletion (1q23.3–24.2) and a de novo translocation of chromosomes 1q and 5q. Am. J. Med. Genet. A 2007, 143A, 2733–2737. [Google Scholar] [CrossRef]
- Lo, L.J.; Noordhoff, M.S.; Huang, C.S.; Chen, K.T.; Chen, Y.R. Proximal deletion of the long arm of chromosome 1: [del(1)(q23–q25)]. Cleft Palate Craniofac. J. 1993, 30, 586–589. [Google Scholar] [CrossRef]
- Libotte, F.; Bizzoco, D.; Gabrielli, I.; Tamburrino, C.; Ernandez, C.; Carpineto, L.; D’Aleo, M.P.; Cima, A.; Mesoraca, A.; Cignini, P.; et al. A new case of interstitial 1q 25.3-32.1 deletion: Cytogenetic analysis molecular characterization and ultrasound findings. J. Prenat. Med. 2015, 9, 8–11. [Google Scholar] [CrossRef]
- Al Saad, R.; El Mansoury, J.; Al Hazzaa, S.A.F.; Dirar, Q.S. Chromosome 1q Terminal Deletion and Congenital Glaucoma: A Case Report. Am. J. Case Rep. 2020, 21, e918128. [Google Scholar] [PubMed]
- Cioclu, M.C.; Mosca, I.; Ambrosino, P.; Puzo, D.; Bayat, A.; Wortmann, S.B.; Koch, J.; Strehlow, V.; Shirai, K.; Matsumoto, N.; et al. KCNT2-Related Disorders: Phenotypes, Functional, and Pharmacological Properties. Ann. Neurol. 2023, 94, 332–349. [Google Scholar] [CrossRef]
- Jackson, A.; Banka, S.; Stewart, H.; Genomics England Research Consortium; Robinson, H.; Lovell, S.; Clayton-Smith, J. Recurrent KCNT2 missense variants affecting p.Arg190 result in a recognizable phenotype. Am. J. Med. Genet. A 2021, 185, 3083–3091. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, N.; Murakami, H.; Saito, T.; Masuno, M.; Kurosawa, K. Tumor predisposition in an individual with chromosomal rearrangements of 1q31.2-q41 encompassing cell division cycle protein 73. Congenit. Anom. 2020, 60, 128–130. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, Y.; Kato, H.; Nagasaki, M.; Yoshida, Y.; Fujisawa, M.; Minegishi, N.; Yamamoto, M.; Nangaku, M. CFH-CFHR1 hybrid genes in two cases of atypical hemolytic uremic syndrome. J. Hum. Genet. 2023, 68, 427–430. [Google Scholar] [CrossRef]
- Fei, Z.G.; Zhen, K.; Zhang, F.J.; Liu, P.Y.; Xu, H.Y.; Chen, H.H. Clinical research advances of CFHR5 nephropathy: A recent review. Eur. Rev. Med. Pharmacol. Sci. 2023, 27, 9987–10000. [Google Scholar]
- Biswas, A.; Ivaskevicius, V.; Thomas, A.; Oldenburg, J. Coagulation factor XIII deficiency. Diagnosis, prevalence and management of inherited and acquired forms. Hamostaseologie 2014, 34, 160–166. [Google Scholar]
- Verloes, A.; Drunat, S.; Passemard, S. ASPM Primary Microcephaly. In GeneReviews® [Internet]; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2020. [Google Scholar]
- Daher, A.; Banjak, M.; Noureldine, J.; Nehme, J.; El Shamieh, S. Genotype-phenotype associations in CRB1 bi-allelic patients: A novel mutation, a systematic review and meta-analysis. BMC Ophthalmol. 2024, 24, 167. [Google Scholar] [CrossRef]
- Van Bergen, N.J.; Bell, K.M.; Carey, K.; Gear, R.; Massey, S.; Murrell, E.K.; Gallacher, L.; Pope, K.; Lockhart, P.J.; Kornberg, A.; et al. Pathogenic variants in nucleoporin TPR (translocated promoter region, nuclear basket protein) cause severe intellectual disability in humans. Hum. Mol. Genet. 2022, 31, 362–375. [Google Scholar] [CrossRef]
- Berkowcz, S.R.; Featherby, T.J.; Whisstock, J.C.; Bird, P.I. Mice Lacking Brinp2 or Brinp3, or both, exhibit behaviors consistent with neurodevelopmental disorders. Front. Behav. Neurosci. 2016, 10, 196. [Google Scholar]
| Authors | Deletion (Breakpoints, GRCh37) | Dysmorphic Features | Involved Genes | Genetic Test | Cardio-Vascular System | Respiratory System | Neuropsychomotor Profile | Hands and/or Feet Anomalies | Outcome |
|---|---|---|---|---|---|---|---|---|---|
| Milani et al. (2012) [14] | Del 1q 31.1-q32.1 (187,437,627–203,015,924) | broad forehead, laterally sparse eyebrows, slightly downward-slanted palpebral fissures, broad and high nasal bridge, hypoplastic nostrils, long philtrum, thin upper lip and slightly protruding lower lip, retroverted ears | F13B, ASPM, CRB1, PTPRC, PKP1, CDC73, CACNA 1S, TNNT2 | Array-CGH | n.r. | n.r. | Motor, social and cognitive developmental delay | At age 6 years, normal physical growth parameters; mild motor and cognitive developmental delay, hyperactivity and behavioral disorders. | |
| Hyder et al. (2019) [15] | del 1q 31.2-q32.1 (191,590,110–201,139,395) | frontal upsweep, hypertelorism, epicanthic folds, broad nasal bridge, prominent nose, low columella, thin upper lip and everted lower lip, prominent ears, short chin. | DDX59, ASPM, CRB1, F13B, CDC73, CFHR5, CACNA1S, UCHL5, TROVE2, B3GALT, ZBTB41, CAMSA2, KIF21B, TMEM9 | Array-CGH | n.r. | n.r. | At birth, hypotonia and feeding difficulties. Subsequently, developmental delay, hyperactivity, aggression, disinhibition, and sleep disturbances. | Clinodactyly, single palmar crease on left hand, tapering fingers, deep-set small nails. | At 31 years, head circumference 57.4 cm (50th–75th centile), height 174.8 cm (25th–50th centile) and weight 140.6 kg (>99th centile). Downslanting palpebral fissures, broad nasal bridge, low-hanging columella, thin upper lip, thick lower lip, deep-set small nails and tapering fingers. He currently lives independently in a flat with supported living. His main difficulties are with arithmetic and finances, but his memory is good, and he is able to read and write independently |
| Carter et al. (2016) [16] | Del. 1q32.1 (199,985,888–203,690,832) | Long face, narrow jaw, down-slanted palpebral fissures, highly arched eyebrows, low-set ears, thick lower lip. | KDM5B, NAV1, KIF21B, GPR37L, SYT2 | Array-CGH | n.r. | n.r. | Global developmental delay, social skills and language difficulties, reduced IQ. Generalized hypotonia and decreased deep tendon reflexes | Bilateral clinodactyly of the fifth finger and proximal positioning of the thumb | Neuropsychological evaluation at 7 years of age: full scale IQ of about 50 (Woodcock-Johnson Tests of Cognitive Abilities), difficulties in visual-motor coordination. Significant difficulties with receptive and expressive language; slow improvement in language acquisition. At 10 years, he requires special education and support in everyday life |
| Our patient | Del.1q31.1-q32.1 (187,95,640–199,996,777) | Broad and sloping forehead, hypertelorism, wide nasal bridge, bulbous nasal tip, anteverted nares, long and thick philtrum, thin lips, dysplastic auricles with thickened helices, low-set and posteriorly rotated ears, complete cleft palate, microretrognathia. | CDC73, KCNT2, CFH, CFHR1, CFHR5, F13B, ASPM, CRB1, PTPRC | Array-CGH | ostium primum-type atrial septal defect | Congenital diaphragmatic hernia | Generalized hypotonia, diminished deep tendon reflexes, reduced primitive reflexes and reactivity, poor cortical gyration, and millimeter-sized cystic lesions in the periventricular white matter | clinodactyly of the fifth finger | At age 8 months, generalized mild hypotonia. Nearly completely acquired head control, not that of the trunk. In the supine position, tendency towards flexion and external rotation of the lower limbs. Normal muscular trophism, feet in varus attitude. Good manual grip and lively free motor skills. Normally elicitable osteo-tendineous reflexes and Landau reaction, not the Babinski sign. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serra, G.; Nardello, R.; Antona, V.; Di Pace, M.R.; Giliberti, A.; Giuffrè, M.; Morreale, D.M.; Piro, E.; Schierz, I.A.M.; Sergio, M.; et al. Interstitial 1q Deletion Syndrome: A New Patient with Congenital Diaphragmatic Hernia and Multiple Midline Anomalies. Genes 2025, 16, 319. https://doi.org/10.3390/genes16030319
Serra G, Nardello R, Antona V, Di Pace MR, Giliberti A, Giuffrè M, Morreale DM, Piro E, Schierz IAM, Sergio M, et al. Interstitial 1q Deletion Syndrome: A New Patient with Congenital Diaphragmatic Hernia and Multiple Midline Anomalies. Genes. 2025; 16(3):319. https://doi.org/10.3390/genes16030319
Chicago/Turabian StyleSerra, Gregorio, Rosaria Nardello, Vincenzo Antona, Maria Rita Di Pace, Alessandra Giliberti, Mario Giuffrè, Daniela Mariarosa Morreale, Ettore Piro, Ingrid Anne Mandy Schierz, Maria Sergio, and et al. 2025. "Interstitial 1q Deletion Syndrome: A New Patient with Congenital Diaphragmatic Hernia and Multiple Midline Anomalies" Genes 16, no. 3: 319. https://doi.org/10.3390/genes16030319
APA StyleSerra, G., Nardello, R., Antona, V., Di Pace, M. R., Giliberti, A., Giuffrè, M., Morreale, D. M., Piro, E., Schierz, I. A. M., Sergio, M., Valenti, G., Pensabene, M., & Corsello, G. (2025). Interstitial 1q Deletion Syndrome: A New Patient with Congenital Diaphragmatic Hernia and Multiple Midline Anomalies. Genes, 16(3), 319. https://doi.org/10.3390/genes16030319