Multi-Transcriptome-Informed Network Pharmacology Reveals Novel Biomarkers and Therapeutic Candidates for Parkinson’s Disease
Abstract
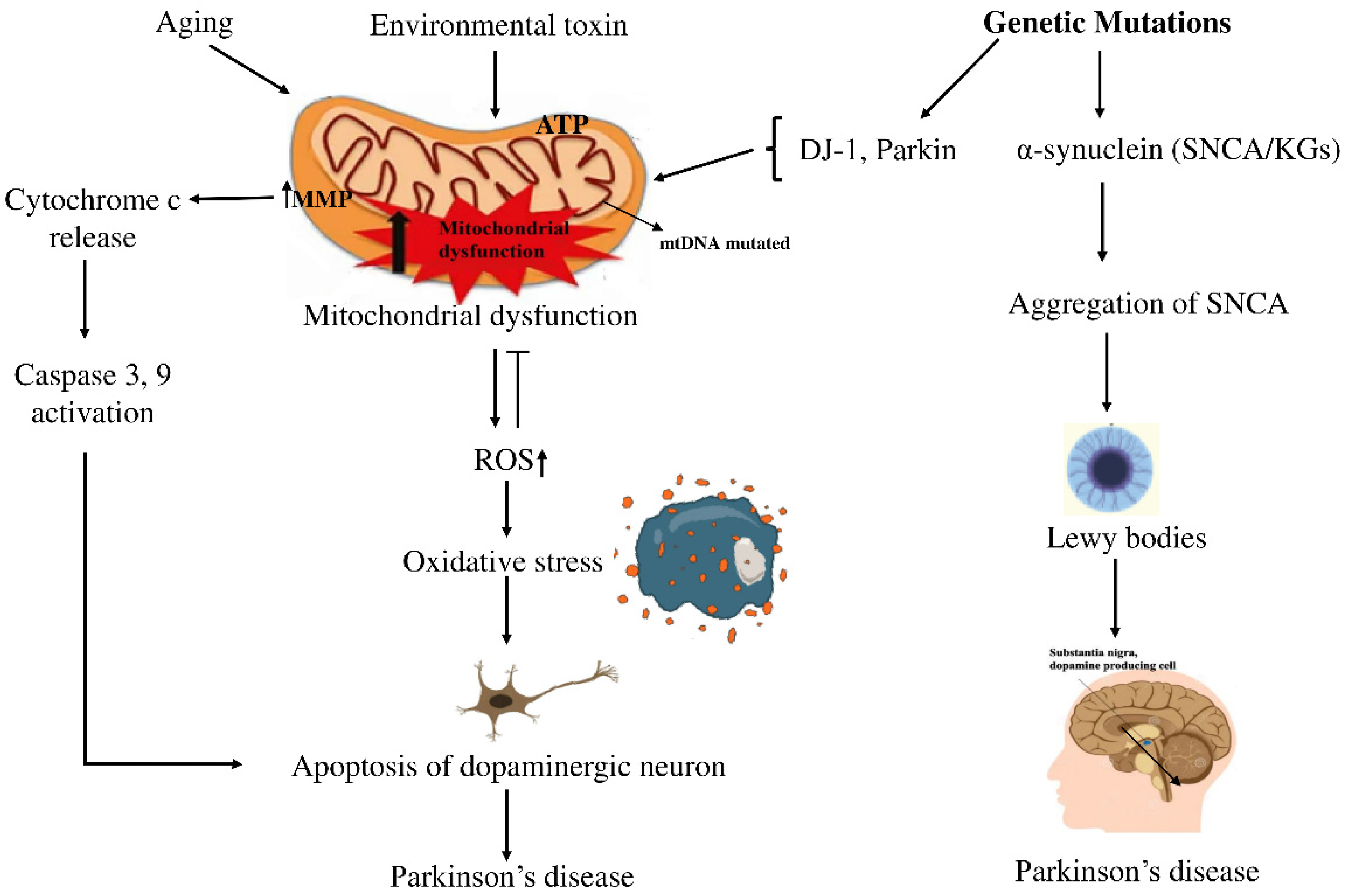
1. Introduction
2. Methods and Materials
2.1. Data Acquisition
2.2. Integration of Transcriptomics Datasets and Identification of DEGs
2.3. Identification of KGs from DEGs
2.4. Validation of KG Expression Profiles and Their Association with PD
2.5. Detection of Key Regulators of KGs
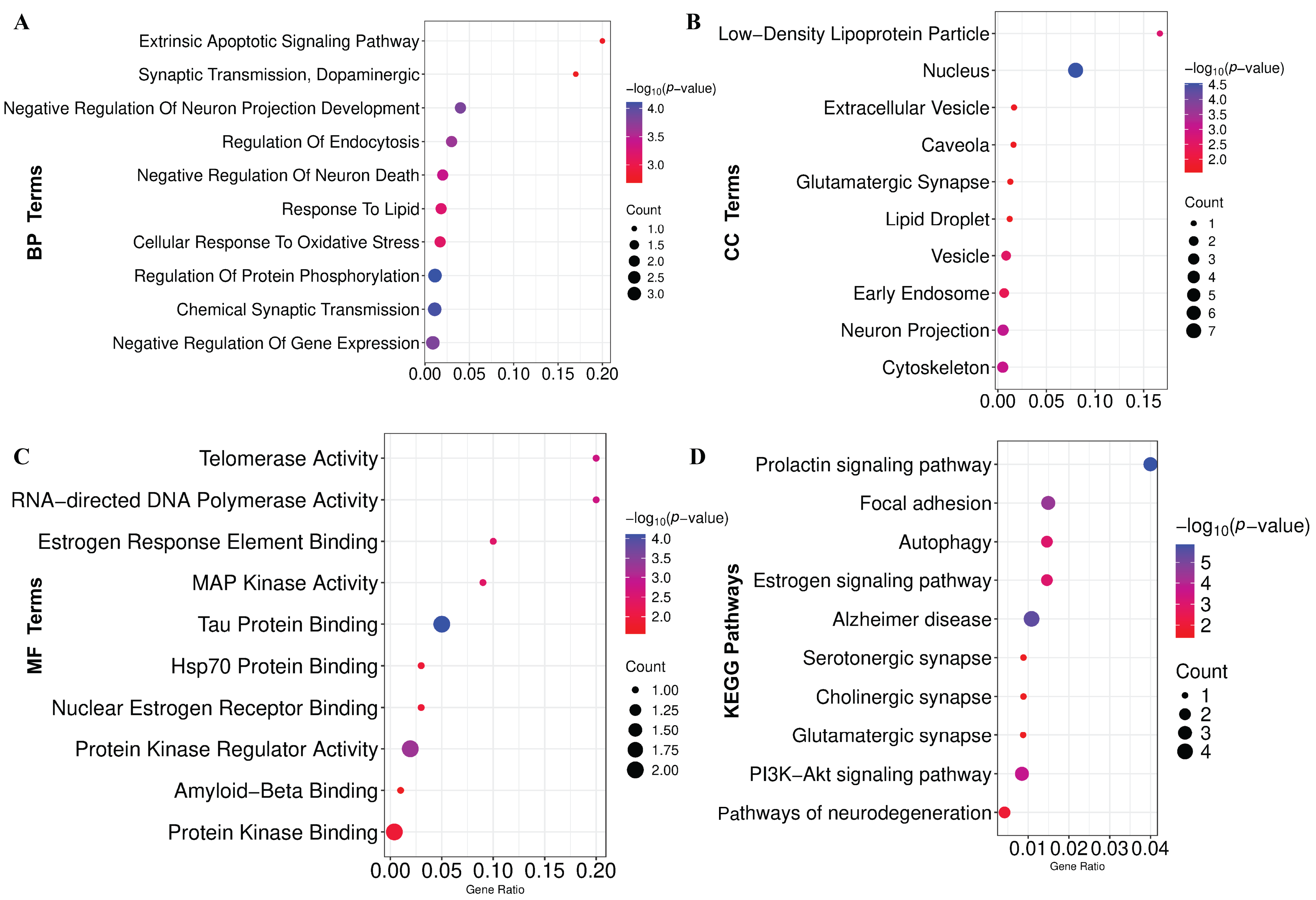
2.6. Detection of GO-Terms and KEGG-Pathways Associated with PD
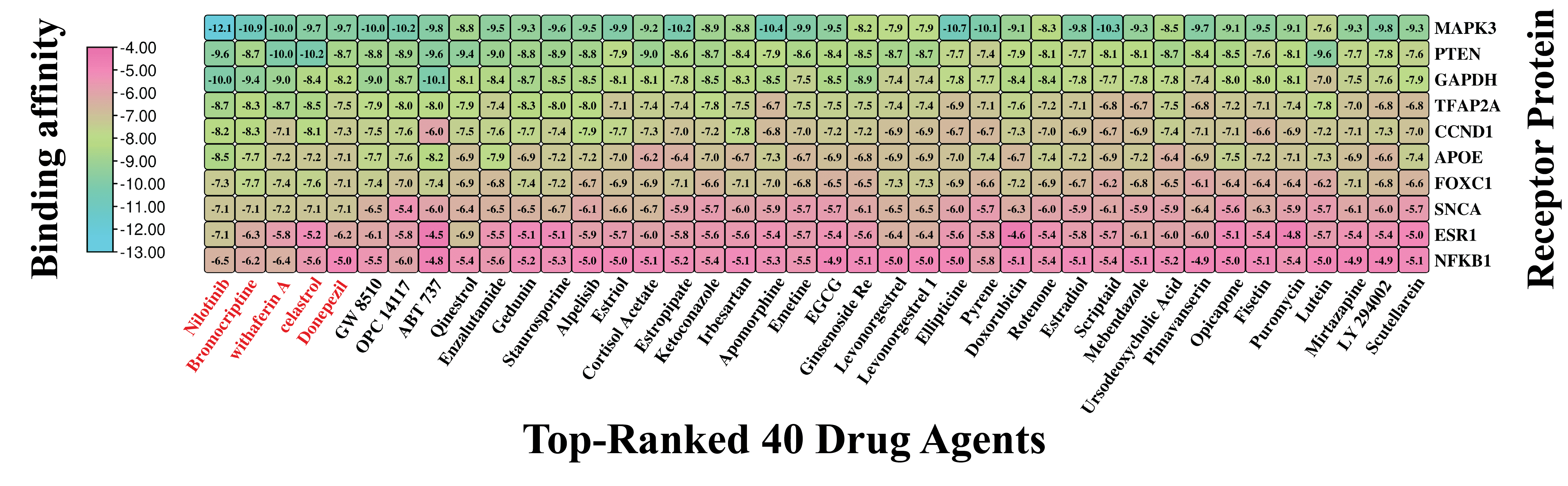
2.7. Exploring KGs-Guided Repurposable Drug Molecules Against PD
2.8. In Silico Validation of Top-Ranked Drug Molecules
2.8.1. ADME/T Analysis
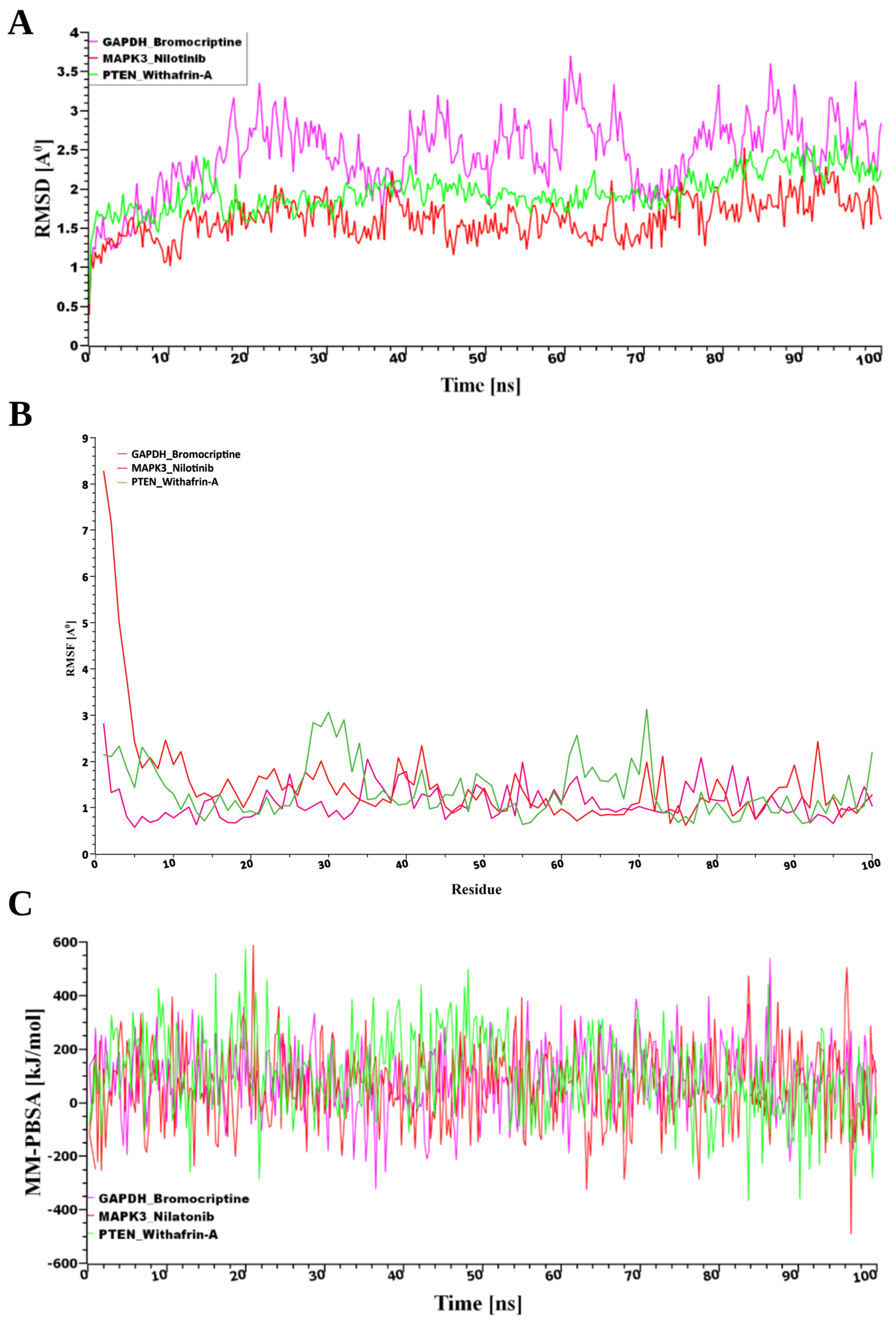
2.8.2. Molecular Dynamics (MD) Simulations
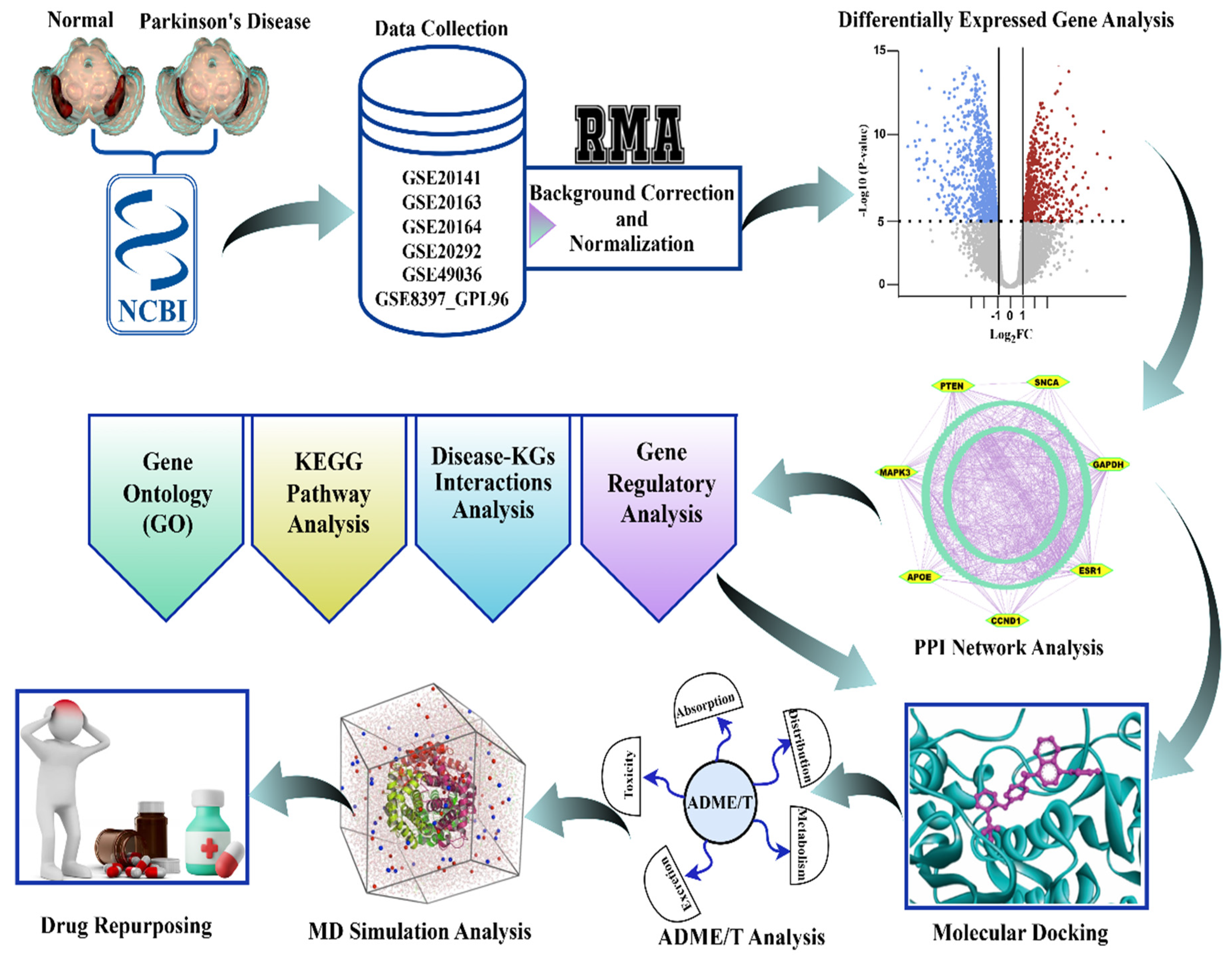
2.9. The Workflow of the Study
3. Result
3.1. Quality Control and Batch Effect Integration
3.2. Differential Gene Expression Patterns Identified from Integrated Transcriptomes
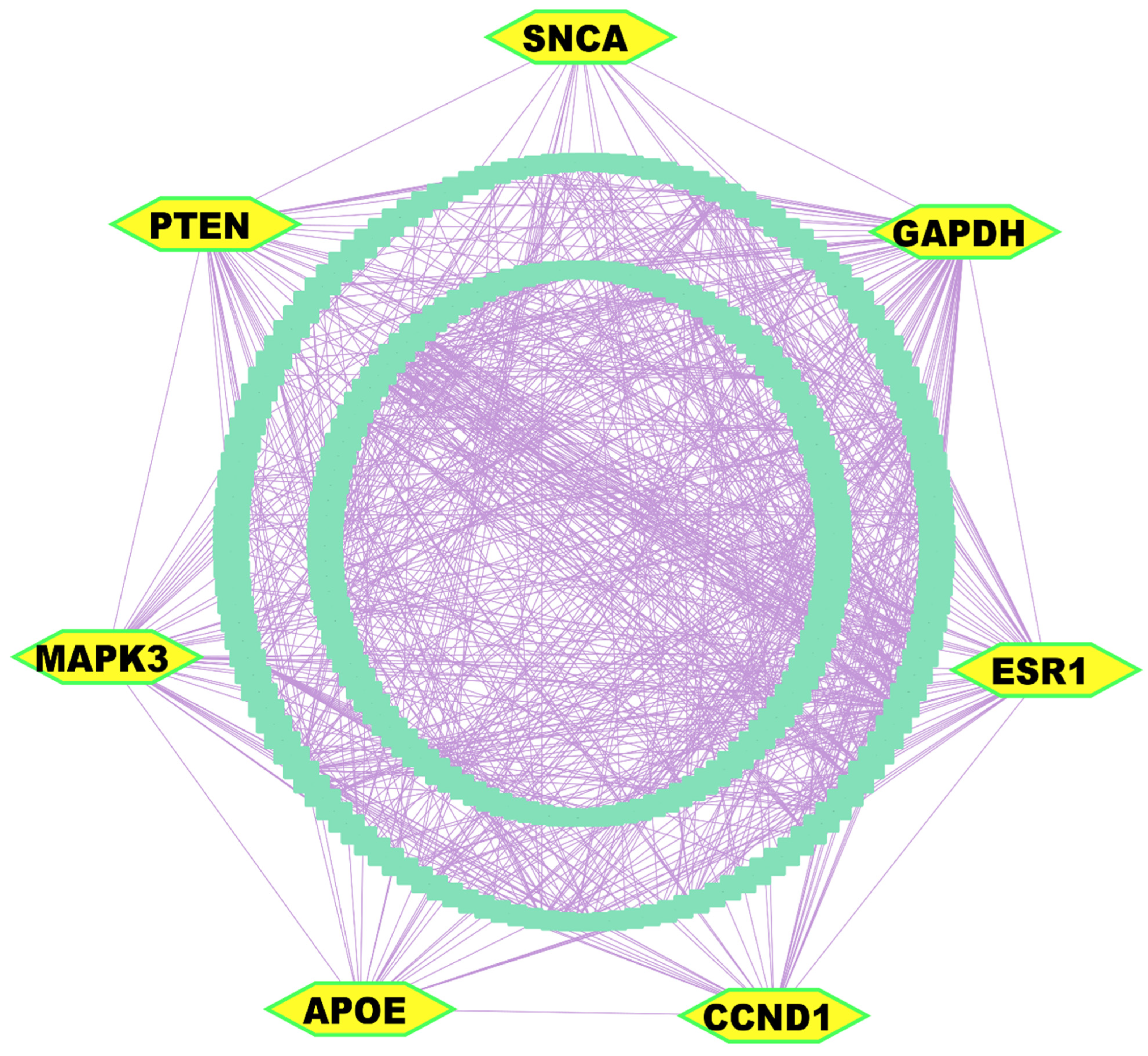
3.3. Key Genes Identified from Differentially Expressed Gene Analysis
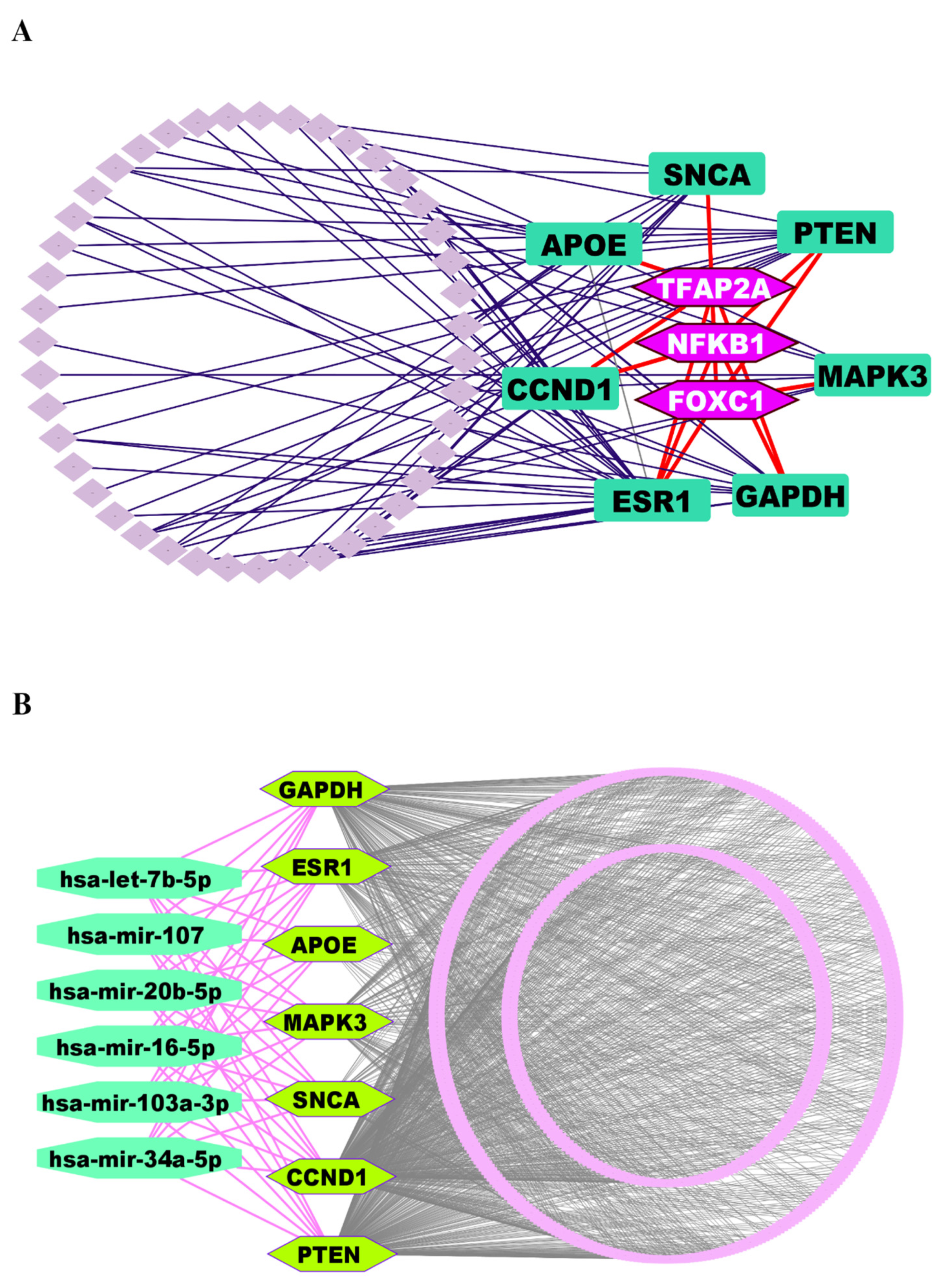
3.4. Regulatory Network Analysis Reveals Key Regulators of Key Genes
3.5. KGs Validation of KG Expression Profiles and Their Association with PD
3.6. Functional Enrichment Analysis Reveals GO Terms and KEGG Pathways Associated with PD
3.7. Exploring KGs-Guided Drug Molecules by Molecular Docking Analysis
3.8. In Silico Validation Confirms the Potential of Top-Ranked Drug Molecules
3.8.1. Pharmacokinetic Analysis
3.8.2. MD Simulations Validate the Binding Stability of Selected Drug Molecules
4. Discussion
4.1. KGs, Regulatory Networks, and Pathways in PD Pathogenesis
4.2. Drug Discovery and Therapeutic Insights
4.2.1. New In Silico Results
4.2.2. A Previously Validated Drug
4.3. Limitations and Future Direction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PD | Parkinson’s Disease |
| AD | Alzheimer’s Disease |
| SN | Substantia Nigra |
| LB | Lewy Bodies |
| LN | Lewy Neurites |
| MMP | Mitochondrial Membrane Potential |
| ROS | Reactive Oxygen Species |
| RMA | Robust Multi-Array Average |
| KGs | Key Genes |
| DEGs | Differentially Expressed Genes |
| cDEGs | Common Differentially Expressed Genes |
| GRN | Gene Regulatory Network |
| PPI | Protein–Protein Interaction |
| GDAs | Gene–Disease Associations |
| VDAs | Variant–Disease Associations |
| TFs | Transcription Factors |
| miRNAs | microRNAs |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PDB | Protein Data Bank |
| BA | Binding Affinity |
| MM-PBSA | MM–Poisson–Boltzmann Surface Area |
| MD | Molecular Dynamics |
| ADME | Absorption, Distribution, Metabolism, and Excretion |
| BBB | Blood-Brain Barrier |
References
- Pagan, F.L.; Hebron, M.L.; Wilmarth, B.; Torres-Yaghi, Y.; Lawler, A.; Mundel, E.E.; Yusuf, N.; Starr, N.J.; Anjum, M.; Arellano, J. Nilotinib Effects on Safety, Tolerability, and Potential Biomarkers in Parkinson Disease: A Phase 2 Randomized Clinical Trial. JAMA Neurol. 2020, 77, 309–317. [Google Scholar] [CrossRef]
- DeMarco, E.C.; Zhang, Z.; Robinson, H.; Hinyard, L. Anxiety in Parkinson’s Patients: What’s Timing Got to Do with It? J. Geriatr. Psychiatry Neurol. 2023, 36, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Rong, S.; Xu, G.; Liu, B.; Sun, Y.; Snetselaar, L.G.; Wallace, R.B.; Li, B.; Liao, J.; Bao, W. Trends in Mortality from Parkinson Disease in the United States, 1999–2019. Neurology 2021, 97, e1986–e1993. [Google Scholar] [CrossRef]
- Armstrong, M.J.; Okun, M.S. Diagnosis and Treatment of Parkinson Disease: A Review. JAMA 2020, 323, 548–560. [Google Scholar] [CrossRef] [PubMed]
- Lew, M. Overview of Parkinson’s Disease. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2007, 27, 155S–160S. [Google Scholar] [CrossRef]
- Vacchi, E.; Burrello, J.; Burrello, A.; Bolis, S.; Monticone, S.; Barile, L.; Kaelin-Lang, A.; Melli, G. Profiling Inflammatory Extracellular Vesicles in Plasma and Cerebrospinal Fluid: An Optimized Diagnostic Model for Parkinson’s Disease. Biomedicines 2021, 9, 230. [Google Scholar] [CrossRef]
- Kalia, L.V.; Lang, A.E. Parkinson’s Disease. Lancet 2015, 386, 896–912. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Liu, X.; Chen, J. Microarray Analysis of the Molecular Mechanism Involved in Parkinson’s Disease. Parkinsons Dis. 2018, 2018, 1590465. [Google Scholar] [CrossRef]
- Whittle, B.J.; Izuogu, O.G.; Lowes, H.; Deen, D.; Pyle, A.; Coxhead, J.; Lawson, R.A.; Yarnall, A.J.; Jackson, M.S.; Santibanez-Koref, M. Early-Stage Idiopathic Parkinson’s Disease Is Associated with Reduced Circular RNA Expression. npj Parkinsons Dis. 2024, 10, 25. [Google Scholar] [CrossRef]
- Diao, H.; Li, X.; Hu, S.; Liu, Y. Gene Expression Profiling Combined with Bioinformatics Analysis Identify Biomarkers for Parkinson Disease. PLoS ONE 2012, 7, e52319. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, Z.; Chi, C.; Li, C.; Yang, M.; Liu, B. Identification of Hub Genes and Potential Molecular Pathogenesis in Substantia Nigra in Parkinson’s Disease via Bioinformatics Analysis. Parkinsons Dis. 2023, 2023, 6755569. [Google Scholar] [CrossRef]
- Xin, G.; Niu, J.; Tian, Q.; Fu, Y.; Chen, L.; Yi, T.; Tian, K.; Sun, X.; Wang, N.; Wang, J. Identification of Potential Immune-Related Hub Genes in Parkinson’s Disease Based on Machine Learning and Development and Validation of a Diagnostic Classification Model. PLoS ONE 2023, 18, e0294984. [Google Scholar] [CrossRef]
- Su, L.; Wang, C.; Zheng, C.; Wei, H.; Song, X. A Meta-Analysis of Public Microarray Data Identifies Biological Regulatory Networks in Parkinson’s Disease. BMC Med. Genom. 2018, 11, 40. [Google Scholar] [CrossRef]
- George, G.; Parambath, S.V.; Lokappa, S.B.; Varkey, J. Construction of Parkinson’s Disease Marker-Based Weighted Protein-Protein Interaction Network for Prioritization of Co-Expressed Genes. Gene 2019, 697, 67–77. [Google Scholar] [CrossRef]
- Javali, S.B.; Gudaganavar, N.V.; Raj, S.M. Effect of Varying Sample Size in Estimation of Coefficients of Internal Consistency. WEbmed Cent. Biostat. 2011, 2, WMC001649. [Google Scholar]
- Chakrabarti, A.; Verma, S. Identifying Potential Genes Driving Ferroptosis in the Substantia Nigra and Dopaminergic Neurons in Parkinson’s Disease. Mol. Cell. Neurosci. 2025, 132, 103993. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jia, W.; Chen, C.; Chen, C.; Chen, J.; Yang, X.; Liu, P. Identification of Biomarkers Associated with Inflammatory Response in Parkinson’s Disease by Bioinformatics and Machine Learning. PLoS ONE 2025, 20, e0320257. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Huang, Y.; Li, J. Bioinformatic Analysis for the Identification of Key Candidate Genes and Pathways in the Substantia Nigra in Parkinson’s Disease. J. Integr. Neurosci. 2018, 17, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Li, B.; Wei, H.; Li, C.; Liu, C.; Mi, H.; Chen, S. Integrative Analysis of Gene Expression Profiles of Substantia Nigra Identifies Potential Diagnosis Biomarkers in Parkinson’s Disease. Sci. Rep. 2024, 14, 2167. [Google Scholar] [CrossRef]
- Jin, X.; Li, J.; Li, W.; Wang, X.; Du, C.; Geng, Z.; Geng, Y.; Kang, L.; Zhang, X.; Wang, M. Weighted Gene Co-Expression Network Analysis Reveals Specific Modules and Biomarkers in Parkinson’s Disease. Neurosci. Lett. 2020, 728, 134950. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Y.; Huang, J.; Hu, B.; Huang, W. Identification of Immune-Related Hub Genes in Parkinson’s Disease. Front. Genet. 2022, 13, 914645. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 Update: Improved Access to Chemical Data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, R.A.; Hobbs, B.; Collin, F.; Beazer-Barclay, Y.D.; Antonellis, K.J.; Scherf, U.; Speed, T.P. Exploration, Normalization, and Summaries of High Density Oligonucleotide Array Probe Level Data. Biostatistics 2003, 4, 249–264. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic. Acids. Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Ben-Hur, A.; Noble, W.S. Kernel Methods for Predicting Protein-Protein Interactions. Bioinformatics 2005, 2, i38–i46. [Google Scholar] [CrossRef]
- Mahmud, S.; Ajadee, A.; Sarker, A.; Ahmmed, R.; Noor, T.; Pappu, M.A.A.; Islam, M.S.; Mollah, M.N.H. Exploring Common Genomic Biomarkers to Disclose Common Drugs for the Treatment of Colorectal Cancer and Hepatocellular Carcinoma with Type-2 Diabetes through Transcriptomics Analysis. PLoS ONE 2025, 20, e0319028. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Lesnick, T.G.; Papapetropoulos, S.; Mash, D.C.; Ffrench-Mullen, J.; Shehadeh, L.; de Andrade, M.; Henley, J.R.; Rocca, W.A.; Ahlskog, J.E.; Maraganore, D.M. A Genomic Pathway Approach to a Complex Disease: Axon Guidance and Parkinson Disease. PLoS Genet. 2007, 3, e98. [Google Scholar] [CrossRef]
- Pappu, M.A.A.; Alamin, M.; Sultana, M.H.; Azad, A.K.M.; Auwul, M.R.; Mahmud, S.; Ajadee, A.; Sarker, A.; Alyami, S.A.; Mollah, M.N.H. Single Nucleus RNA Sequencing Profile Analysis to Reveal Cell Type Specific Common Molecular Drivers of Parkinson’s Disease and Therapeutic Agents. Sci. Rep. 2025, 15, 27086. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.F.; Al Noman, M.; Ahmed, M.F.; Latif, M.A.; Pappu, M.A.A.; Islam, M.S.; Hossain, M.S.; Hossen, M.B.; Faysal, M.F.; Hasan, M.M.; et al. In-Silico Discovery of Pediatric Acute-Myeloid-Leukemia (PAML) Causing Druggable Molecular Signatures Highlighting Their Pathogenetic Processes and Therapeutic Agents through Single-Cell RNA-Seq Profile Analysis. PLoS ONE 2025, 20, e0335410. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET Knowledge Platform for Disease Genomics: 2019 Update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar] [CrossRef] [PubMed]
- Fornes, O.; Castro-Mondragon, J.A.; Khan, A.; van der Lee, R.; Zhang, X.; Richmond, P.A.; Modi, B.P.; Correard, S.; Gheorghe, M.; Baranašić, D.; et al. JASPAR 2020: Update of the Open-Access Database of Transcription Factor Binding Profiles. Nucleic Acids Res. 2020, 48, D87–D92. [Google Scholar] [CrossRef]
- Xia, J.; Gill, E.E.; Hancock, R.E.W. NetworkAnalyst for Statistical, Visual and Network-Based Meta-Analysis of Gene Expression Data. Nat. Protoc. 2015, 10, 823–844. [Google Scholar] [CrossRef]
- Sethupathy, P.; Corda, B.; Hatzigeorgiou, A.G. TarBase: A Comprehensive Database of Experimentally Supported Animal MicroRNA Targets. Rna 2006, 12, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Doms, A.; Schroeder, M. GoPubMed: Exploring PubMed with the Gene Ontology. Nucleic Acids Res. 2005, 33, W783–W786. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for Linking Genomes to Life and the Environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Wang, T.; Jiang, X.; Ruan, Y.; Zhuang, J.; Yin, Y. Based on Network Pharmacology and in Vitro Experiments to Prove the Effective Inhibition of Myocardial Fibrosis by Buyang Huanwu Decoction. Bioengineered 2022, 13, 13767–13783. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An Automated Protein Homology-Modeling Server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef]
- Baroroh, U.; Biotek, M.; Muscifa, Z.S.; Destiarani, W.; Rohmatullah, F.G.; Yusuf, M. Molecular Interaction Analysis and Visualization of Protein-Ligand Docking Using Biovia Discovery Studio Visualizer. Indones. J. Comput. Biol. (IJCB) 2023, 2, 22–30. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Jayaram, B.; Singh, T.; Mukherjee, G.; Mathur, A.; Shekhar, S.; Shekhar, V. Sanjeevini: A Freely Accessible Web-Server for Target Directed Lead Molecule Discovery. BMC Bioinform. 2012, 13, S7. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. PkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the Precision of Comparative Models with YASARA NOVA—A Self-Parameterizing Force Field. Proteins Struct. Funct. Bioinform. 2002, 47, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Cheatham, T.E., III; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber Biomolecular Simulation Programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. New Ways to Boost Molecular Dynamics Simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef]
- Krieger, E.; Nielsen, J.E.; Spronk, C.A.E.M.; Vriend, G. Fast Empirical PKa Prediction by Ewald Summation. J. Mol. Graph. Model 2006, 25, 481–486. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Mitra, S.; Dash, R. Structural Dynamics and Quantum Mechanical Aspects of Shikonin Derivatives as CREBBP Bromodomain Inhibitors. J. Mol. Graph. Model 2018, 83, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, E.; Rajasekaran, R. Computational Investigation of Curcumin, a Natural Polyphenol That Inhibits the Destabilization and the Aggregation of Human SOD1 Mutant (Ala4Val). RSC Adv. 2016, 6, 102744–102753. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING Database in 2011: Functional Interaction Networks of Proteins, Globally Integrated and Scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef]
- Chen, X.; Li, H.; Tian, L.; Li, Q.; Luo, J.; Zhang, Y. Analysis of the Physicochemical Properties of Acaricides Based on Lipinski’s Rule of Five. J. Comput. Biol. 2020, 27, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
- Gabathuler, R. Approaches to Transport Therapeutic Drugs across the Blood–Brain Barrier to Treat Brain Diseases. Neurobiol. Dis. 2010, 37, 48–57. [Google Scholar] [CrossRef]
- Wong, A.D.; Ye, M.; Levy, A.F.; Rothstein, J.D.; Bergles, D.E.; Searson, P.C. The Blood-Brain Barrier: An Engineering Perspective. Front. Neuroeng. 2013, 6, 7. [Google Scholar] [CrossRef]
- Batista, P.; Pereira, A. Impact and Prevention of Neurodegenerative Diseases in Society: Alzheimer and Parkinson. In Neurodegenerative Diseases; SM Group Open Access eBooks: Brentford, UK, 2016. [Google Scholar]
- Lee, T.K.; Yankee, E.L. A Review on Parkinson’s Disease Treatment. Neuroimmunol. Neuroinflamm. 2021, 8, 222. [Google Scholar] [CrossRef]
- Wood-Kaczmar, A.; Gandhi, S.; Wood, N.W. Understanding the Molecular Causes of Parkinson’s Disease. Trends Mol. Med. 2006, 12, 521–528. [Google Scholar] [CrossRef]
- Ping, Z.; Xiaomu, W.; Xufang, X.; Wenfeng, C.; Liang, S.; Tao, W. GAPDH Rs1136666 SNP Indicates a High Risk of Parkinson’s Disease. Neurosci. Lett. 2018, 685, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, K.; Tajima, H.; Kuwae, T.; Takeshima, T.; Nakano, T.; Tanaka, M.; Sunaga, K.; Fukuhara, Y.; Nakashima, K.; Ohama, E. Pro-apoptotic Protein Glyceraldehyde-3-phosphate Dehydrogenase Promotes the Formation of Lewy Body-like Inclusions. Eur. J. Neurosci. 2005, 21, 317–326. [Google Scholar] [CrossRef]
- Liu, L.; Xiong, N.; Zhang, P.; Chen, C.; Huang, J.; Zhang, G.; Xu, X.; Shen, Y.; Lin, Z.; Wang, T. Genetic Variants in GAPDH Confer Susceptibility to Sporadic Parkinson’s Disease in a Chinese Han Population. PLoS ONE 2015, 10, e0135425. [Google Scholar] [CrossRef]
- Erekat, N.S. Apoptosis and Its Role in Parkinson’s Disease. Exon. Publ. 2018, 65–82. [Google Scholar] [CrossRef]
- Primiani, C.T.; Ryan, V.H.; Rao, J.S.; Cam, M.C.; Ahn, K.; Modi, H.R.; Rapoport, S.I. Coordinated Gene Expression of Neuroinflammatory and Cell Signaling Markers in Dorsolateral Prefrontal Cortex during Human Brain Development and Aging. PLoS ONE 2014, 9, e110972. [Google Scholar] [CrossRef]
- Kaltschmidt, B.; Helweg, L.P.; Greiner, J.F.W.; Kaltschmidt, C. NF-ΚB in Neurodegenerative Diseases: Recent Evidence from Human Genetics. Front. Mol. Neurosci. 2022, 15, 954541. [Google Scholar] [CrossRef]
- Santos-Lobato, B.L.; Vidal, A.F.; Ribeiro-dos-Santos, Â. Regulatory MiRNA–MRNA Networks in Parkinson’s Disease. Cells 2021, 10, 1410. [Google Scholar] [CrossRef] [PubMed]
- Findeiss, E.; Schwarz, S.C.; Evsyukov, V.; Rösler, T.W.; Höllerhage, M.; Chakroun, T.; Nykänen, N.P.; Shen, Y.; Wurst, W.; Kohl, M. Comprehensive MiRNome-Wide Profiling in a Neuronal Cell Model of Synucleinopathy Implies Involvement of Cell Cycle Genes. Front. Cell Dev. Biol. 2021, 9, 561086. [Google Scholar] [CrossRef]
- Zhang, H. CCND1 Silencing Suppresses Liver Cancer Stem Cell Differentiation through Inhibiting Autophagy. Hum. Cell 2020, 33, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Ogino, M.; Ichimura, M.; Nakano, N.; Minami, A.; Kitagishi, Y.; Matsuda, S. Roles of PTEN with DNA Repair in Parkinson’s Disease. Int. J. Mol. Sci. 2016, 17, 954. [Google Scholar] [CrossRef] [PubMed]
- Pivtoraiko, V.N.; Stone, S.L.; Roth, K.A.; Shacka, J.J. Oxidative Stress and Autophagy in the Regulation of Lysosome-Dependent Neuron Death. Antioxid Redox Signal 2009, 11, 481–496. [Google Scholar] [CrossRef]
- Singh, N.K.; Banerjee, B.D.; Bala, K.; Mitrabasu; Dung Dung, A.A.; Chhillar, N. APOE and LRPAP1 Gene Polymorphism and Risk of Parkinson’s Disease. Neurol. Sci. 2014, 35, 1075–1081. [Google Scholar] [CrossRef] [PubMed]
- Emamzadeh, F.N. Role of Apolipoproteins and α-Synuclein in Parkinson’s Disease. J. Mol. Neurosci. 2017, 62, 344–355. [Google Scholar] [CrossRef]
- Han, S.; Fang, J.; Yu, L.; Li, B.; Hu, Y.; Chen, R.; Li, C.; Zhao, C.; Li, J.; Wang, Y. Serum-derived Exosomal Hsa-let-7b-5p as a Biomarker for Predicting the Severity of Coronary Stenosis in Patients with Coronary Heart Disease and Hyperglycemia. Mol. Med. Rep. 2023, 28, 203. [Google Scholar] [CrossRef]
- Liu, J.; Yuan, S.; Niu, X.; Kelleher, R.; Sheridan, H. ESR1 Dysfunction Triggers Neuroinflammation as a Critical Upstream Causative Factor of the Alzheimer’s Disease Process. Aging 2022, 14, 8595. [Google Scholar] [CrossRef]
- Sulhan, S.; Lyon, K.A.; Shapiro, L.A.; Huang, J.H. Neuroinflammation and Blood–Brain Barrier Disruption Following Traumatic Brain Injury: Pathophysiology and Potential Therapeutic Targets. J. Neurosci. Res. 2020, 98, 19–28. [Google Scholar] [CrossRef]
- Yao, Z.; Li, J.; Bian, L.; Li, Q.; Wang, X.; Yang, X.; Wei, X.; Wan, G.; Wang, Y.; Shi, J. Nootkatone Alleviates Rotenone-induced Parkinson’s Disease Symptoms through Activation of the PI3K/Akt Signaling Pathway. Phytother. Res. 2022, 36, 4183–4200. [Google Scholar] [CrossRef]
- Sun, A.; Li, Y.; Miao, Y.; Wang, H.; Zhang, L. Research on the Mechanism of Ursolic Acid for Treating Parkinson’s Disease by Network Pharmacology and Experimental Verification. Heliyon 2024, 10, e34113. [Google Scholar] [CrossRef]
- das Campêlo, C.L.C.; Silva, R.H. Genetic Variants in SNCA and the Risk of Sporadic Parkinson’s Disease and Clinical Outcomes: A Review. Parkinsons Dis. 2017, 2017, 4318416. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, C.C.; Lange, J.; Førland, M.G.G.; Macleod, A.D.; Alves, G.; Maple-Grødem, J. A Systematic Review of Associations between Common SNCA Variants and Clinical Heterogeneity in Parkinson’s Disease. npj Parkinsons Dis. 2021, 7, 54. [Google Scholar] [CrossRef]
- Irene Litvan, M.D.; Parkkinen, L. α-Synuclein Genetic Variability: A Biomarker for Dementia in Parkinson’s Disease. Ann. Neurol. 2016, 79, 991–999. [Google Scholar] [CrossRef]
- Goris, A.; Williams-Gray, C.H.; Clark, G.R.; Foltynie, T.; Lewis, S.J.G.; Brown, J.; Ban, M.; Spillantini, M.G.; Compston, A.; Burn, D.J. Tau and A-synuclein in Susceptibility to, and Dementia in, Parkinson’s Disease. Ann. Neurol. 2007, 62, 145–153. [Google Scholar] [CrossRef]
- Chu, Y.; Hirst, W.D.; Federoff, H.J.; Harms, A.S.; Stoessl, A.J.; Kordower, J.H. Nigrostriatal Tau Pathology in Parkinsonism and Parkinson’s Disease. Brain 2024, 147, 444–457. [Google Scholar] [CrossRef]
- Lovering, A.L.; Lee, S.S.; Kim, Y.W.; Withers, S.G.; Strynadka, N.C.J. Mechanistic and Structural Analysis of a Family 31 α-Glycosidase and Its Glycosyl-Enzyme Intermediate. J. Biol. Chem. 2005, 280, 2105–2115. [Google Scholar] [CrossRef]
- Pagan, F.L.; Hebron, M.L.; Wilmarth, B.; Torres-Yaghi, Y.; Lawler, A.; Mundel, E.E.; Yusuf, N.; Starr, N.J.; Arellano, J.; Howard, H.H. Pharmacokinetics and Pharmacodynamics of a Single Dose Nilotinib in Individuals with Parkinson’s Disease. Pharmacol. Res. Perspect. 2019, 7, e00470. [Google Scholar] [CrossRef] [PubMed]
- Lieberman, A.; Kupersmith, M.; Estey, E.; Goldstein, M. Treatment of Parkinson’s Disease with Bromocriptine. New Engl. J. Med. 1976, 295, 1400–1404. [Google Scholar] [CrossRef]
- Van Hilten, J.J.; Ramaker, C.C.; Stowe, R.; Ives, N.J. Bromocriptine versus Levodopa in Early Parkinson’s Disease. Cochrane Database Syst. Rev. 2007, 2009, CD002258. [Google Scholar] [CrossRef]
- Birla, H.; Keswani, C.; Rai, S.N.; Singh, S.S.; Zahra, W.; Dilnashin, H.; Rathore, A.S.; Singh, S.P. Neuroprotective Effects of Withania Somnifera in BPA Induced-Cognitive Dysfunction and Oxidative Stress in Mice. Behav. Brain Funct. 2019, 15, 9. [Google Scholar] [CrossRef] [PubMed]
- Dar, N.J.; Ahmad, M. Neurodegenerative Diseases and Withania Somnifera (L.): An Update. J. Ethnopharmacol. 2020, 256, 112769. [Google Scholar] [CrossRef]
- Lin, M.W.; Lin, C.C.; Chen, Y.H.; Yang, H.B.; Hung, S.Y. Celastrol Inhibits Dopaminergic Neuronal Death of Parkinson’s Disease through Activating Mitophagy. Antioxidants 2019, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Fu, R.J.; Zhang, S.; Yue, S.J.; Chen, Y.Y.; Xu, D.Q.; Tang, Y.P. Potential Medicinal Value of Celastrol and Its Synthesized Analogues for Central Nervous System Diseases. Biomed. Pharmacother. 2021, 139, 111551. [Google Scholar] [CrossRef]
- Mori, S. Responses to Donepezil in Alzheimer’s Disease and Parkinson’s Disease. Ann. N. Y. Acad. Sci. 2002, 977, 493–500. [Google Scholar] [CrossRef]
- Dubois, B.; Tolosa, E.; Katzenschlager, R.; Emre, M.; Lees, A.J.; Schumann, G.; Pourcher, E.; Gray, J.; Thomas, G.; Swartz, J. Donepezil in Parkinson’s Disease Dementia: A Randomized, Double-blind Efficacy and Safety Study. Mov. Disord. 2012, 27, 1230–1238. [Google Scholar] [CrossRef] [PubMed]
- Morgan, J.C.; Sethi, K.D. Emerging drugs for Parkinson’s disease. Expert Opin. Emerg. Drugs 2006, 11, 403–417. [Google Scholar] [CrossRef]
- Whitney, C.M. Medications for Parkinson’s disease. Neurologist 2007, 13, 387–388. [Google Scholar] [CrossRef]
- Schwab, R.S.; England, A.C., Jr.; Poskanzer, D.C.; Young, R.R. Amantadine in the treatment of Parkinson’s disease. JAMA 1969, 208, 1168–1170. [Google Scholar] [CrossRef]
- Rondón-Villarreal, P.; López, W.O.C. Identification of potential natural neuroprotective molecules for Parkinson’s disease by using chemoinformatics and molecular docking. J. Mol. Graph. Model. 2020, 97, 107547. [Google Scholar] [CrossRef]
- Agúndez, J.A.G.; García-Martín, E.; Alonso-Navarro, H.; Jiménez-Jiménez, F.J. Anti-Parkinson’s disease drugs and pharmacogenetic considerations. Expert Opin. Drug Metab. Toxicol. 2013, 9, 859–874. [Google Scholar] [CrossRef]
- Hubble, J.P. Novel drugs for Parkinson’s disease. Med. Clin. N. Am. 1999, 83, 525–536. [Google Scholar] [CrossRef] [PubMed]
- McOmish, C.; Pavey, G.; McLean, C.; Horne, M.; Dean, B.; Scarr, E. Muscarinic receptor binding changes in postmortem Parkinson’s disease. J. Neural Transm. 2017, 124, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Yoo, M.; Shin, J.; Kim, J.; Ryall, K.A.; Lee, K.; Lee, S.; Jeon, M.; Kang, J.; Tan, A.C. DSigDB: Drug signatures database for gene set analysis. Bioinformatics 2015, 31, 3069–3071. [Google Scholar] [CrossRef] [PubMed]
- Athauda, D.; Foltynie, T. Drug repurposing in Parkinson’s disease. CNS Drugs 2018, 32, 747–761. [Google Scholar] [CrossRef]
- Griffith, M.; Griffith, O.L.; Coffman, A.C.; Weible, J.V.; McMichael, J.F.; Spies, N.C.; Koval, J.; Das, I.; Callaway, M.B.; Eldred, J.M.; et al. DGIdb: Mining the druggable genome. Nat. Methods 2013, 10, 1209–1210. [Google Scholar] [CrossRef] [PubMed]
| GEO Accession | Platform | Sample Size | Country | |
|---|---|---|---|---|
| Control | Disease | |||
| GSE20141 | GPL570 | 8 | 10 | USA |
| GSE49036 | GPL570 | 8 | 20 | Netherlands |
| GSE20164 | GPL96 | 5 | 6 | USA |
| GSE20292 | GPL96 | 18 | 11 | USA |
| GSE20163 | GPL96 | 9 | 8 | USA |
| GSE8397 | GPL96 | 15 | 24 | United Kingdom |
| Total | 63 | 79 | ||
| Lipinski Rule | Surface Area | H-Bond Donor (HBD) | H-Bond Acceptor (HBA) | LogP | Molecular Weight | Compounds | |
|---|---|---|---|---|---|---|---|
| Violation | Follow | ||||||
| 1 | 3 | 221.175 | 2 | 7 | 4.153 | 529.18 | Nilotinib |
| 1 | 3 | 259.451 | 3 | 6 | 3.1928 | 654.606 | Bromocriptine |
| 0 | 4 | 201.317 | 2 | 6 | 3.3529 | 470.6 | Withaferin A |
| 1 | 3 | 197.082 | 2 | 3 | 6.6977 | 450.61 | Celastrol |
| 0 | 4 | 167.005 | 0 | 4 | 4.3611 | 379.5 | Donepezil |
| Toxicity | Excretion | Metabolism | Distribution | Absorption | Compounds | ||||
|---|---|---|---|---|---|---|---|---|---|
| LD50 (mole/kg) | LC50 (log mM) | AMES | TC | CYP3A4 Inhibitor | CNS LogPS | BBB (LogBB) | P-gpI | HIA (%) | |
| (Permeability) | |||||||||
| 2.489 | 1.301 | No | 0.406 | Yes | −2.052 | −0.684 | Yes | 99.538 | Nilotinib |
| 3.739 | 2.448 | No | 0.327 | Yes | −2.601 | −0.711 | Yes | 71.357 | Bromocriptine |
| 2.779 | 0.738 | No | 0.435 | No | −2.72 | −0.03 | Yes | 85.345 | Withaferin A |
| 2.362 | −0.642 | No | −0.094 | Yes | −1.278 | 0.078 | No | 100 | Celastrol |
| 2.753 | −2.011 | No | 0.987 | Yes | −1.464 | 0.157 | Yes | 93.707 | Donepezil |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pappu, M.A.A.; Alamin, M.; Noman, M.A.; Sultana, M.H.; Ahmed, M.F.; Hossain, M.S.; Latif, M.A.; Faysal, M.F.; Azad, A.; Alyami, S.A.; et al. Multi-Transcriptome-Informed Network Pharmacology Reveals Novel Biomarkers and Therapeutic Candidates for Parkinson’s Disease. Genes 2025, 16, 1459. https://doi.org/10.3390/genes16121459
Pappu MAA, Alamin M, Noman MA, Sultana MH, Ahmed MF, Hossain MS, Latif MA, Faysal MF, Azad A, Alyami SA, et al. Multi-Transcriptome-Informed Network Pharmacology Reveals Novel Biomarkers and Therapeutic Candidates for Parkinson’s Disease. Genes. 2025; 16(12):1459. https://doi.org/10.3390/genes16121459
Chicago/Turabian StylePappu, Md. Al Amin, Md. Alamin, Md Al Noman, Most. Humaira Sultana, Md. Foysal Ahmed, Md. Sanoar Hossain, Md. Abdul Latif, Md. Fahim Faysal, AKM Azad, Salem A. Alyami, and et al. 2025. "Multi-Transcriptome-Informed Network Pharmacology Reveals Novel Biomarkers and Therapeutic Candidates for Parkinson’s Disease" Genes 16, no. 12: 1459. https://doi.org/10.3390/genes16121459
APA StylePappu, M. A. A., Alamin, M., Noman, M. A., Sultana, M. H., Ahmed, M. F., Hossain, M. S., Latif, M. A., Faysal, M. F., Azad, A., Alyami, S. A., Alotaibi, N., & Mollah, M. N. H. (2025). Multi-Transcriptome-Informed Network Pharmacology Reveals Novel Biomarkers and Therapeutic Candidates for Parkinson’s Disease. Genes, 16(12), 1459. https://doi.org/10.3390/genes16121459







