In Vitro Gene Expression Profiling of Quantum Molecular Resonance Effects on Human Endometrium Models: A Preliminary Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. QMR Exposure
2.2. Cell Cultures and Endometrial Biopsies
2.3. Real Time PCR
2.4. Microarray Analysis
2.5. Statistical Analysis
3. Results
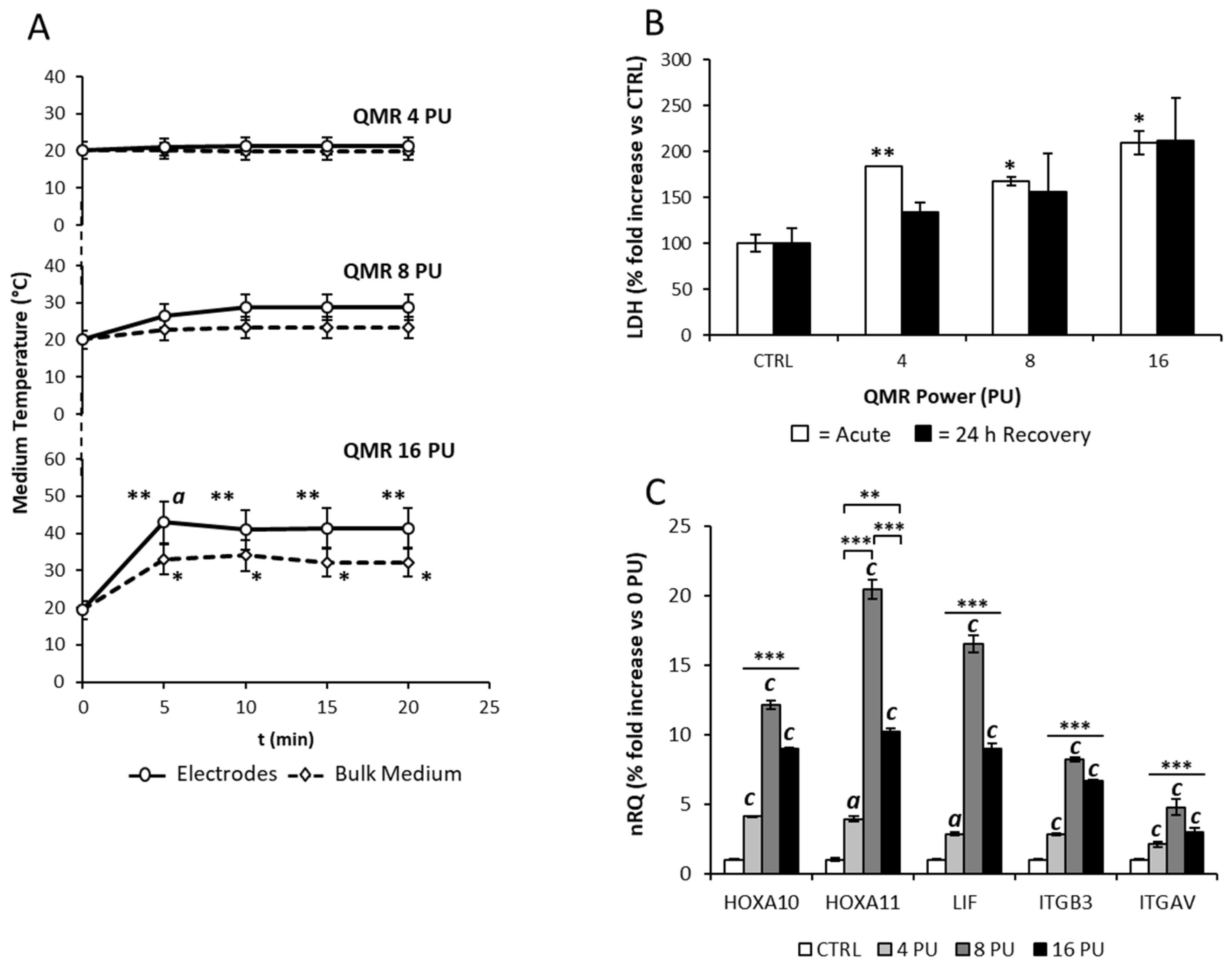
3.1. Effects of the Single Exposure of Ishikawa Cells to QMR
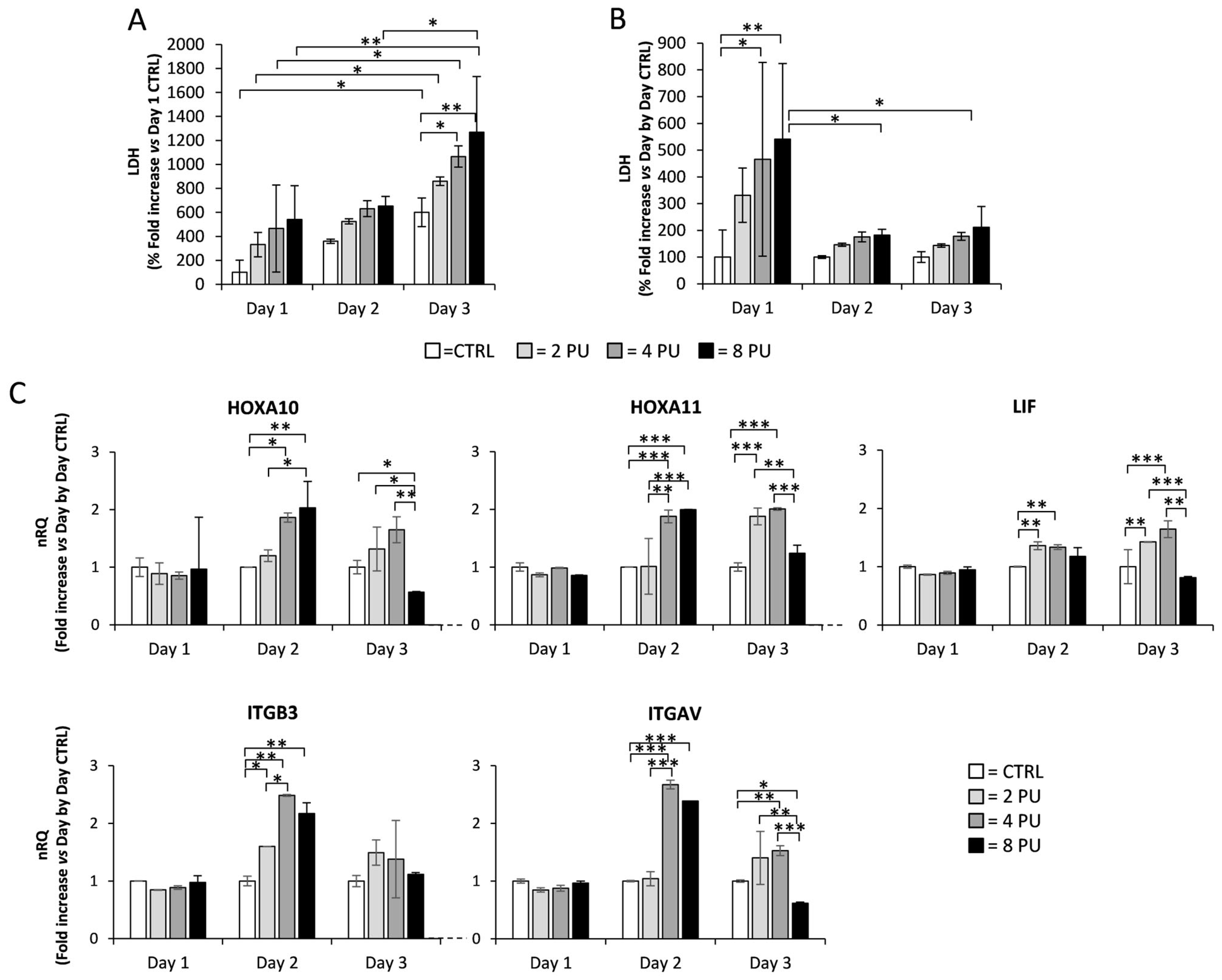
3.2. Effects of Repeated Exposure of Ishikawa Cells to QMR
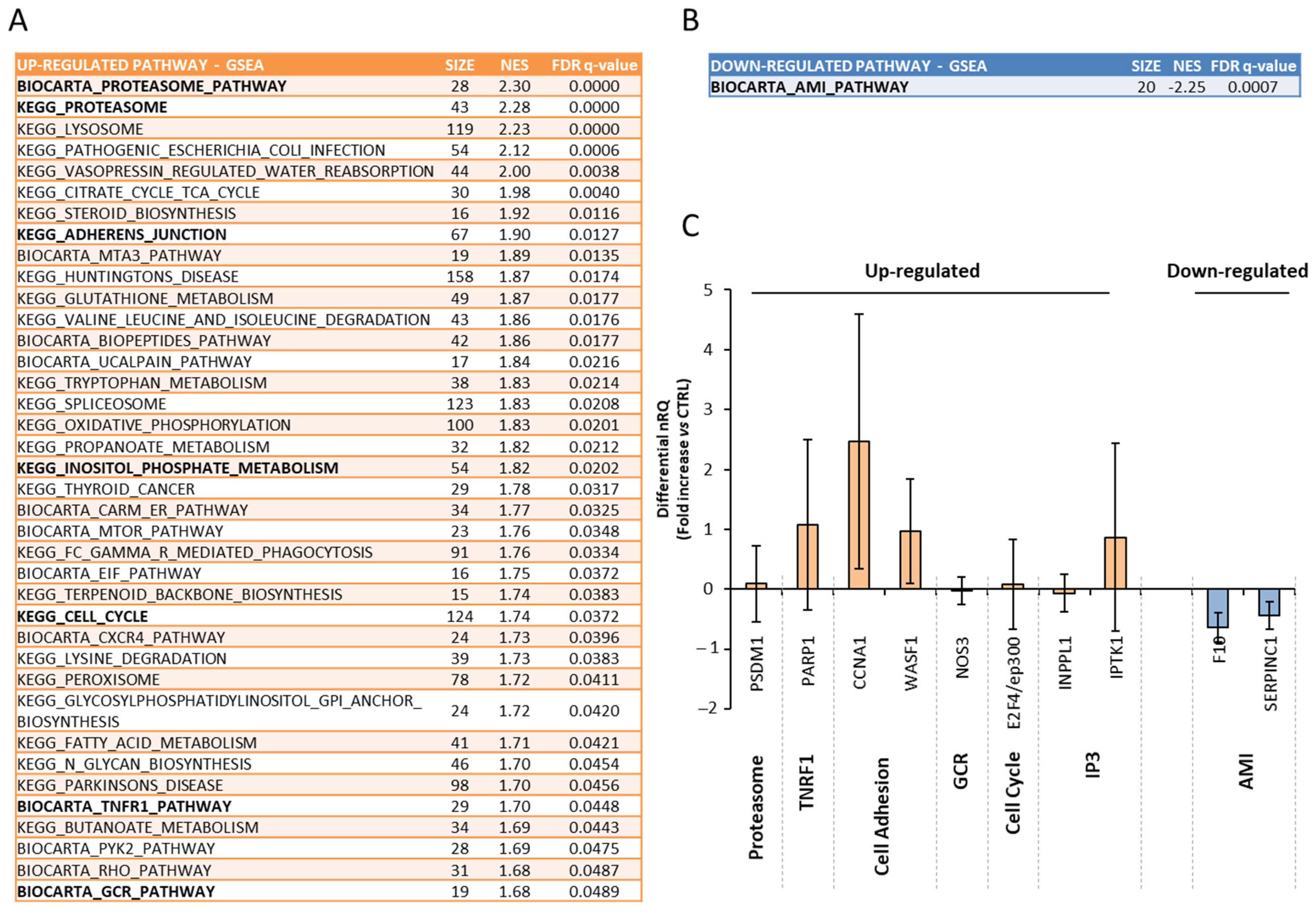
3.3. Effects of Multiple Daily Exposure to QMR on Gene Expression in Endometrial Biopsies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harvey, A.J.; Willson, B.E.; Surrey, E.S.; Gardner, D.K. Ovarian Stimulation Protocols: Impact on Oocyte and Endometrial Quality and Function. Fertil. Steril. 2025, 123, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Gliozheni, O.; Hambartsoumian, E.; Strohmer, H.; Petrovskaya, E.; Tishkevich, O.; De Neubourg, D.; Bogaerts, K.; Balic, D.; Antonova, I.; Cvetkova, E.; et al. ART in Europe, 2019: Results Generated from European Registries by ESHRE. Hum. Reprod. 2023, 38, 2321–2338. [Google Scholar] [CrossRef]
- Adamson, G.D.; de Mouzon, J.; Chambers, G.M.; Zegers-Hochschild, F.; Mansour, R.; Ishihara, O.; Banker, M.; Dyer, S. International Committee for Monitoring Assisted Reproductive Technology: World Report on Assisted Reproductive Technology, 2011. Fertil. Steril. 2018, 110, 1067–1080. [Google Scholar] [CrossRef]
- Kushnir, V.A.; Barad, D.H.; Albertini, D.F.; Darmon, S.K.; Gleicher, N. Systematic Review of Worldwide Trends in Assisted Reproductive Technology 2004–2013. Reprod. Biol. Endocrinol. 2017, 15, 6. [Google Scholar] [CrossRef]
- Edwards, R.G. Human Implantation: The Last Barrier in Assisted Reproduction Technologies? Reprod. Biomed. Online 2006, 13, 887–904. [Google Scholar] [CrossRef]
- Coughlan, C. What to Do When Good-Quality Embryos Repeatedly Fail to Implant. Best Pract. Res. Clin. Obstet. Gynaecol. 2018, 53, 48–59. [Google Scholar] [CrossRef]
- Paria, B.C.; Reese, J.; Das, S.K.; Dey, S.K. Deciphering the Cross-Talk of Implantation: Advances and Challenges. Science 2002, 296, 2185–2188. [Google Scholar] [CrossRef]
- Yoshinaga, K. Uterine Receptivity for Blastocyst Implantation. Ann. N. Y. Acad. Sci. 1988, 541, 424–431. [Google Scholar] [CrossRef]
- Finn, C.A.; Martin, L. The control of implantation. Reproduction 1974, 39, 195–206. [Google Scholar] [CrossRef]
- Noyes, R.W.; Hertig, A.T.; Rock, J. Reprint of: Dating the Endometrial Biopsy. Fertil. Steril. 2019, 112, e93–e115. [Google Scholar] [CrossRef]
- Murray, M.J.; Meyer, W.R.; Zaino, R.J.; Lessey, B.A.; Novotny, D.B.; Ireland, K.; Zeng, D.; Fritz, M.A. A Critical Analysis of the Accuracy, Reproducibility, and Clinical Utility of Histologic Endometrial Dating in Fertile Women. Fertil. Steril. 2004, 81, 1333–1343. [Google Scholar] [CrossRef] [PubMed]
- Talbi, S.; Hamilton, A.E.; Vo, K.C.; Tulac, S.; Overgaard, M.T.; Dosiou, C.; Le Shay, N.; Nezhat, C.N.; Kempson, R.; Lessey, B.A.; et al. Molecular Phenotyping of Human Endometrium Distinguishes Menstrual Cycle Phases and Underlying Biological Processes in Normo-Ovulatory Women. Endocrinology 2006, 147, 1097–1121. [Google Scholar] [CrossRef] [PubMed]
- Ponnampalam, A.P.; Weston, G.C.; Trajstman, A.C.; Susil, B.; Rogers, P.A.W. Molecular Classification of Human Endometrial Cycle Stages by Transcriptional Profiling. MHR Basic Sci. Reprod. Med. 2004, 10, 879–893. [Google Scholar] [CrossRef] [PubMed]
- Riesewijk, A. Gene Expression Profiling of Human Endometrial Receptivity on Days LH+2 versus LH+7 by Microarray Technology. Mol. Hum. Reprod. 2003, 9, 253–264. [Google Scholar] [CrossRef]
- Glujovsky, D.; Lattes, K.; Miguens, M.; Pesce, R.; Ciapponi, A. Personalized Embryo Transfer Guided by Endometrial Receptivity Analysis: A Systematic Review with Meta-Analysis. Hum. Reprod. 2023, 38, 1305–1317. [Google Scholar] [CrossRef]
- Neves, A.R.; Devesa, M.; Martínez, F.; Garcia-Martinez, S.; Rodriguez, I.; Polyzos, N.P.; Coroleu, B. What Is the Clinical Impact of the Endometrial Receptivity Array in PGT-A and Oocyte Donation Cycles? J. Assist. Reprod. Genet. 2019, 36, 1901–1908. [Google Scholar] [CrossRef]
- Bassil, R.; Casper, R.; Samara, N.; Hsieh, T.-B.; Barzilay, E.; Orvieto, R.; Haas, J. Does the Endometrial Receptivity Array Really Provide Personalized Embryo Transfer? J. Assist. Reprod. Genet. 2018, 35, 1301–1305. [Google Scholar] [CrossRef]
- Smith, M.B.; Paulson, R.J. Endometrial Preparation for Third-Party Parenting and Cryopreserved Embryo Transfer. Fertil. Steril. 2019, 111, 641–649. [Google Scholar] [CrossRef]
- Dieamant, F.; Vagnini, L.D.; Petersen, C.G.; Mauri, A.L.; Renzi, A.; Petersen, B.; Mattila, M.C.; Nicoletti, A.; Oliveira, J.B.A.; Baruffi, R.; et al. New Therapeutic Protocol for Improvement of Endometrial Receptivity (PRIMER) for Patients with Recurrent Implantation Failure (RIF)—A Pilot Study. JBRA Assist. Reprod. 2019, 23, 250. [Google Scholar] [CrossRef]
- Liu, Q.; Song, B. Electric Field Regulated Signaling Pathways. Int. J. Biochem. Cell Biol. 2014, 55, 264–268. [Google Scholar] [CrossRef]
- Costin, G.-E.; Birlea, S.A.; Norris, D.A. Trends in Wound Repair: Cellular and Molecular Basis of Regenerative Therapy Using Electromagnetic Fields. Curr. Mol. Med. 2012, 12, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Qian, F.; Ma, Q.; Zhang, P.; Chen, T.; Chen, C.; Zhang, Y.; Deng, P.; Zhou, Z.; Yu, Z. 50 Hz Electromagnetic Field Exposure Promotes Proliferation and Cytokine Production of Bone Marrow Mesenchymal Stem Cells. Int. J. Clin. Exp. Med. 2015, 8, 7394–7404. [Google Scholar] [PubMed]
- Serena, E.; Figallo, E.; Tandon, N.; Cannizzaro, C.; Gerecht, S.; Elvassore, N.; Vunjak-Novakovic, G. Electrical Stimulation of Human Embryonic Stem Cells: Cardiac Differentiation and the Generation of Reactive Oxygen Species. Exp. Cell Res. 2009, 315, 3611–3619. [Google Scholar] [CrossRef]
- Rouabhia, M.; Park, H.; Meng, S.; Derbali, H.; Zhang, Z. Electrical Stimulation Promotes Wound Healing by Enhancing Dermal Fibroblast Activity and Promoting Myofibroblast Transdifferentiation. PLoS ONE 2013, 8, e71660. [Google Scholar] [CrossRef]
- Maioli, M.; Rinaldi, S.; Santaniello, S.; Castagna, A.; Pigliaru, G.; Gualini, S.; Fontani, V.; Ventura, C. Radiofrequency Energy Loop Primes Cardiac, Neuronal, and Skeletal Muscle Differentiation in Mouse Embryonic Stem Cells: A New Tool for Improving Tissue Regeneration. Cell Transpl. 2012, 21, 1225–1233. [Google Scholar] [CrossRef]
- Zhao, Z.; Watt, C.; Karystinou, A.; Roelofs, A.; McCaig, C.; Gibson, I.; De Bari, C. Directed Migration of Human Bone Marrow Mesenchymal Stem Cells in a Physiological Direct Current Electric Field. Eur. Cell Mater. 2011, 22, 344–358. [Google Scholar] [CrossRef]
- Gómez-Ochoa, I.; Gómez-Ochoa, P.; Gómez-Casal, F.; Cativiela, E.; Larrad-Mur, L. Pulsed Electromagnetic Fields Decrease Proinflammatory Cytokine Secretion (IL-1β and TNF-α) on Human Fibroblast-like Cell Culture. Rheumatol. Int. 2011, 31, 1283–1289. [Google Scholar] [CrossRef]
- Vianale, G.; Reale, M.; Amerio, P.; Stefanachi, M.; Di Luzio, S.; Muraro, R. Extremely Low Frequency Electromagnetic Field Enhances Human Keratinocyte Cell Growth and Decreases Proinflammatory Chemokine Production. Br. J. Dermatol. 2008, 158, 1189–1196. [Google Scholar] [CrossRef]
- Lopresti, M.; Tomba, A.; Caserta, A.; Di Domenica, F. Studio Clinico Sull’efficacia Della Risonanza Quantica Molecolare Nel Trattamento Dell’edema Post-Chirurgico in Pazienti Sottoposti a Intervento Di Artroprotesi Di Ginocchio. Arch. Di Ortop. E Reumatol. 2011, 122, 34–35. [Google Scholar] [CrossRef]
- Foo, V.H.X.; Liu, Y.-C.; Tho, B.; Tong, L. Quantum Molecular Resonance Electrotherapy (Rexon-Eye) for Recalcitrant Dry Eye in an Asian Population. Front. Med. 2023, 10, 1209886. [Google Scholar] [CrossRef]
- Sella, S.; Adami, V.; Amati, E.; Bernardi, M.; Chieregato, K.; Gatto, P.; Menarin, M.; Pozzato, A.; Pozzato, G.; Astori, G. In-Vitro Analysis of Quantum Molecular Resonance Effects on Human Mesenchymal Stromal Cells. PLoS ONE 2018, 13, e0190082. [Google Scholar] [CrossRef] [PubMed]
- Nishida, M. The Ishikawa Cells from Birth to the Present. Hum. Cell 2002, 15, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Paolucci, T.; Tommasi, M.; Pozzato, G.; Pozzato, A.; Pezzi, L.; Zuccarini, M.; Di Lanzo, A.; Palumbo, R.; Porto, D.; Messeri, R.; et al. Management and Rehabilitative Treatment in Osteoarthritis with a Novel Physical Therapy Approach: A Randomized Control Study. Diagnostics 2024, 14, 1200. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ishihara, O.; Adamson, G.D.; Dyer, S.; de Mouzon, J.; Nygren, K.G.; Sullivan, E.A.; Zegers-Hochschild, F.; Mansour, R. International Committee for Monitoring Assisted Reproductive Technologies: World Report on Assisted Reproductive Technologies, 2007. Fertil. Steril. 2015, 103, 402–413.e11. [Google Scholar] [CrossRef]
- Vaiarelli, A.; Cimadomo, D.; Patrizio, P.; Venturella, R.; Orlando, G.; Soscia, D.; Giancani, A.; Capalbo, A.; Rienzi, L.; Ubaldi, F.M. Biochemical Pregnancy Loss after Frozen Embryo Transfer Seems Independent of Embryo Developmental Stage and Chromosomal Status. Reprod. Biomed. Online 2018, 37, 349–357. [Google Scholar] [CrossRef]
- Glujovsky, D.; Pesce, R.; Sueldo, C.; Quinteiro Retamar, A.M.; Hart, R.J.; Ciapponi, A. Endometrial Preparation for Women Undergoing Embryo Transfer with Frozen Embryos or Embryos Derived from Donor Oocytes. Cochrane Database Syst. Rev. 2020, 10, CD006359. [Google Scholar] [CrossRef]
- Mahajan, N. Endometrial Receptivity Array: Clinical Application. J. Hum. Reprod. Sci. 2015, 8, 121. [Google Scholar] [CrossRef]
- Díaz-Gimeno, P.; Horcajadas, J.A.; Martínez-Conejero, J.A.; Esteban, F.J.; Alamá, P.; Pellicer, A.; Simón, C. A Genomic Diagnostic Tool for Human Endometrial Receptivity Based on the Transcriptomic Signature. Fertil. Steril. 2011, 95, 50–60.e15. [Google Scholar] [CrossRef]
- Dal Maschio, M.; Canato, M.; Pigozzo, F.; Cipullo, A.; Pozzato, G.; Reggiani, C. Biophysical Effects of High Frequency Electrical Field (4-64 MHz) on Muscle Fibers in Culture. Basic Appl. Myol. 2009, 19, 49–56. [Google Scholar]
- Du, H.; Taylor, H.S. The Role of HOX Genes in Female Reproductive Tract Development, Adult Function, and Fertility. Cold Spring Harb. Perspect. Med. 2016, 6, a023002. [Google Scholar] [CrossRef] [PubMed]
- Taylor, H.S.; Vanden Heuvel, G.B.; Igarashi, P. A Conserved HOX Axis in the Mouse and Human Female Reproductive System: Late Establishment and Persistent Adult Expression of the Hoxa Cluster Genes. Biol. Reprod. 1997, 57, 1338–1345. [Google Scholar] [CrossRef] [PubMed]
- Taylor, H.S. HOX Gene Expression Is Altered in the Endometrium of Women with Endometriosis. Hum. Reprod. 1999, 14, 1328–1331. [Google Scholar] [CrossRef]
- Graf, U.; Casanova, E.A.; Cinelli, P. The Role of the Leukemia Inhibitory Factor (LIF)—Pathway in Derivation and Maintenance of Murine Pluripotent Stem Cells. Genes 2011, 2, 280–297. [Google Scholar] [CrossRef]
- Daikoku, T.; Cha, J.; Sun, X.; Tranguch, S.; Xie, H.; Fujita, T.; Hirota, Y.; Lydon, J.; DeMayo, F.; Maxson, R.; et al. Conditional Deletion of MSX Homeobox Genes in the Uterus Inhibits Blastocyst Implantation by Altering Uterine Receptivity. Dev. Cell 2011, 21, 1014–1025. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, L.; Xie, H.; Wan, H.; Magella, B.; Whitsett, J.A.; Dey, S.K. Kruppel-like Factor 5 (KLF5) Is Critical for Conferring Uterine Receptivity to Implantation. Proc. Natl. Acad. Sci. USA 2012, 109, 1145–1150. [Google Scholar] [CrossRef]
- Szczepańska, M.; Wirstlein, P.; Luczak, M.; Jagodzinski, P.P.; Skrzypczak, J. Expression of HOXA-10 and HOXA-11 in the Endometria of Women with Idiopathic Infertility. Folia Histochem. Cytobiol. 2011, 49, 111–118. [Google Scholar] [CrossRef]
- Elnaggar, A.; Farag, A.H.; Gaber, M.E.; Hafeez, M.A.; Ali, M.S.; Atef, A.M. AlphaVBeta3 Integrin Expression within Uterine Endometrium in Unexplained Infertility: A Prospective Cohort Study. BMC Womens Health 2017, 17, 90. [Google Scholar] [CrossRef]
- Lessey, B.A.; Castelbaum, A.J.; Wolf, L.; Greene, W.; Paulson, M.; Meyer, W.R.; Fritz, M.A. Use of Integrins to Date the Endometrium. Fertil. Steril. 2000, 73, 779–787. [Google Scholar] [CrossRef]
- Manohar, M.; Khan, H.; Sirohi, V.K.; Das, V.; Agarwal, A.; Pandey, A.; Siddiqui, W.A.; Dwivedi, A. Alteration in Endometrial Proteins during Early- and Mid-Secretory Phases of the Cycle in Women with Unexplained Infertility. PLoS ONE 2014, 9, e111687. [Google Scholar] [CrossRef]
- Whirledge, S.; Kisanga, E.P.; Taylor, R.N.; Cidlowski, J.A. Pioneer Factors FOXA1 and FOXA2 Assist Selective Glucocorticoid Receptor Signaling in Human Endometrial Cells. Endocrinology 2017, 158, 4076–4092. [Google Scholar] [CrossRef] [PubMed]
- Rakeman, A.S.; Anderson, K.V. Axis Specification and Morphogenesis in the Mouse Embryo Require Nap1, a Regulator of WAVE-Mediated Actin Branching. Development 2006, 133, 3075–3083. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, J.-D.; Novoa, E.M.; Vejnar, C.E.; Yartseva, V.; Takacs, C.M.; Kellis, M.; Giraldez, A.J. Analyses of MRNA Structure Dynamics Identify Embryonic Gene Regulatory Programs. Nat. Struct. Mol. Biol. 2018, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Elbaz, M.; Hadas, R.; Bilezikjian, L.M.; Gershon, E. Uterine Foxl2 Regulates the Adherence of the Trophectoderm Cells to the Endometrial Epithelium. Reprod. Biol. Endocrinol. 2018, 16, 12. [Google Scholar] [CrossRef]
- Wang, P.; Zhu, Q.; Peng, H.; Du, M.; Dong, M.; Wang, H. Fatty Acid-Binding Protein 4 in Endometrial Epithelium Is Involved in Embryonic Implantation. Cell Physiol. Biochem. 2017, 41, 501–509. [Google Scholar] [CrossRef]
- Canosa, S.; Moggio, A.; Brossa, A.; Pittatore, G.; Marchino, G.L.; Leoncini, S.; Benedetto, C.; Revelli, A.; Bussolati, B. Angiogenic Properties of Endometrial Mesenchymal Stromal Cells in Endothelial Co-Culture: An in Vitro Model of Endometriosis. MHR Basic Sci. Reprod. Med. 2017, 23, 187–198. [Google Scholar] [CrossRef]
- Celik, O.; Unlu, C.; Otlu, B.; Celik, N.; Caliskan, E. Laparoscopic Endometrioma Resection Increases Peri-Implantation Endometrial HOXA-10 and HOXA-11 MRNA Expression. Fertil. Steril. 2015, 104, 356–365. [Google Scholar] [CrossRef]

| Donor ID | Age (Years) | Clinical Diagnosis | Pharmacological Treatment |
|---|---|---|---|
| 1 | 42 | suspected endometriosis | none |
| 2 | 37 | previous endometriosis | none |
| 3 | 48 | myomas | none |
| 4 | 34 | menometrorrahgia | progesterone |
| 5 | 48 | menometrorrahgia | estrogen–progesterone |
| 6 | 42 | endometriosis | none |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grassi, A.; Rocca, M.S.; Noventa, M.; Pozzato, G.; Pozzato, A.; Scioscia, M.; Andrisani, A.; Pontrelli, G.; Foresta, C.; De Toni, L. In Vitro Gene Expression Profiling of Quantum Molecular Resonance Effects on Human Endometrium Models: A Preliminary Study. Genes 2025, 16, 290. https://doi.org/10.3390/genes16030290
Grassi A, Rocca MS, Noventa M, Pozzato G, Pozzato A, Scioscia M, Andrisani A, Pontrelli G, Foresta C, De Toni L. In Vitro Gene Expression Profiling of Quantum Molecular Resonance Effects on Human Endometrium Models: A Preliminary Study. Genes. 2025; 16(3):290. https://doi.org/10.3390/genes16030290
Chicago/Turabian StyleGrassi, Angela, Maria Santa Rocca, Marco Noventa, Gianantonio Pozzato, Alessandro Pozzato, Marco Scioscia, Alessandra Andrisani, Giovanni Pontrelli, Carlo Foresta, and Luca De Toni. 2025. "In Vitro Gene Expression Profiling of Quantum Molecular Resonance Effects on Human Endometrium Models: A Preliminary Study" Genes 16, no. 3: 290. https://doi.org/10.3390/genes16030290
APA StyleGrassi, A., Rocca, M. S., Noventa, M., Pozzato, G., Pozzato, A., Scioscia, M., Andrisani, A., Pontrelli, G., Foresta, C., & De Toni, L. (2025). In Vitro Gene Expression Profiling of Quantum Molecular Resonance Effects on Human Endometrium Models: A Preliminary Study. Genes, 16(3), 290. https://doi.org/10.3390/genes16030290









