Breeding Selection for U.S. Siberian Huskies Has Altered Genes Regulating Metabolism, Endurance, Development, Body Conformation, Immune Function, and Behavior
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Genotyping
2.2. Principal Component Analysis (PCA)
2.3. Signatures of Selection—Marker-Based FST
2.4. Signatures of Selection—Runs of Homozygosity (ROH)
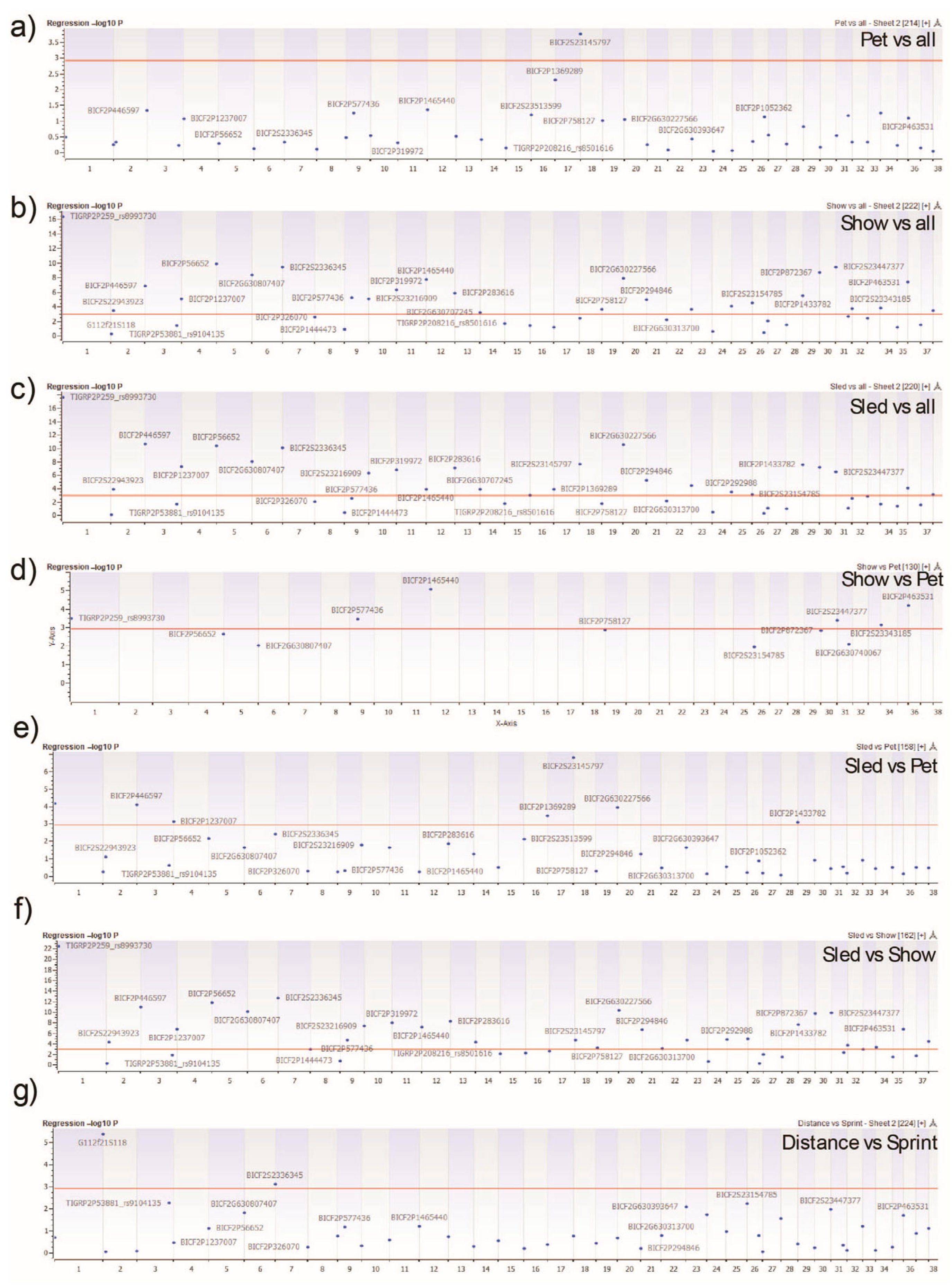
2.5. Genome-Wide Homozygosity Association Analysis
2.6. Gene Annotation and Pathway Enrichment Analysis
3. Results
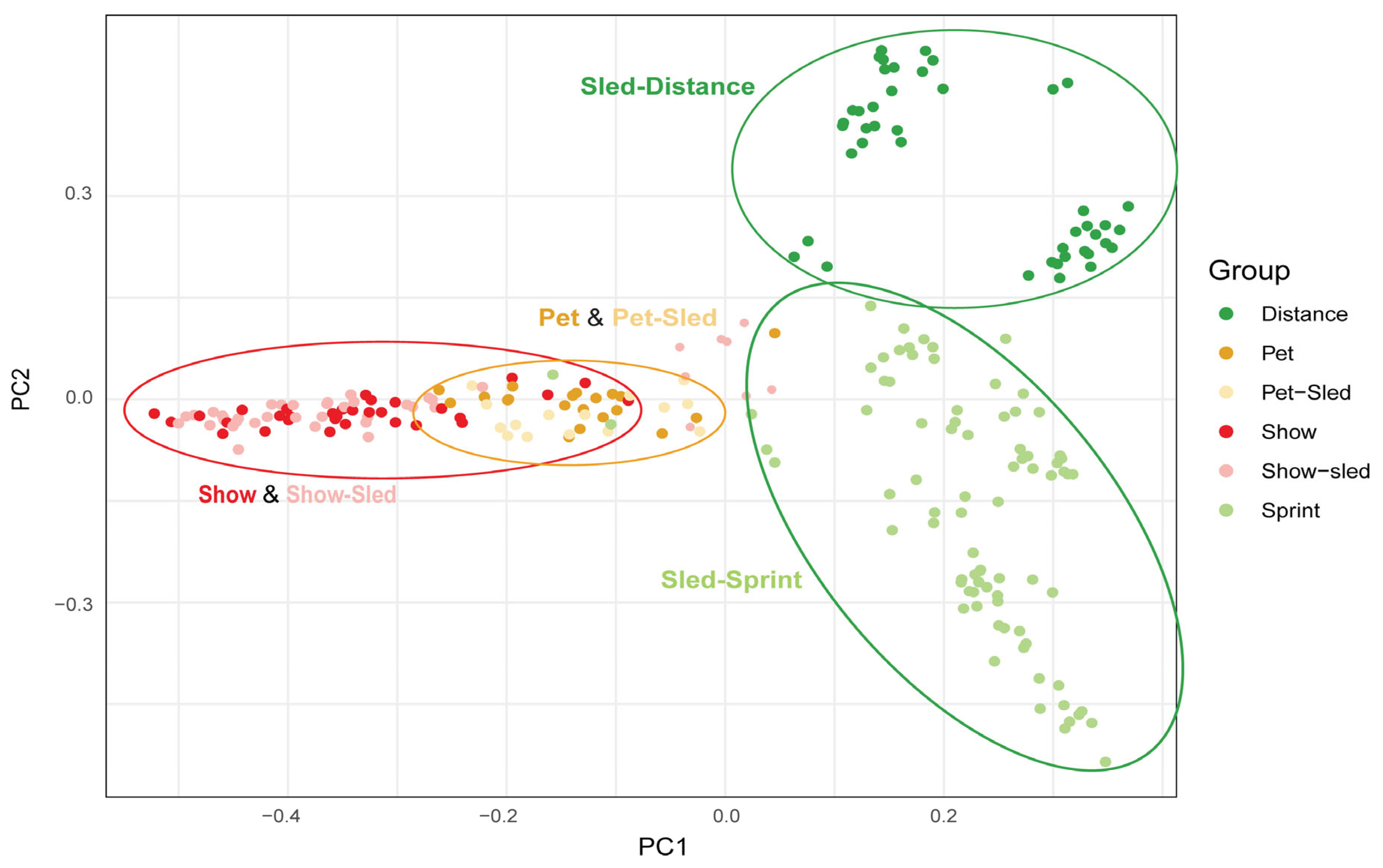
3.1. Genetic Population Structure
3.2. Morphological Variation
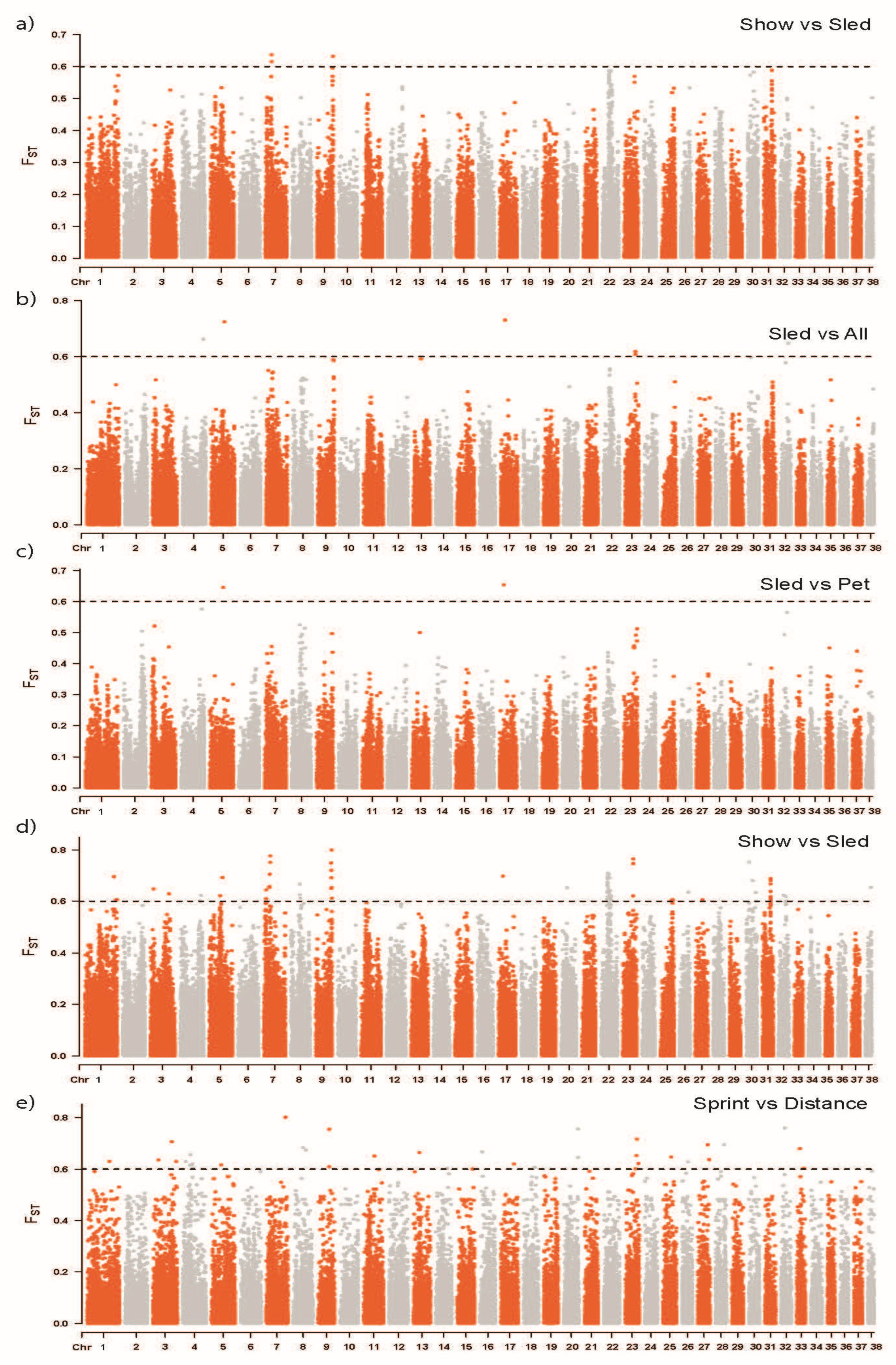
3.3. Marker-Based FST
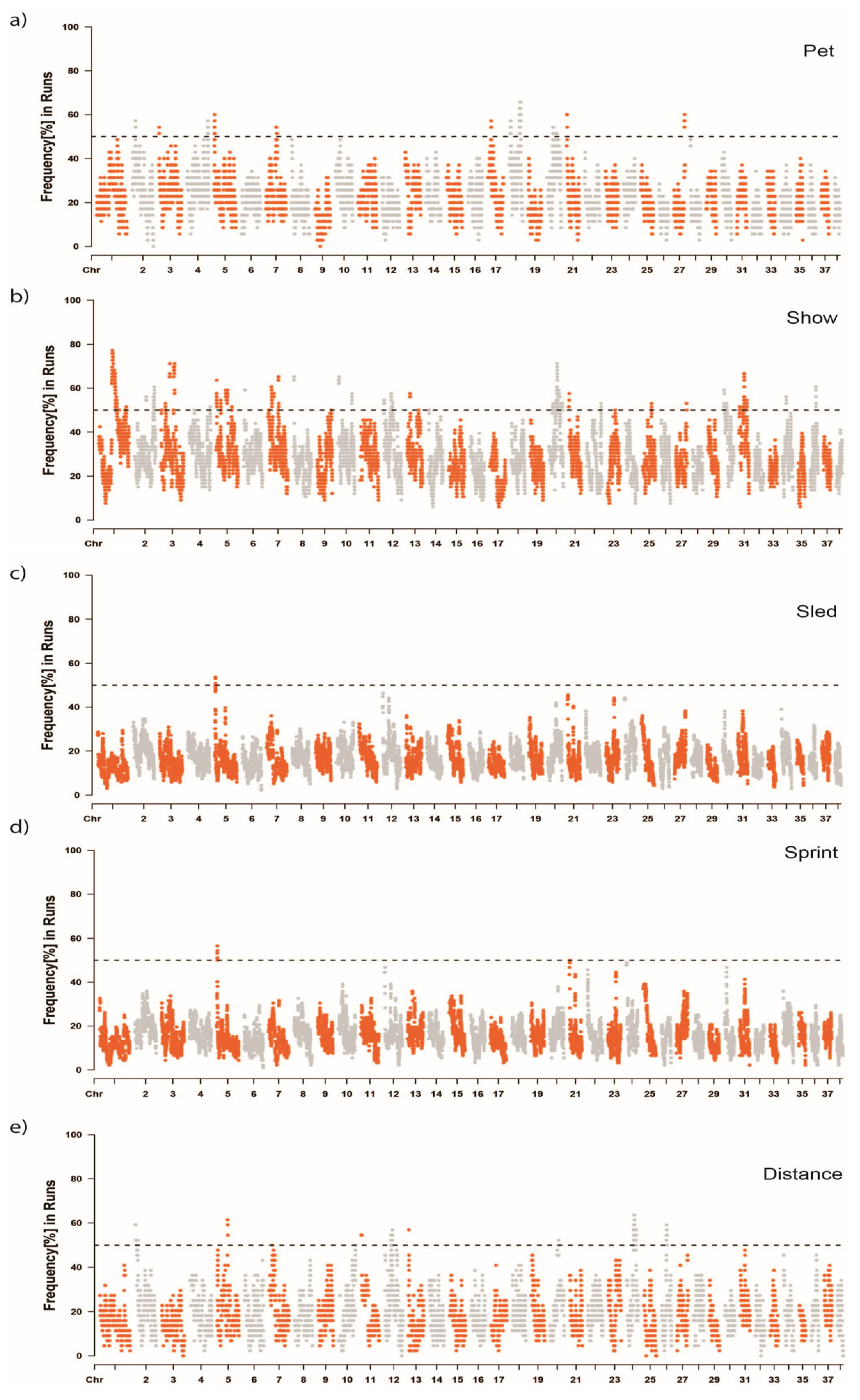
3.4. Runs of Homozygosity
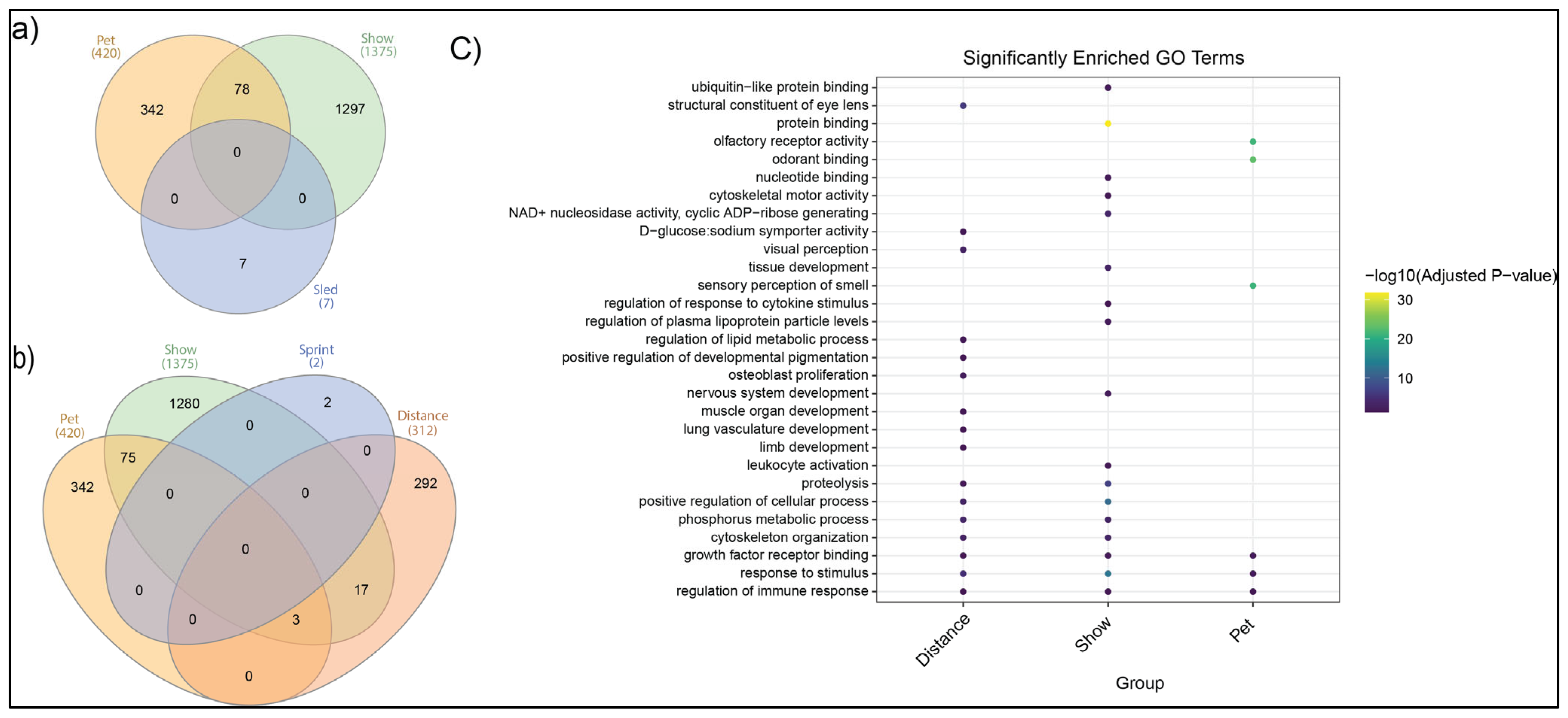
3.5. Gene Ontology and Pathway Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AKC | American Kennel Club |
| ROH | Runs of homozygosity |
| SHCA | Siberian Husky Club of America |
| SNP | Single-nucleotide polymorphism |
References
- Thomas, B.P. Leonhard Seppala; Pictorial Histories Publishing Company: Missoula, MT, USA, 2015. [Google Scholar]
- SHCA. Siberian Husky Club of America, Inc. Working Program. Available online: https://www.shca.org/specialty-sled-dog-class (accessed on 23 September 2025).
- Smith, T.A.; Srikanth, K.; Huson, H.J. Comparative Population Genomics of Arctic Sled Dogs Reveals a Deep and Complex History. Genome Biol. Evol. 2024, 16, evae190. [Google Scholar] [CrossRef]
- AKC. American Kennel Club-AKC.org. 2011. Available online: https://www.akc.org/dog-breeds/siberian-husky/ (accessed on 30 September 2025).
- Iditarod. Iditarod, the Last Great Race. 2025. Available online: https://iditarod.com/ (accessed on 30 September 2025).
- ISDRA; International Sled Dog Racing Association. 2011. Available online: http://www.isdra.org/ (accessed on 30 September 2025).
- Parker, H.G.; Dreger, D.L.; Rimbault, M.; Davis, B.W.; Mullen, A.B.; Carpintero-Ramirez, G.; Ostrander, E.A. Genomic Analyses Reveal the Influence of Geographic Origin, Migration, and Hybridization on Modern Dog Breed Development. Cell Rep. 2017, 19, 697–708. [Google Scholar] [CrossRef]
- Parker, H.G.; Ostrander, E.A. Canine genomics and genetics: Running with the pack. PLoS Genet. 2005, 1, e58. [Google Scholar] [CrossRef] [PubMed]
- Sutter, N.B.; Mosher, D.S.; Gray, M.M.; Ostrander, E.A. Morphometrics within dog breeds are highly reproducible and dispute Rensch’s rule. Mamm. Genome 2008, 19, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, K.; von Pfeil, D.J.; Stanley, B.J.; Griffitts, C.; Huson, H.J. Genome wide association study with imputed whole genome sequence data identifies a 431 kb risk haplotype on CFA18 for congenital laryngeal paralysis in Alaskan sled dogs. Genes 2022, 13, 1808. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Hoeppner, M.P.; Lundquist, A.; Pirun, M.; Meadows, J.R.; Zamani, N.; Johnson, J.; Sundström, G.; Cook, A.; FitzGerald, M.G.; Swofford, R. An improved canine genome and a comprehensive catalogue of coding genes and non-coding transcripts. PLoS ONE 2014, 9, e91172. [Google Scholar] [CrossRef] [PubMed]
- Marras, G.; Gaspa, G.; Sorbolini, S.; Dimauro, C.; Ajmone-Marsan, P.; Valentini, A.; Williams, J.L.; Macciotta, N.P. Analysis of runs of homozygosity and their relationship with inbreeding in five cattle breeds farmed in Italy. Anim. Genet. 2015, 46, 110–121. [Google Scholar] [CrossRef]
- Biscarini, F.; Cozzi, P.; Gaspa, G.; Marras, G. Detect Runs of Homozygosity and Runs of Heterozygosity in Diploid Genomes. 2019. Available online: https://cran.r-project.org/web/packages/detectRUNS/index.html (accessed on 2 September 2025).
- Gorssen, W.; Meyermans, R.; Janssens, S.; Buys, N. A publicly available repository of ROH islands reveals signatures of selection in different livestock and pet species. Genet. Sel. Evol. 2021, 53, 2. [Google Scholar] [CrossRef] [PubMed]
- Purfield, D.C.; Berry, D.P.; McParland, S.; Bradley, D.G. Runs of homozygosity and population history in cattle. BMC Genet. 2012, 13, 70. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X. rMVP: A memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef]
- Huson, H.J.; Sonstegard, T.S.; Godfrey, J.; Hambrook, D.; Wolfe, C.; Wiggans, G.; Blackburn, H.; VanTassell, C.P. A genetic investigation of Island Jersey cattle, the foundation of the Jersey breed: Comparing population structure and selection to Guernsey, Holstein, and United States Jersey cattle. Front. Genet. 2020, 11, 366. [Google Scholar] [CrossRef]
- Lencz, T.; Lambert, C.; DeRosse, P.; Burdick, K.E.; Morgan, T.V.; Kane, J.M.; Kucherlapati, R.; Malhotra, A.K. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc. Natl. Acad. Sci. USA 2007, 104, 19942–19947. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Lampi, S.; Donner, J.; Anderson, H.; Pohjoismaki, J. Variation in breeding practices and geographic isolation drive subpopulation differentiation, contributing to the loss of genetic diversity within dog breed lineages. Canine Med. Genet. 2020, 7, 5. [Google Scholar] [CrossRef]
- Pohjoismaki, J.L.O.; Lampi, S.; Donner, J.; Anderson, H. Origins and wanderings of the Finnish hunting spitzes. PLoS ONE 2018, 13, e0199992. [Google Scholar] [CrossRef] [PubMed]
- Schoenebeck, J.J.; Ostrander, E.A. The genetics of canine skull shape variation. Genetics 2013, 193, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Zhou, X.; Cao, Z.; Pan, J. Latest Findings on the Role of RUNX1 in Bone Development and Disorders. Sichuan Da Xue Xue Bao Yi Xue Ban 2024, 55, 256–262. [Google Scholar] [PubMed]
- Carroll, S.H.; Schafer, S.; Kawasaki, K.; Tsimbal, C.; Jule, A.M.; Hallett, S.A.; Li, E.; Liao, E.C. Genetic requirement of dact1/2 to regulate noncanonical Wnt signaling and calpain 8 during embryonic convergent extension and craniofacial morphogenesis. Elife 2024, 13, RP91648. [Google Scholar] [CrossRef]
- UniProt. UniProt Consortium EMBL-EBI: Creative Commons Attribution 4.0 International; UniProt: Hinxton, UK, 2025. [Google Scholar]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- NCBI. National Center for Biotechnology Information. 2011. Available online: http://www.ncbi.nlm.nih.gov/ (accessed on 26 September 2025).
- Leung, A.W.Y.; Backstrom, I.; Bally, M.B. Sulfonation, an underexploited area: From skeletal development to infectious diseases and cancer. Oncotarget 2016, 7, 55811–55827. [Google Scholar] [CrossRef]
- D’Acquisto, F.; Perretti, M.; Flower, R.J. Annexin-A1: A pivotal regulator of the innate and adaptive immune systems. Brit J. Pharmacol. 2008, 155, 152–169. [Google Scholar] [CrossRef]
- Nadarajan, S.; Azmi, N.; Jasamai, M.; Kumolosasi, E. Annexin A1 (ANXA1): A Systematic Review of Its Role in Inflammation. Sains Malays. 2021, 50, 207–226. [Google Scholar] [CrossRef]
- Park, S.M.; Roache, C.E.; Ii, P.H.I.; Moldenhauer, H.J.; Matychak, K.K.; Plante, A.E.; Lieberman, A.G.; Crino, P.B.; Meredith, A. BK channel properties correlate with neurobehavioral severity in three-linked channelopathy mouse models. Elife 2022, 11, e77953. [Google Scholar] [CrossRef]
- Sutter, N.B.; Bustamante, C.D.; Chase, K.; Gray, M.M.; Zhao, K.; Zhu, L.; Padhukasahasram, B.; Karlins, E.; Davis, S.; Jones, P.G.; et al. A single IGF1 allele is a major determinant of small size in dogs. Science 2007, 316, 112–115. [Google Scholar] [CrossRef]
- Deane-Coe, P.E.; Chu, E.T.; Slavney, A.; Boyko, A.R.; Sams, A.J. Direct-to-consumer DNA testing of 6,000 dogs reveals 98.6-kb duplication associated with blue eyes and heterochromia in Siberian Huskies. PLoS Genet. 2018, 14, e1007648. [Google Scholar] [CrossRef] [PubMed]
- vonHoldt, B.M.; Shuldiner, E.; Koch, I.J.; Kartzinel, R.Y.; Hogan, A.; Brubaker, L.; Wanser, S.; Stahler, D.; Wynne, C.D.L.; Ostrander, E.A.; et al. Structural variants in genes associated with human Williams-Beuren syndrome underlie stereotypical hypersociability in domestic dogs. Sci. Adv. 2017, 3, e1700398. [Google Scholar] [CrossRef] [PubMed]
- Plassais, J.; Kim, J.; Davis, B.W.; Karyadi, D.M.; Hogan, A.N.; Harris, A.C.; Decker, B.; Parker, H.G.; Ostrander, E.A. Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat. Commun. 2019, 10, 1489. [Google Scholar] [CrossRef] [PubMed]
- Stattin, E.L.; Lindblom, K.; Struglics, A.; Önnerfjord, P.; Goldblatt, J.; Dixit, A.; Sarkar, A.; Randell, T.; Suri, M.; Raggio, C.; et al. Novel missense gene variants linked to familial osteochondritis dissecans cluster in the C-terminal globular domain of aggrecan. Sci. Rep. 2022, 12, 5215. [Google Scholar] [CrossRef]
- Drackley, A.; Somerville, C.; Arnaud, P.; Baudhuin, L.M.; Hanna, N.; Kluge, M.L.; Kotzer, K.; Boileau, C.; Bronicki, L.; Callewaert, B.; et al. Interpretation and classification of variants associated with Marfan syndrome: Consensus recommendations from the Clinical Genome Resource’s variant curation expert panel. Genome Med. 2024, 16, 154. [Google Scholar] [CrossRef]
- Kosho, T.; Mizumoto, S.; Watanabe, T.; Yoshizawa, T.; Miyake, N.; Yamada, S. Recent Advances in the Pathophysiology of Musculocontractural Ehlers-Danlos Syndrome. Genes 2020, 11, 43. [Google Scholar] [CrossRef]
- Kim, J.; Williams, F.J.; Dreger, D.L.; Plassais, J.; Davis, B.W.; Parker, H.G.; Ostrander, E.A. Genetic selection of athletic success in sport-hunting dogs. Proc. Natl. Acad. Sci. USA 2018, 115, E7212–E7221. [Google Scholar] [CrossRef]
- Kokocinska-Kusiak, A.; Woszczylo, M.; Zybala, M.; Maciocha, J.; Barlowska, K.; Dzieciol, M. Canine Olfaction: Physiology, Behavior, and Possibilities for Practical Applications. Animals 2021, 11, 2463. [Google Scholar] [CrossRef]
- Johnson, E.C.; Evans, L.M.; Keller, M.C. Relationships between estimated autozygosity and complex traits in the UK Biobank. PLoS Genet. 2018, 14, e1007556. [Google Scholar] [CrossRef]


| Trait (Measured in cm) | Pet | Show | Sled |
|---|---|---|---|
| Eye Width * | 4.45 ± 0.53 a | 4.42 ± 0.64 a | 4.11 ± 0.81 b |
| Snout Length | 9.45 ± 0.97 a | 9.19 ± 0.91 a | 9.42 ± 1.17 a |
| Outside Ear Length | 8.79 ± 1.30 a | 8.43 ± 0.89 a | 8.38 ± 1.22 a |
| Chest Width * | 13.94 ± 1.98 a | 12.29 ± 1.40 b | 12.65 ± 1.35 b |
| Neck Girth | 35.99 ± 3.81 a | 37.41 ± 3.86 a | 36.42 ± 3.81 a |
| Chest Girth * | 67.23 ± 6.78 a | 62.56 ± 4.55 b | 64.85 ± 4.47 ab |
| Wither Height * | 56.92 ± 3.84 a | 56.85 ± 3.66 a | 60.22 ± 3.43 b |
| Height at Base of Tail * | 55.32 ± 4.01 a | 54.03 ± 4.04 a | 58.37 ± 5.97 b |
| Body Length * | 61.85 ± 6.68 a | 54.69 ± 4.52 b | 57.68 ± 4.34 c |
| Tail Length | 35.71 ± 4.67 a | 35.84 ± 2.87 a | 34.42 ± 4.42 a |
| Upper Foreleg Length * | 20.04 ± 2.41 a | 19.10 ± 1.91 a | 17.83 ± 2.64 b |
| Lower Foreleg Length | 21.41 ± 2.03 a | 20.93 ± 1.55 a | 21.29 ± 1.83 a |
| Fore Foot Length * | 13.56 ± 1.60 a | 12.50 ± 1.55 b | 12.83 ± 1.50 ab |
| Fore Foot Circumference | 9.91 ± 0.86 a | 10.24 ± 1.24 a | 10.13 ± 0.86 a |
| Upper Hind Leg Length * | 30.51 ± 3.33 a | 27.30 ± 2.36 b | 30.15 ± 3.38 a |
| Lower Hind Leg Length * | 24.21 ± 2.92 a | 23.60 ± 2.24 a | 22.00 ± 3.20 b |
| Hind Foot Length * | 21.23 ± 2.01 a | 20.80 ± 1.60 ab | 20.24 ± 2.24 b |
| Hind Foot Circumference | 9.47 ± 0.86 a | 9.63 ± 1.04 a | 9.68 ± 0.86 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huson, H.J.; Srikanth, K.; Ellis, K.M. Breeding Selection for U.S. Siberian Huskies Has Altered Genes Regulating Metabolism, Endurance, Development, Body Conformation, Immune Function, and Behavior. Genes 2025, 16, 1355. https://doi.org/10.3390/genes16111355
Huson HJ, Srikanth K, Ellis KM. Breeding Selection for U.S. Siberian Huskies Has Altered Genes Regulating Metabolism, Endurance, Development, Body Conformation, Immune Function, and Behavior. Genes. 2025; 16(11):1355. https://doi.org/10.3390/genes16111355
Chicago/Turabian StyleHuson, Heather J., Krishnamoorthy Srikanth, and Karolynn M. Ellis. 2025. "Breeding Selection for U.S. Siberian Huskies Has Altered Genes Regulating Metabolism, Endurance, Development, Body Conformation, Immune Function, and Behavior" Genes 16, no. 11: 1355. https://doi.org/10.3390/genes16111355
APA StyleHuson, H. J., Srikanth, K., & Ellis, K. M. (2025). Breeding Selection for U.S. Siberian Huskies Has Altered Genes Regulating Metabolism, Endurance, Development, Body Conformation, Immune Function, and Behavior. Genes, 16(11), 1355. https://doi.org/10.3390/genes16111355






