Cost-Effective Method for Using Cross-Species Spike-In RNA for Normalization and Quantification in Polysome Profiling Experiments
Abstract
1. Introduction
2. Materials and Methods
- Minimizing RNA degradation during experimentation
- Cell culture and siRNA treatment
- Hypertonic stress and cell lysate preparation
- Sucrose gradient preparation and fractionation
- Preparation of spike-in RNA
- Addition of Spike-in RNA, RNA isolation, cDNA preparation, and RT-qPCR
- Polysome profiling data analysis
- Statistical analysis
3. Results
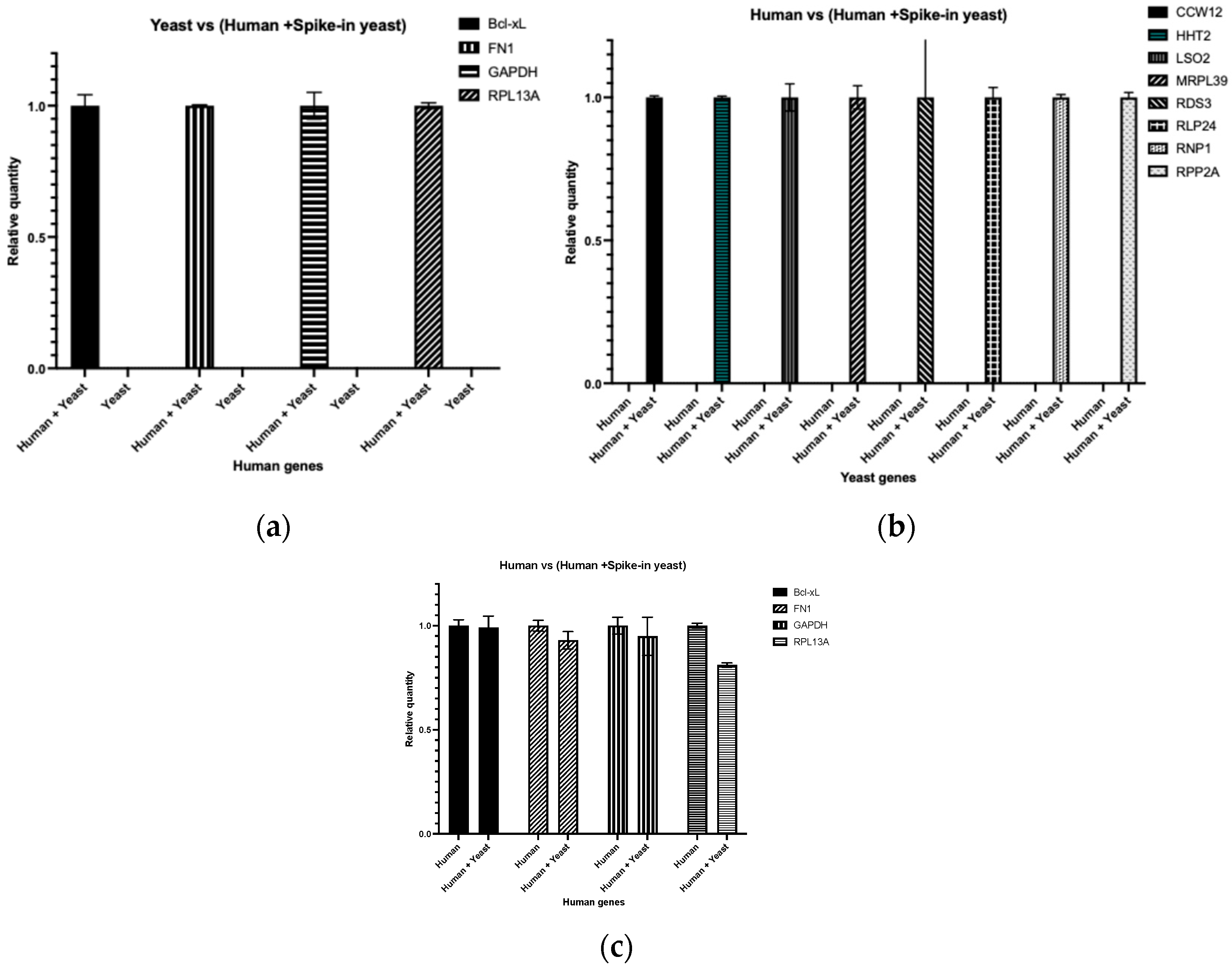
3.1. Cross-Species Spike-In RNA Does Not Interfere with Target mRNA Quantification
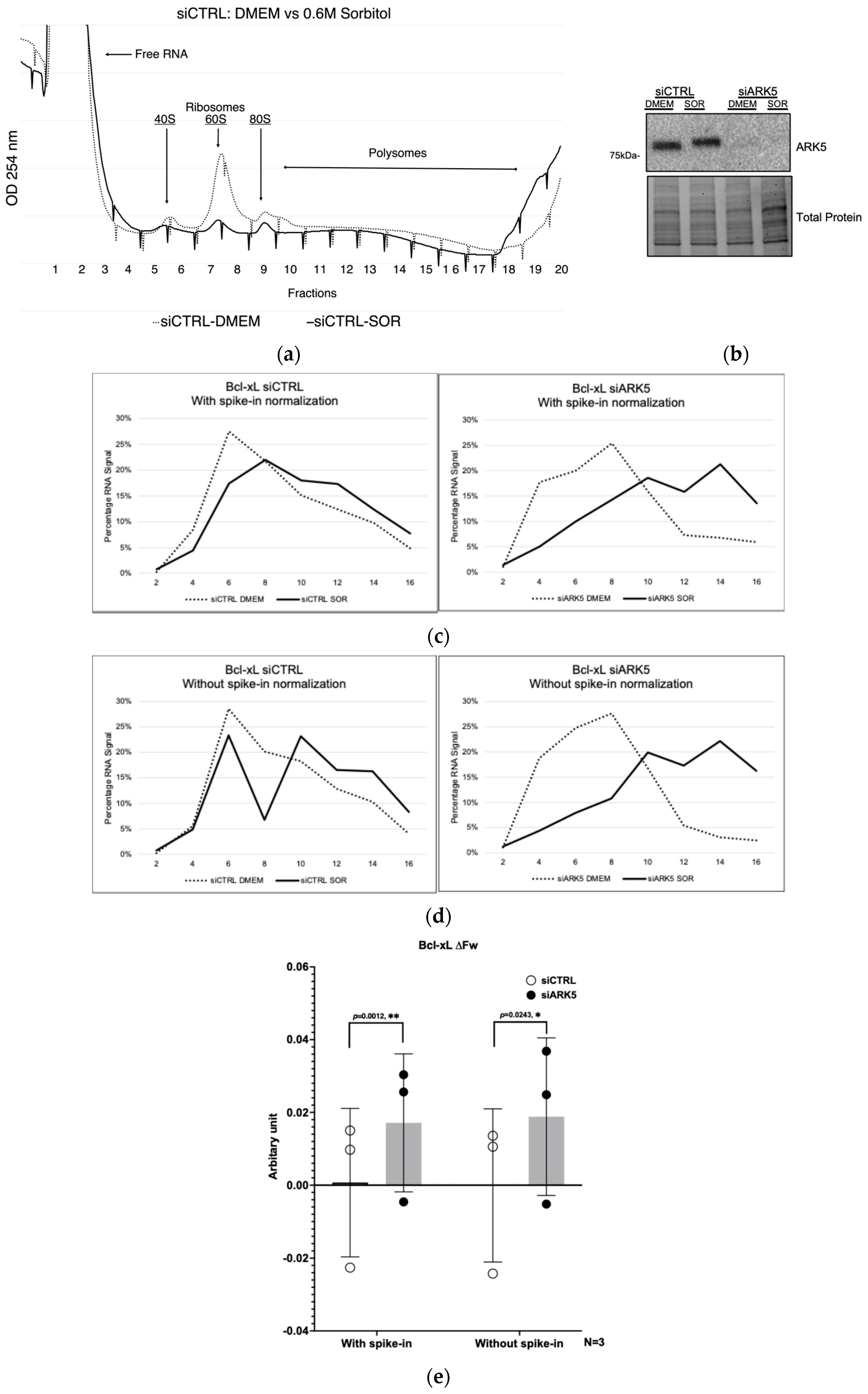
3.2. Spike-In RNA Reveals Inconsistencies in RNA Isolation from Polysome Fractions
3.3. Hypertonic Stress Increases the Translation Efficiency of Bcl-xL in siARK5 Treated Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| DEPC | Diethylpyrocarbonate |
| DTT | Dithiothreitol |
| HEPES | 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid |
| MgCl2 | Magnesium chloride |
| PBS | Phosphate-buffered saline |
| SDS | Sodium dodecyl sulfate |
| RNase | Ribonuclease |
| CAT | Chloramphenicol acetyltransferase |
| ERCC | External RNA Controls Consortium |
| RNA | Ribonucleic acid |
| mRNA | Messenger RNA |
| cDNA | Complementary DNA |
| PCR | Polymerase chain reaction |
| qPCR | Quantitative polymerase chain reaction |
| RT-qPCR | Reverse transcription quantitative polymerase chain reaction |
| RNA-Seq | RNA sequencing |
| Cq | Quantification cycle |
| ARK5 | AMP-activated protein kinase family member 5 |
| Bcl-xL | B-cell lymphoma-extra large |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| FN1 | Fibronectin 1 |
| RPL13A | Ribosomal protein L13a |
| siRNA | Small interfering RNA |
| U2OS | Human osteosarcoma cell line |
Appendix A
| Gene (Species) | Catalogue Number/ Target Sequence: 5′-3′ (Use) | Company |
|---|---|---|
| siCTRL | 1027281 (siRNA) | Qiagen (Toronto, ON, CA) |
| siARK5 | J-004931-12-0020 (siRNA) | Horizon (Cambridge, UK) |
| RPL13A (Human) | QT00000721 (RT-qPCR) | Qiagen (Toronto, ON, CA) |
| GAPDH (Human) | QT01192646 (RT-qPCR) | Qiagen (Toronto, ON, CA) |
| Bcl-xL/BCL2L1 (Human) | QT00236712 (RT-qPCR) | Qiagen (Toronto, ON, CA) |
| FN1 (Human) | QT00038024 (RT-qPCR) | Qiagen (Toronto, ON, CA) |
| CCW12 (Yeast) | GTTTCCACCGCTACCGTCACCG(Forward) GGGGCTTCAGTGGTCAATGGGC(Reverse) (RT-qPCR) | IDT (Coralville, IA, USA) |
| HHT2 (Yeast) | AAGGCTGCCAGAAAATCCGCCC(Forward) TCT CAA GGCAACAGTACCTGGCT(Reverse) (RT-qPCR) | IDT (Coralville, IA, USA) |
| LSO2 (Yeast) | ATCCGCCGCAAAGAAAGCAGCT(Forward) TCCTCCGCTTCTAGCTGCTCCA(Reverse) (RT-qPCR) | IDT (Coralville, IA, USA) |
| MRPL39 (Yeast) | ACAGCGGCAAGTGGATACTCTCGT(Forward) GCACATGCCGTTTCACCACAGG(Reverse) (RT-qPCR) | IDT (Coralville, IA, USA) |
| RDS3 (Yeast) | GCAGCCTGGTGTACAGACAGGT(Forward) AAGCGGCAGCACTCCCAACAAT(Reverse) (RT-qPCR) | IDT (Coralville, IA, USA) |
| RLP24 (Yeast) | TGCCGTTCTAAGTGTCACAAGGCA(Forward) TCCGGCAGCCTTTCTGAAAGCT(Reverse) (RT-qPCR) | IDT (Coralville, IA, USA) |
| RNP1 (Yeast) | ACTGCTCCTGATCTCAAGCCACT(Forward) TTCGCCGGTGTGAAACTCGTCG(Reverse) (RT-qPCR) | IDT (Coralville, IA, USA) |
| RPP2A (Yeast) | GCCTCTGGTGATGCTGCTGCTG(Forward) ACCGAAACCCATGTCGTCGTCA(Reverse) (RT-qPCR) | IDT (Coralville, IA, USA) |
| ARK5 | 4458S (Primary antibody, Western blot) | Cell signaling (NEB, Whitby, ON, CA) |
| Anti-Rabbit | 7074S (Secondary antibody, Western blot) | Cell signaling (NEB, Whitby, ON, CA) |
References
- Crick, F. Central dogma of molecular biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Holcik, M.; Sonenberg, N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005, 6, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K. RNA therapy: Rich history, various applications and unlimited future prospects. Exp. Mol. Med. 2022, 54, 455–465. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhu, L.; Wang, X.; Jin, H. RNA-based therapeutics: An overview and prospectus. Cell Death Dis. 2022, 13, 644. [Google Scholar] [CrossRef]
- Vermeulen, J.; De Preter, K.; Lefever, S.; Nuytens, J.; De Vloed, F.; Derveaux, S.; Hellemans, J.; Speleman, F.; Vandesompele, J. Measurable impact of RNA quality on gene expression results from quantitative PCR. Nucleic Acids Res. 2011, 39, e63. [Google Scholar] [CrossRef]
- Luo, G.; Zhang, J.; Zhang, S.; Hu, B.; Hu, L.; Huang, Z. High-quality RT-PCR with chemically modified RNA controls. Talanta 2021, 224, 121850. [Google Scholar] [CrossRef]
- Jahn, C.E.; Charkowski, A.O.; Willis, D.K. Evaluation of isolation methods and RNA integrity for bacterial RNA quantitation. J. Microbiol. Methods 2008, 75, 318–324. [Google Scholar] [CrossRef]
- Rodríguez, A.; Vaneechoutte, M. Comparison of the efficiency of different cell lysis methods and different commercial methods for RNA extraction from Candida albicans stored in RNAlater. BMC Microbiol. 2019, 19, 223. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Young, S.; Russo, V.; Amsden, B.G.; Flynn, L.E. Techniques for the isolation of high-quality RNA from cells encapsulated in chitosan hydrogels. Tissue Eng. Part C Methods 2013, 19, 829–838. [Google Scholar] [CrossRef]
- Kuang, J.; Yan, X.; Genders, A.J.; Granata, C.; Bishop, D.J. An overview of technical considerations when using quantitative real-time PCR analysis of gene expression in human exercise research. PLoS ONE 2018, 13, e0196438. [Google Scholar] [CrossRef]
- Faye, M.D.; Graber, T.E.; Holcik, M. Assessment of selective mRNA translation in mammalian cells by polysome profiling. J. Vis. Exp. 2014, 92, e52295. [Google Scholar] [CrossRef]
- Riley, A.; Jordan, L.E.; Holcik, M. Distinct 5′ UTRs regulate XIAP expression under normal growth conditions and during cellular stress. Nucleic Acids Res. 2010, 38, 4665–4674. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pine, P.S.; Munro, S.A.; Parsons, J.R.; McDaniel, J.; Lucas, A.B.; Lozach, J.; Myers, T.G.; Su, Q.; Jacobs-Helber, S.M.; Salit, M. Evaluation of the External RNA Controls Consortium (ERCC) reference material using a modified Latin square design. BMC Biotechnol. 2016, 16, 54. [Google Scholar] [CrossRef]
- Jiang, L.; Schlesinger, F.; Davis, C.A.; Zhang, Y.; Li, R.; Salit, M.; Gingeras, T.R.; Oliver, B. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 2011, 21, 1543–1551. [Google Scholar] [CrossRef]
- Bhattarai, K.; Richard, T.; Fatica, T.; Frangione, B.; Willmore, W.G.; Holcik, M. AMPK-related protein kinase ARK5 regulates subcellular localization of RNA-binding protein hnRNP A1 during hypertonic stress. J. Biol. Chem. 2022, 298, 102364. [Google Scholar] [CrossRef]
- Liang, S.; Bellato, H.M.; Lorent, J.; Lupinacci, F.C.S.; Oertlin, C.; van Hoef, V.; Andrade, V.P.; Roffé, M.; Masvidal, L.; Hajj, G.N.M.; et al. Polysome-profiling in small tissue samples. Nucleic Acids Res. 2018, 46, e3. [Google Scholar] [CrossRef]
- Chiluiza, D.; Bargo, S.; Callahan, R.; Rhoads, R.E. Expression of truncated eukaryotic initiation factor 3e (EIF3e) resulting from integration of mouse mammary tumor virus (MMTV) causes a shift from cap-dependent to cap-independent translation. J. Biol. Chem. 2011, 286, 31288–31296. [Google Scholar] [CrossRef]
- Mendelevich, A.; Gupta, S.; Pakharev, A.; Teodosiadis, A.; Mironov, A.A.; Gimelbrant, A.A. Foreign RNA spike-ins enable accurate allele-specific expression analysis at scale. Bioinformatics 2023, 39, i431–i439. [Google Scholar] [CrossRef] [PubMed]
- Spies, N.; Burge, C.B.; Bartel, D.P. 3′ UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res. 2013, 23, 2078–2090. [Google Scholar] [CrossRef]
- Botstein, D.; Chervitz, S.A.; Cherry, M. Yeast as a model organism. Science 1997, 277, 1259–1260. [Google Scholar] [CrossRef] [PubMed]
- Wheelan, S.J.; Boguski, M.S.; Duret, L.; Makałowski, W. Human and nematode orthologs—lessons from the analysis of 1800 human genes and the proteome of Caenorhabditis elegans. Gene 1999, 238, 163–170. [Google Scholar] [CrossRef]
- Mirzoyan, Z.; Sollazzo, M.; Allocca, M.; Valenza, A.M.; Grifoni, D.; Bellosta, P. Drosophila melanogaster: A model organism to study cancer. Front. Genet. 2019, 10, 51. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Hahn, Y.; Park, S.R.; Chung, S.K.; Jeong, S.; Yang, I. Normalization of human RNA-seq experiments using chimpanzee RNA as a spike-in standard. Sci. Rep. 2016, 6, 31923. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Hu, Z.; Xia, Z.; Zhao, D.; Li, W.; Tyler, J.K. The overlooked fact: Fundamental need for spike-in control for virtually all genome-wide analyses. Mol. Cell. Biol. 2016, 36, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Purification of total RNA from mammalian cells and tissues. Cold Spring Harb. Protoc. 2020, 2020, 101659. [Google Scholar] [CrossRef]
- Ridgeway, J.A.; Timm, A.E. Comparison of RNA isolation methods from insect larvae. J. Insect Sci. 2014, 14, 268. [Google Scholar] [CrossRef]
- Johnson, M.T.J.; Carpenter, E.J.; Tian, Z.; Bruskiewich, R.; Burris, J.N.; Carrigan, C.T.; Chase, M.W.; Clarke, N.D.; Covshoff, S.; dePamphilis, C.W.; et al. Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. PLoS ONE 2012, 7, e50226. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhattarai, K.; Slade, A.; Holcik, M. Cost-Effective Method for Using Cross-Species Spike-In RNA for Normalization and Quantification in Polysome Profiling Experiments. Genes 2025, 16, 1354. https://doi.org/10.3390/genes16111354
Bhattarai K, Slade A, Holcik M. Cost-Effective Method for Using Cross-Species Spike-In RNA for Normalization and Quantification in Polysome Profiling Experiments. Genes. 2025; 16(11):1354. https://doi.org/10.3390/genes16111354
Chicago/Turabian StyleBhattarai, Krishna, Angelo Slade, and Martin Holcik. 2025. "Cost-Effective Method for Using Cross-Species Spike-In RNA for Normalization and Quantification in Polysome Profiling Experiments" Genes 16, no. 11: 1354. https://doi.org/10.3390/genes16111354
APA StyleBhattarai, K., Slade, A., & Holcik, M. (2025). Cost-Effective Method for Using Cross-Species Spike-In RNA for Normalization and Quantification in Polysome Profiling Experiments. Genes, 16(11), 1354. https://doi.org/10.3390/genes16111354







