The Mitochondrial Genome of the Springtail Semicerura bryophila (Collembola): New Data Call into Question the Relevance of the Subfamilies of the Isotomidae
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction
2.2. Mitogenome Sequencing, Assembly, and Annotation
2.3. Phylogenetic Analysis
3. Results
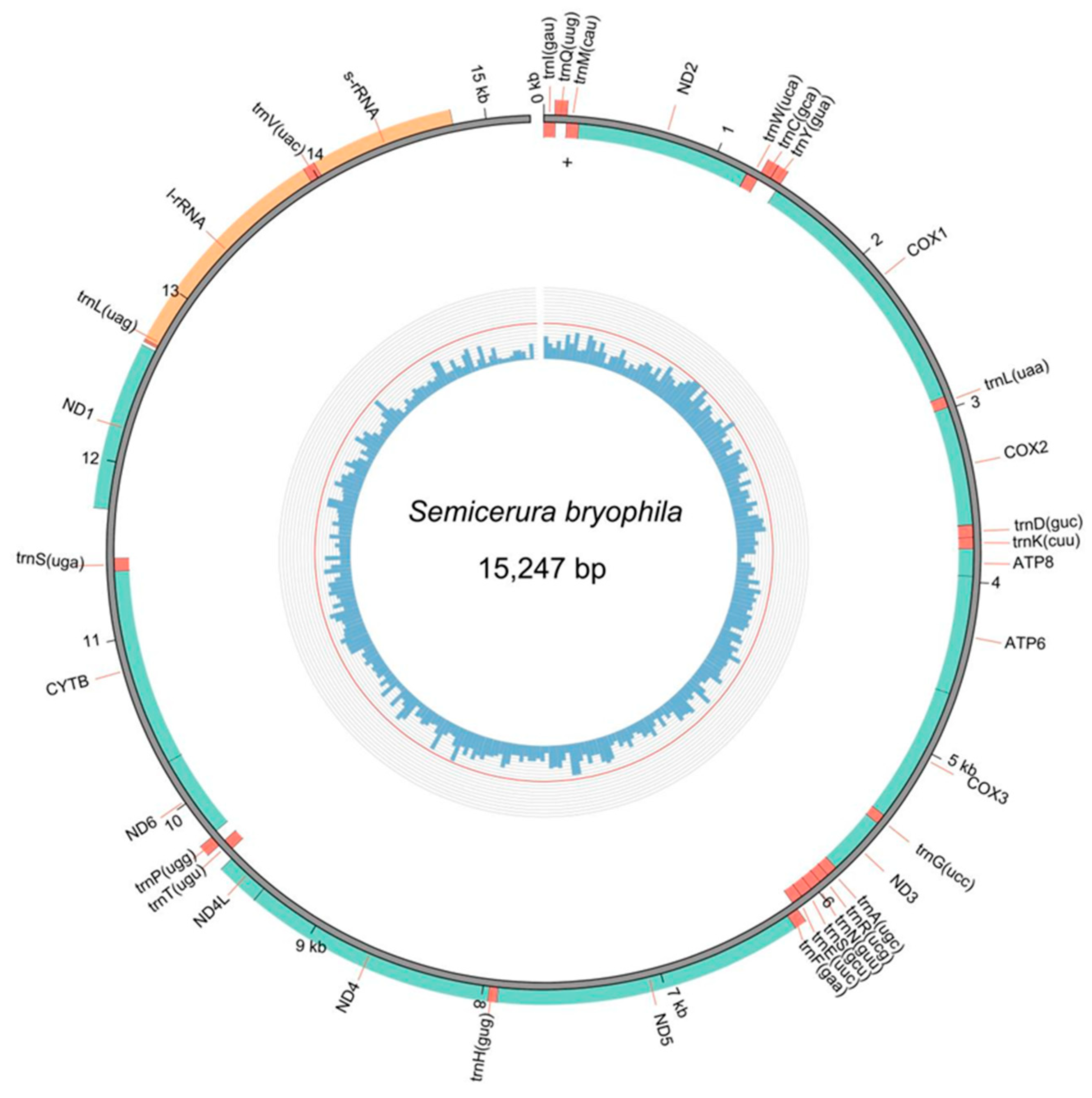
3.1. Genome Organization and Composition
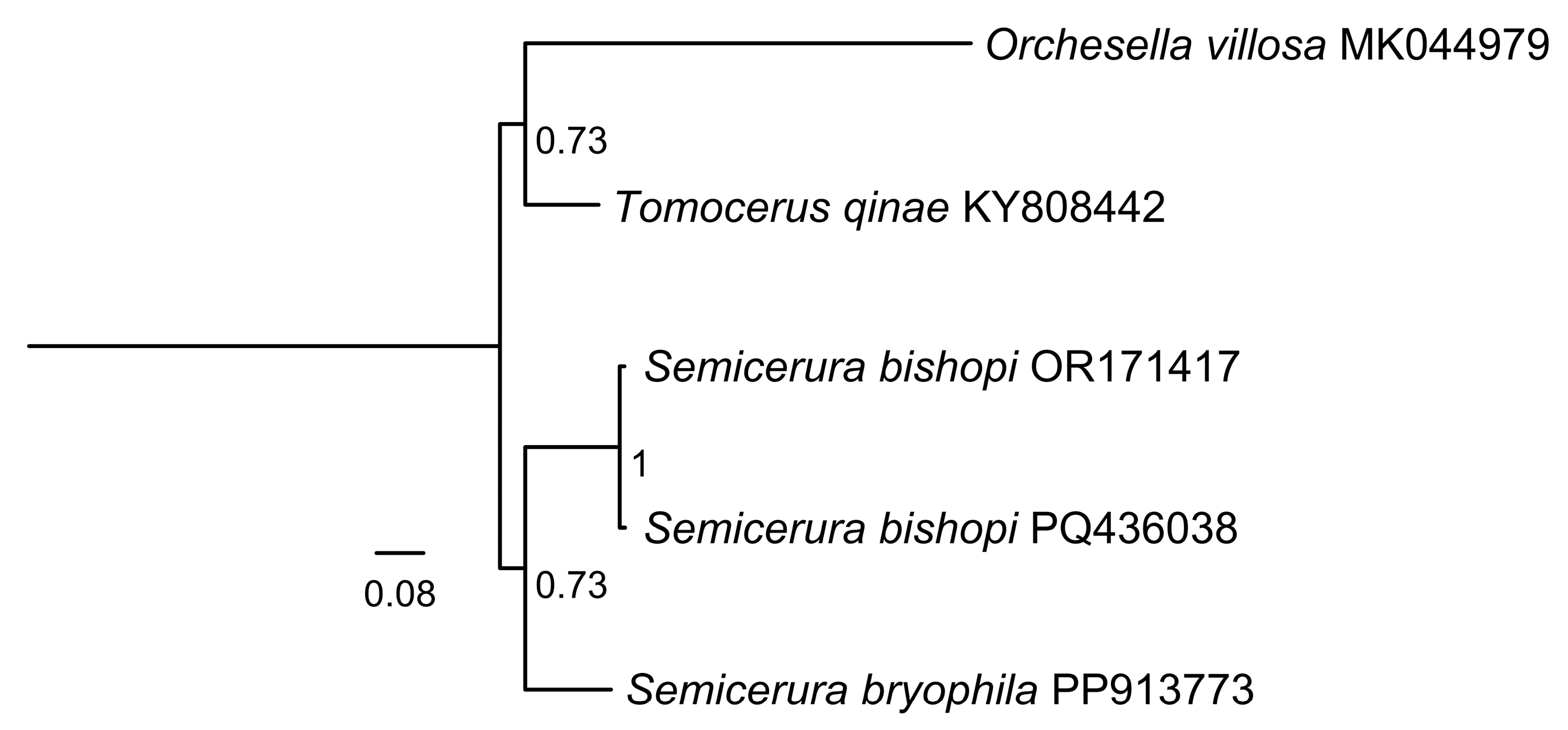
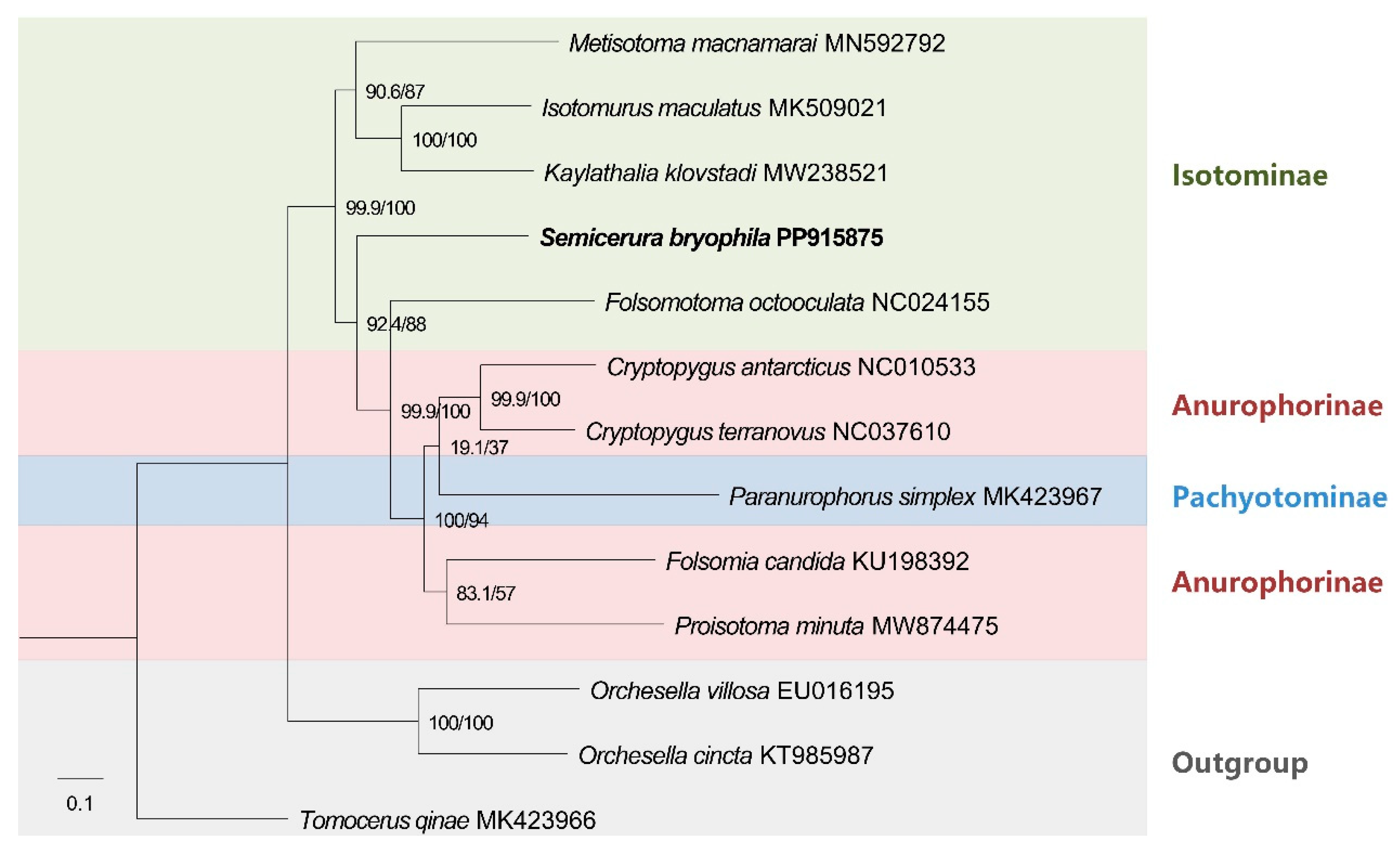
3.2. Phylogeny of Collembola
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hopkin, S.P. Biology of the Springtails: (Insecta: Collembola); OUP Oxford: Oxford, UK, 1997. [Google Scholar]
- Rusek, J. Biodiversity of Collembola and Their Functional Role in the Ecosystem. Biodivers. Conserv. 1998, 7, 1207–1219. [Google Scholar] [CrossRef]
- Deharveng, L. Recent Advances in Collembola Systematics. Pedobiologia 2004, 48, 415–433. [Google Scholar] [CrossRef]
- Bellinger, P.F.; Christiansen, K.; Janssens, F. Checklist of the Collembola of the World. Available online: https://www.collembola.org/ (accessed on 2 February 2025).
- Carl, B. Die Familien Der Collembolen. Zool. Anz. 1913, 41, 315–322. [Google Scholar]
- Potapov, M. Synopses on Palaearctic Collembola: Isotomidae; Senckenberg Museum of Natural History Görlitz: Görlitz, Germany, 2001. [Google Scholar]
- Potapov, M.; Xie, Z.; Kuprin, A.; Sun, X. The Genus Semicerura (Collembola; Isotomidae) in Asia. Zootaxa 2020, 4751, 105–118. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Zhang, F.; Yu, D.; Luo, Y.; Ho, S.Y.W.W.; Wang, B.; Zhu, C. Cryptic Diversity, Diversification and Vicariance in Two Species Complexes of Tomocerus (Collembola, Tomoceridae) from China. Zool. Scr. 2014, 43, 393–404. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Sedlazeck, F.J.; Rescheneder, P.; Von Haeseler, A. NextGenMap: Fast and Accurate Read Mapping in Highly Polymorphic Genomes. Bioinformatics 2013, 29, 2790–2791. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De Novo Assembly of Organelle Genomes from Whole Genome Data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. MitoZ: A Toolkit for Animal Mitochondrial Genome Assembly, Annotation and Visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for Large-Scale Multiple Sequence Alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Kück, P.; Longo, G.C. FASconCAT-G: Extensive Functions for Multiple Sequence Alignment Preparations Concerning Phylogenetic Studies. Front. Zool. 2014, 11, 81. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R.; Teeling, E. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Chen, T.-W.; Potapov, M.; Zhang, F.; Wu, D.; Scheu, S.; Sun, X. Ecological and Evolutionary Processes Shape Below-ground Springtail Communities along an Elevational Gradient. J. Biogeogr. 2022, 49, 469–482. [Google Scholar] [CrossRef]
- Xie, Z.; Yao, H.; Potapov, M.; Dong, J.; Wu, D.; Scheu, S.; Sun, X. The Complete Mitochondrial Genome of an Enigmatic Predaceous Springtail Metisotoma Macnamarai from Northeast China. Mitochondrial DNA Part B Resour. 2020, 5, 506–508. [Google Scholar] [CrossRef]
- Jagatap, H.; Monsanto, D.M.; van Vuuren, B.J.; Parbhu, S.P.; Dinoi, A.; Janion-Scheepers, C.; Sekar, S.; Teske, P.R.; Emami-Khoyi, A. The Complete Mitogenome of Isotomurus Maculatus: A Widespread Species That Is Invading the Sub-Antarctic Region. Mitochondrial DNA Part B Resour. 2019, 4, 1706–1708. [Google Scholar] [CrossRef]
- Cucini, C.; Fanciulli, P.P.; Frati, F.; Convey, P.; Nardi, F.; Carapelli, A. Re-evaluating the Internal Phylogenetic Relationships of Collembola by Means of Mitogenome Data. Genes 2020, 12, 44. [Google Scholar] [CrossRef]
- Carapelli, A.; Convey, P.; Nardi, F.; Frati, F. The Mitochondrial Genome of the Antarctic Springtail Folsomotoma Octooculata (Hexapoda; Collembola), and an Update on the Phylogeny of Collembolan Lineages Based on Mitogenomic Data. Entomologia 2014, 2, 46–55. [Google Scholar] [CrossRef]
- Monsanto, D.M.; Jansen van Vuuren, B.; Jagatap, H.; Jooste, C.M.; Janion-Scheepers, C.; Teske, P.R.; Emami-Khoyi, A. The Complete Mitogenome of the Springtail Cryptopygus Antarcticus Travei Provides Evidence for Speciation in the Sub-Antarctic Region. Mitochondrial DNA Part B Resour. 2019, 4, 1195–1197. [Google Scholar] [CrossRef]
- Carapelli, A.; Paolo, P.; Francesco, F.; Chiara, F. Mitogenomic Data to Study the Taxonomy of Antarctic Springtail Species (Hexapoda: Collembola) and Their Adaptation to Extreme Environments. Polar Biol. 2019, 42, 715–732. [Google Scholar] [CrossRef]
- Sun, X.; Yu, D.; Xie, Z.; Dong, J.; Ding, Y.; Yao, H.; Greenslade, P. Phylomitogenomic Analyses on Collembolan Higher Taxa with Enhanced Taxon Sampling and Discussion on Method Selection. PLoS ONE 2020, 15, e0230827. [Google Scholar] [CrossRef]
- Faddeeva-Vakhrusheva, A.; Kraaijeveld, K.; Derks, M.F.L.; Anvar, S.Y.; Agamennone, V.; Suring, W.; Kampfraath, A.A.; Ellers, J.; Le Ngoc, G.; van Gestel, C.A.M.; et al. Coping with Living in the Soil: The Genome of the Parthenogenetic Springtail Folsomia Candida. BMC Genom. 2017, 18, 493. [Google Scholar] [CrossRef]
- Tian, Y.; Miao, X.; Song, S.; Zhang, Z.; Hu, S.; Wei, D.; Wang, M. Complete Mitochondrial Genome of Proisotoma Minuta (Collembola: Proisotominae). Mitochondrial DNA Part B Resour. 2022, 7, 727–728. [Google Scholar] [CrossRef]
- Carapelli, A.; Liò, P.; Nardi, F.; Van Der Wath, E.; Frati, F. Phylogenetic Analysis of Mitochondrial Protein Coding Genes Confirms the Reciprocal Paraphyly of Hexapoda and Crustacea. BMC Evol. Biol. 2007, 7, S8. [Google Scholar] [CrossRef]
- Faddeeva-Vakhrusheva, A.; Derks, M.F.L.; Anvar, S.Y.; Agamennone, V.; Suring, W.; Smit, S.; van Straalen, N.M.; Roelofs, D. Gene Family Evolution Reflects Adaptation to Soil Environmental Stressors in the Genome of the Collembolan Orchesella Cincta. Genome Biol. Evol. 2016, 8, 2106–2117. [Google Scholar] [CrossRef]
- Boore, J.L. Animal Mitochondrial Genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Lang, B.F. Poriferan MtDNA and Animal Phylogeny Based on Mitochondrial Gene Arrangements. Syst. Biol. 2005, 54, 651–659. [Google Scholar] [CrossRef]
- Janion-Scheepers, C.; Potapov, M.; Deharveng, L. New and Little-Known Isotominae (Collembola, Isotomidae) from South Africa. Zootaxa 2023, 5346, 337–347. [Google Scholar] [CrossRef]
- Yu, D.; Deharveng, L.; Lukić, M.; Wei, Y.; Hu, F.; Liu, M. Molecular Phylogeny and Trait Evolution in an Ancient Terrestrial Arthropod Lineage: Systematic Revision and Implications for Ecological Divergence (Collembola, Tomocerinae). Mol. Phylogenet. Evol. 2020, 154, 106995. [Google Scholar] [CrossRef]
- Leo, C.; Carapelli, A.; Cicconardi, F.; Frati, F.; Nardi, F. Mitochondrial Genome Diversity in Collembola: Phylogeny, Dating and Gene Order. Diversity 2019, 11, 169. [Google Scholar] [CrossRef]
- Leo, C.; Nardi, F.; Frati, F.; Fanciulli, P.P.; Cucini, C.; Vitale, M.; Brunetti, C.; Carapelli, A. The Mitochondrial Genome of the Springtail Bourletiella Arvalis (Symphypleona, Collembola). Mitochondrial DNA Part B 2019, 4, 2978–2979. [Google Scholar] [CrossRef]
- Potapov, M.B.; Lobkova, L.E.; Shrubovych, J.E. New and Little Known Palaearctic Pachyotominae (Collembola: Isotomidae). Russ. Entomol. J. 2005, 18, 75–82. [Google Scholar]
- Deharveng, L. Chétotaxie Sensillaire et Phylogenese Chez les Collemboles Arthropleona; Travaux du Laboratoire d’Ecobiologie des Arthropodes Édaphiques; Université P. Sabatier: Toulouse, France, 1979; pp. 1–15. [Google Scholar]
- Potapov, M.; Babenko, A.; Fjellberg, A.; Greenslade, P. Taxonomy of the Proisotoma Complex. II. A Revision of the Genus Subisotoma and a Description of Isotopenola Gen. Nov. (Collembola: Isotomidae). Zootaxa 2009, 2314, 1–40. [Google Scholar] [CrossRef]

| Family | Subfamily | Species | Size (bp) | Accession No. | Reference |
|---|---|---|---|---|---|
| Isotomidae | Isotominae | Metisotoma macnamarai | 15,177 | MN592792 | [26] |
| Isotomidae | Isotominae | Isotomurus maculatus | 15,263 | MK509021 | [27] |
| Isotomidae | Isotominae | Kaylathalia klovstadi | 15,485 | MW238521 | [28] |
| Isotomidae | Isotominae | Semicerura bryophila | 15,247 | PP915875 | This study |
| Isotomidae | Isotominae | Folsomotoma octooculata | 15,338 | NC024155 | [29] |
| Isotomidae | Anurophorinae | Cryptopygus antarcticus | 15,297 | NC010533 | [30] |
| Isotomidae | Anurophorinae | Cryptopygus terranovus | 15,352 | NC037610 | [31] |
| Isotomidae | Pachyotominae | Paranurophorus simplex | 9518 | MK423967 | [32] |
| Isotomidae | Anurophorinae | Folsomia candida | 15,147 | KU198392 | [33] |
| Isotomidae | Anurophorinae | Proisotoma minuta | 15,930 | MW874475 | [34] |
| Tomoceridae | Tomocerinae | Tomocerus qinae | 15,045 | MK423966 | [32] |
| Orchesellidae | Orchesellinae | Orchesella villosa | 14,924 | EU016195 | [35] |
| Orchesellidae | Orchesellinae | Orchesella Cincta | 15,728 | KT985987 | [36] |
| Gene | Start | Stop | Strand | Size (bp) |
|---|---|---|---|---|
| trnI (gau) | 1 | 64 | + | 64 |
| trnQ (uug) | 63 | 130 | + | 68 |
| trnM (cau) | 130 | 199 | + | 70 |
| ND2 | 200 | 1198 | + | 999 |
| trnW (uca) | 1202 | 1268 | + | 67 |
| trnC (gca) | 1268 | 1328 | - | 61 |
| trnY (gua) | 1331 | 1392 | - | 62 |
| COX1 | 1393 | 2926 | + | 1534 |
| trnL (uaa) | 2927 | 2989 | + | 63 |
| COX2 | 2990 | 3673 | + | 684 |
| trnK (cuu) | 3676 | 3746 | + | 71 |
| trnD (guc) | 3747 | 3809 | + | 63 |
| ATP8 | 3810 | 3974 | + | 165 |
| ATP6 | 3968 | 4648 | + | 681 |
| COX3 | 4648 | 5434 | + | 787 |
| trnG (ucc) | 5435 | 5496 | + | 62 |
| ND3 | 5497 | 5841 | + | 345 |
| trnA (ugc) | 5840 | 5900 | + | 61 |
| trnR (ucg) | 5902 | 5964 | + | 63 |
| trnN (guu) | 5960 | 6023 | + | 64 |
| trnS (gcu) | 6024 | 6091 | + | 68 |
| trnE (uuc) | 6093 | 6155 | + | 63 |
| trnF (gaa) | 6155 | 6217 | - | 63 |
| ND5 | 6218 | 7919 | - | 1702 |
| trnH (gug) | 7920 | 7982 | - | 63 |
| ND4 | 7983 | 9354 | - | 1372 |
| ND4L | 9357 | 9623 | - | 267 |
| trnT (ugu) | 9641 | 9702 | + | 62 |
| trnP (ugg) | 9703 | 9764 | - | 62 |
| ND6 | 9776 | 10,255 | + | 480 |
| CYTB | 10,252 | 11,388 | + | 1137 |
| trnS (uga) | 11,387 | 11,458 | + | 72 |
| ND1 | 11,735 | 12,655 | - | 921 |
| trnL (uag) | 12,677 | 12,741 | - | 65 |
| rrnL | 12,742 | 14,003 | - | 1262 |
| trnV (uac) | 13,958 | 14,022 | - | 65 |
| rrnS | 14,023 | 14,812 | - | 790 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Z.; Zheng, M.; Li, Y.; Du, S.; Saifutdinov, R.; Potapov, M.; Sun, X.; Wu, D. The Mitochondrial Genome of the Springtail Semicerura bryophila (Collembola): New Data Call into Question the Relevance of the Subfamilies of the Isotomidae. Genes 2025, 16, 315. https://doi.org/10.3390/genes16030315
Xie Z, Zheng M, Li Y, Du S, Saifutdinov R, Potapov M, Sun X, Wu D. The Mitochondrial Genome of the Springtail Semicerura bryophila (Collembola): New Data Call into Question the Relevance of the Subfamilies of the Isotomidae. Genes. 2025; 16(3):315. https://doi.org/10.3390/genes16030315
Chicago/Turabian StyleXie, Zhijng, Mingxin Zheng, Yueying Li, Shiyu Du, Ruslan Saifutdinov, Mikhail Potapov, Xin Sun, and Donghui Wu. 2025. "The Mitochondrial Genome of the Springtail Semicerura bryophila (Collembola): New Data Call into Question the Relevance of the Subfamilies of the Isotomidae" Genes 16, no. 3: 315. https://doi.org/10.3390/genes16030315
APA StyleXie, Z., Zheng, M., Li, Y., Du, S., Saifutdinov, R., Potapov, M., Sun, X., & Wu, D. (2025). The Mitochondrial Genome of the Springtail Semicerura bryophila (Collembola): New Data Call into Question the Relevance of the Subfamilies of the Isotomidae. Genes, 16(3), 315. https://doi.org/10.3390/genes16030315







