Genetic Insights into Familial Hypospadias Identifying Rare Variants and Their Potential Role in Urethral Development
Abstract
1. Introduction
2. Material and Methods
2.1. Ethical Approval
2.2. Study Participants
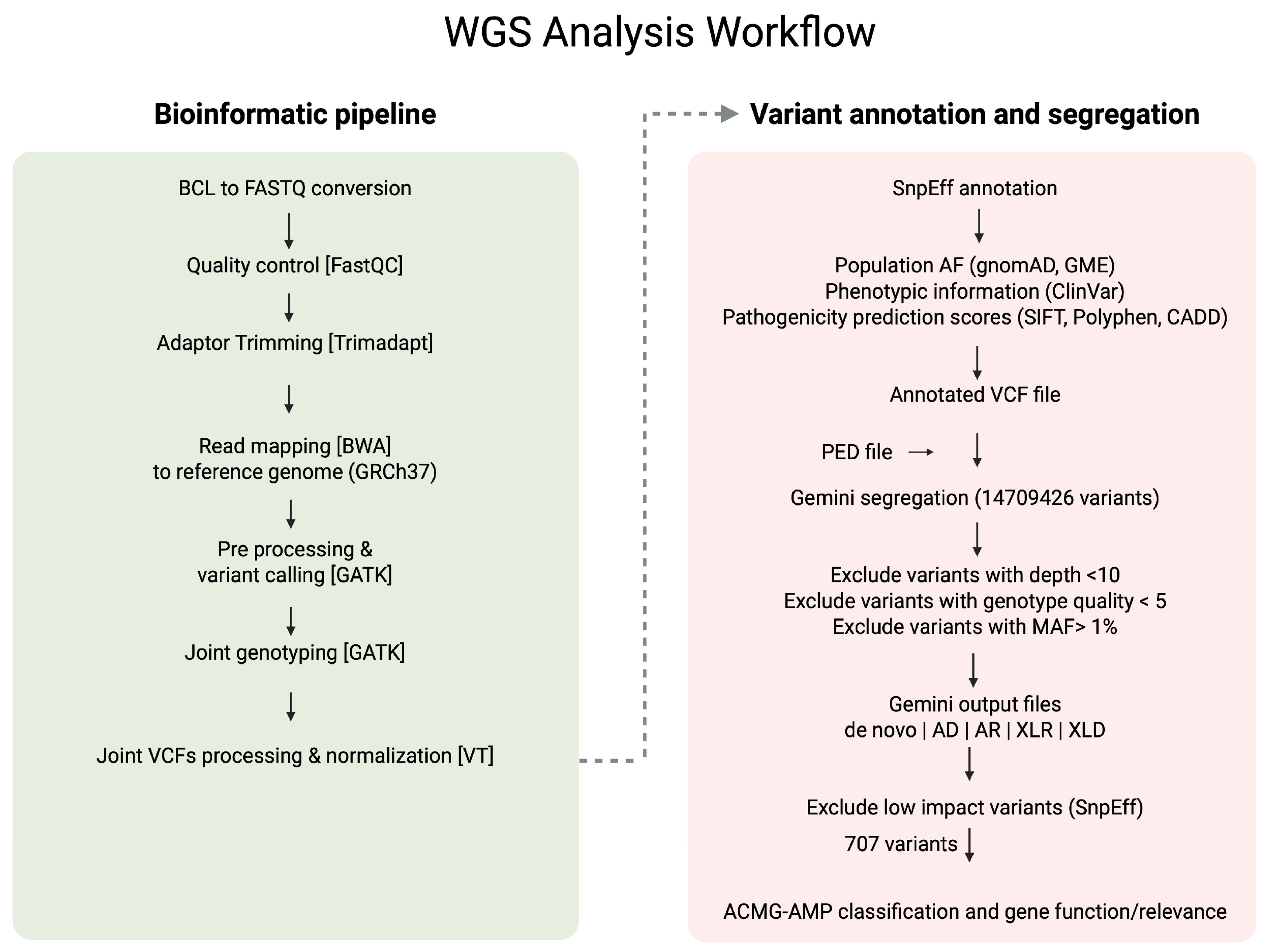
2.3. Whole Genome Sequencing (WGS) Data Generation and Processing
2.4. Single-Nucleotide Variant (SNV) Segregation Analysis and Filtration
3. Results
3.1. Demographic and Clinical Characteristics
3.2. Variants Detected from WGS Analysis
3.3. Pathogenic and Likely Pathogenic Gene Variants
3.4. Variants of Uncertain Significance (VUS)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yu, X.; Nassar, N.; Mastroiacovo, P.; Canfield, M.; Groisman, B.; Bermejo-Sánchez, E.; Ritvanen, A.; Kiuru-Kuhlefelt, S.; Benavides, A.; Sipek, A.; et al. Hypospadias Prevalence and Trends in International Birth Defect Surveillance Systems, 1980–2010. Eur. Urol. 2019, 76, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Stadler, H.S.; Peters, C.A.; Sturm, R.M.; Baker, L.A.; Best, C.J.M.; Bird, V.Y.; Geller, F.; Hoshizaki, D.K.; Knudsen, T.B.; Norton, J.M.; et al. Meeting report on the NIDDK/AUA Workshop on Congenital Anomalies of External Genitalia: Challenges and opportunities for translational research. J. Pediatr. Urol. 2020, 16, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Abbas, T. Management of Distal Hypospadias: New Insights and Stepwise Management Algorithm. In Hypospadiology: Current Challenges and Future Perspectives; Abbas, T., Ed.; Springer Nature: Singapore, 2023; pp. 67–80. [Google Scholar]
- Geller, F.; Feenstra, B.; Carstensen, L.; Pers, T.H.; van Rooij, I.A.; Körberg, I.B.; Choudhry, S.; Karjalainen, J.M.; Schnack, T.H.; Hollegaard, M.V.; et al. Genome-wide association analyses identify variants in developmental genes associated with hypospadias. Nat. Genet. 2014, 46, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Schnack, T.H.; Zdravkovic, S.; Myrup, C.; Westergaard, T.; Christensen, K.; Wohlfahrt, J.; Melbye, M. Familial aggregation of hypospadias: A cohort study. Am. J. Epidemiol. 2008, 167, 251–256. [Google Scholar] [CrossRef]
- Abbas, T.; Vallasciani, S. Toward an Ecosystem Model of Hypospadiology. In Hypospadiology: Current Challenges and Future Perspectives; Abbas, T., Ed.; Springer Nature: Singapore, 2023; pp. 1–23. [Google Scholar]
- Chen, Z.; Lei, Y.; Finnell, R.H.; Ding, Y.; Su, Z.; Wang, Y.; Xie, H.; Chen, F. Whole-exome sequencing study of hypospadias. iScience 2023, 26, 106663. [Google Scholar] [CrossRef]
- Matsushita, S.; Suzuki, K.; Murashima, A.; Kajioka, D.; Acebedo, A.R.; Miyagawa, S.; Haraguchi, R.; Ogino, Y.; Yamada, G. Regulation of masculinization: Androgen signalling for external genitalia development. Nat. Rev. Urol. 2018, 15, 358–368. [Google Scholar] [CrossRef]
- Chang, J.; Wang, S.; Zheng, Z. Etiology of Hypospadias: A Comparative Review of Genetic Factors and Developmental Processes Between Human and Animal Models. Res. Rep. Urol. 2020, 12, 673–686. [Google Scholar] [CrossRef]
- Abbas, T.O.; Al-Shafai, K.; Jamil, A.; Mancha, M.; Azzah, A.; Arar, S.; Kumar, S.; Al Massih, A.; Mackeh, R.; Tomei, S.; et al. Establishing a Structured Hypospadias Biobank Cohort for Integrated Research: Methodology, Comprehensive Database Integration, and Phenotyping. Diagnostics 2025, 15, 561. [Google Scholar] [CrossRef]
- Allouba, M.; Walsh, R.; Afify, A.; Hosny, M.; Halawa, S.; Galal, A.; Fathy, M.; Theotokis, P.I.; Boraey, A.; Ellithy, A.; et al. Ethnicity, consanguinity, and genetic architecture of hypertrophic cardiomyopathy. Eur. Heart J. 2023, 44, 5146–5158. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Tan, A.; Abecasis, G.R.; Kang, H.M. Unified representation of genetic variants. Bioinformatics 2015, 31, 2202–2204. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang le, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.M.; Halees, A.; Itan, Y.; Spencer, E.G.; He, Y.; Azab, M.A.; Gabriel, S.B.; Belkadi, A.; Boisson, B.; Abel, L.; et al. Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat. Genet. 2016, 48, 1071–1076. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef]
- Vaser, R.; Adusumalli, S.; Leng, S.N.; Sikic, M.; Ng, P.C. SIFT missense predictions for genomes. Nat. Protoc. 2016, 11, 1–9. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7–20. [Google Scholar] [CrossRef]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef]
- Paila, U.; Chapman, B.A.; Kirchner, R.; Quinlan, A.R. GEMINI: Integrative exploration of genetic variation and genome annotations. PLoS Comput. Biol. 2013, 9, e1003153. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. Off. J. Am. Coll. Med. Genet. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2018, 35, 1978–1980. [Google Scholar] [CrossRef] [PubMed]
- Ahmed Gunaid, A.; Ali Hummad, N.; Abdallah Tamim, K. Consanguineous Marriage in the Capital City Sana’a, Yemen. J. Biosoc. Sci. 2004, 36, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Hussain, R.; Bittles, A.H. The Prevalence and Demographic Characteristics of Consanguineous Marriages in Pakistan. J. Biosoc. Sci. 1998, 30, 261–275. [Google Scholar] [CrossRef]
- Kögel, A.; Keidel, A.; Bonneau, F.; Schäfer, I.B.; Conti, E. The human SKI complex regulates channeling of ribosome-bound RNA to the exosome via an intrinsic gatekeeping mechanism. Mol. Cell 2022, 82, 756–769.e8. [Google Scholar] [CrossRef]
- Kinnear, C.; Glanzmann, B.; Banda, E.; Schlechter, N.; Durrheim, G.; Neethling, A.; Nel, E.; Schoeman, M.; Johnson, G.; van Helden, P.D.; et al. Exome sequencing identifies a novel TTC37 mutation in the first reported case of Trichohepatoenteric syndrome (THE-S) in South Africa. BMC Med. Genet. 2017, 18, 26. [Google Scholar] [CrossRef]
- Jaganathan, K.; Kyriazopoulou Panagiotopoulou, S.; McRae, J.F.; Darbandi, S.F.; Knowles, D.; Li, Y.I.; Kosmicki, J.A.; Arbelaez, J.; Cui, W.; Schwartz, G.B.; et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell 2019, 176, 535–548.e24. [Google Scholar] [CrossRef]
- Leegwater, P.A.; Vermeulen, G.; Könst, A.A.; Naidu, S.; Mulders, J.; Visser, A.; Kersbergen, P.; Mobach, D.; Fonds, D.; van Berkel, C.G.; et al. Subunits of the translation initiation factor eIF2B are mutant in leukoencephalopathy with vanishing white matter. Nat. Genet. 2001, 29, 383–388. [Google Scholar] [CrossRef]
- Koprulu, M.; Shabbir, R.M.K.; Mumtaz, S.; Tolun, A.; Malik, S. Expanding OBSL1 Mutation Phenotype: Disproportionate Short Stature, Barrel Chest, Thoracic Kyphoscoliosis, Hypogonadism, and Hypospadias. Yale J. Biol. Med. 2023, 96, 367–382. [Google Scholar] [CrossRef]
- Qiu, Z.; Elsayed, Z.; Peterkin, V.; Alkatib, S.; Bennett, D.; Landry, J.W. Ino80 is essential for proximal-distal axis asymmetry in part by regulating Bmp4 expression. BMC Biol. 2016, 14, 18. [Google Scholar] [CrossRef]
- Chen, T.; Li, Q.; Xu, J.; Ding, K.; Wang, Y.; Wang, W.; Li, S.; Shen, Y. Mutation screening of BMP4, BMP7, HOXA4 and HOXB6 genes in Chinese patients with hypospadias. Eur. J. Hum. Genet. 2007, 15, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Zhou, J.; Li, R.; Dudley, E.A.; Ye, X. Novel function of LHFPL2 in female and male distal reproductive tract development. Sci. Rep. 2016, 6, 23037. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Hussain, H.M.J.; Khan, M.; Muhammad, Z.; Zhou, J.; Ma, A.; Huang, X.; Ye, J.; Chen, M.; Zhi, A.; et al. Deficiency in DNAH12 causes male infertility by impairing DNAH1 and DNALI1 recruitment in humans and mice. eLife 2025, 13, RP100350. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Francioli, L.C.; Goodrich, J.K.; Collins, R.L.; Kanai, M.; Wang, Q.; Alföldi, J.; Watts, N.A.; Vittal, C.; Gauthier, L.D.; et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature 2024, 625, 92–100. [Google Scholar] [CrossRef]
- Scott, C.H.; Amarillo, I.E. Identification of Small Regions of Overlap from Copy Number Variable Regions in Patients with Hypospadias. Int. J. Mol. Sci. 2022, 23, 4246. [Google Scholar] [CrossRef]
- Ajayi, A.F.; Oyowvi, M.O.; Akanbi, G.B.; Ajayi, L.O.; Ayomide, J.J. Genetic Susceptibility and Pathogenesis of Hypospadias. Biochem. Genet. 2025. [Google Scholar] [CrossRef]
- Giordano, F.; Abballe, A.; De Felip, E.; di Domenico, A.; Ferro, F.; Grammatico, P.; Ingelido, A.M.; Marra, V.; Marrocco, G.; Vallasciani, S.; et al. Maternal exposures to endocrine disrupting chemicals and hypospadias in offspring. Birth Defects Res. A Clin. Mol. Teratol. 2010, 88, 241–250. [Google Scholar] [CrossRef]
- Vottero, A.; Minari, R.; Viani, I.; Tassi, F.; Bonatti, F.; Neri, T.M.; Bertolini, L.; Bernasconi, S.; Ghizzoni, L. Evidence for Epigenetic Abnormalities of the Androgen Receptor Gene in Foreskin from Children with Hypospadias. J. Clin. Endocrinol. Metab. 2011, 96, E1953–E1962. [Google Scholar] [CrossRef]
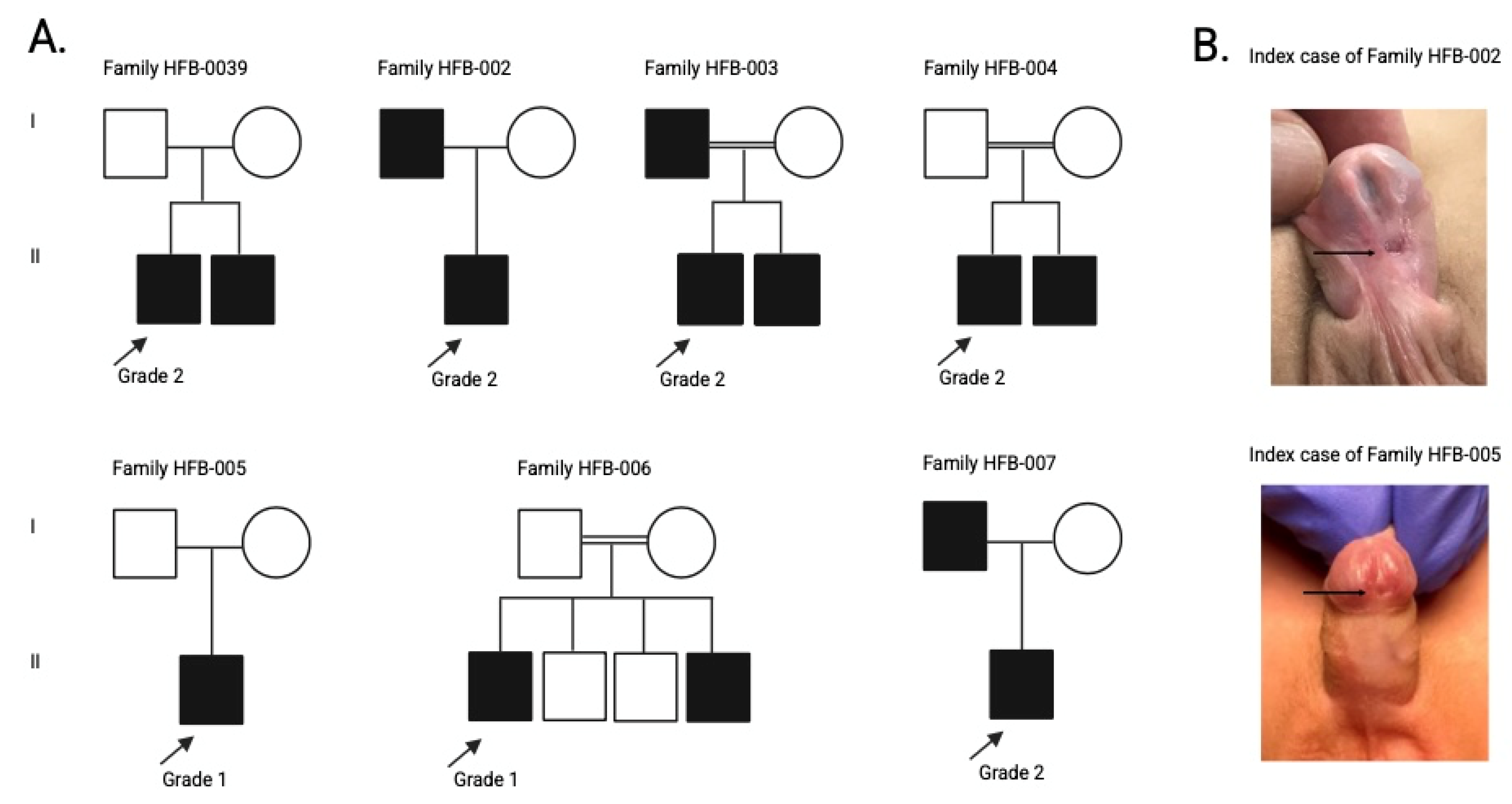
| Feature | HFB-0039-I | HFB-002-I | HFB-003-I | HFB-004-I | HFB-005-I | HFB-006-I | HFB-007-I |
|---|---|---|---|---|---|---|---|
| Age | 3 years | 2 years | 2 years | 6 years | 3 years | 7 years | 10 years |
| Nationality | Sudanese | Indian | Pakistani | Palestinian | Yemeni | Yemeni | Egyptian |
| Consanguinity (Yes/No/Not reported) | No | No | yes | yes | Not reported | Yes | No |
| Grade of Hypospadias | 2 | 2 | 2 | 2 | 1 | 1 | 2 |
| Treatment Given (Surgery/Medication/None) | Surgery/ Testosterone | Surgery | Surgery | Surgery | Surgery | Surgery | Surgery/ Testosterone |
| Family ID | Gene | Nucleotide Change | AA Change | Variant Impact | RS ID | GnomAD Genome AF (v4.1.0) | CADD Score | ACMG/AMP Classification | Zygosity | Variant Inheritance in Our Families |
|---|---|---|---|---|---|---|---|---|---|---|
| HFB-0039 | LHFP | c.-176A>G | - | Splice region | rs866455435 | 0.0002036 | 9.2 | VUS | Hom | Inherited from unaffected het. parents Affected brother also homozygous |
| HFB-002 | TTC37 | c.642+1G>A | - | Splice donor | rs538894487 | 0.00001313 | 34 | P | Hett | Inherited from affected het. father |
| EIF2B5 | c.821G>A | p.Arg274Gln | Missense | rs1460067929 | 0.000006574 | 33 | LP | Het | Inherited from affected het. father | |
| DNAH12 | c.7604C>T | p.Pro2535Leu | Missense | rs529341489 | 0.00006570 | 31 | VUS | Het | Inherited from affected het. father | |
| HFB-003 | OBSL1 | c.458dupG | p.Leu154fs | Frameshift | rs767237510 | 0.00003959 | - | P | Het | Inherited from affected het. father Affected brother also heterozygous |
| INO80 | c.445G>C | p.Glu149Gln | Missense | rs749039242 | 0.00001315 | 18.9 | LP | Het | Inherited from affected het. father Affected brother also heterozygous | |
| HFB-004 | COL6A3 | c.3223C>T | p.Arg1075Trp | Missense | rs201962257 | 0.0004079 | 25.6 | VUS | Hom | Inherited from unaffected het. parents Affected brother also homozygous |
| HFB-005 | - | - | - | |||||||
| HFB-006 | - | - | - | |||||||
| HFB-007 | ACADVL | c.67C>T | p.Gln23 * | Stop gain | rs1237915800 | 0.00000658 | 24.9 | LP | Het | Inherited from affected het. father |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Shafai, K.N.; Arar, S.; Jamil, A.; Azzah, A.; Mancha, M.; Saraiva, L.R.; Abbas, T. Genetic Insights into Familial Hypospadias Identifying Rare Variants and Their Potential Role in Urethral Development. Genes 2025, 16, 1340. https://doi.org/10.3390/genes16111340
Al-Shafai KN, Arar S, Jamil A, Azzah A, Mancha M, Saraiva LR, Abbas T. Genetic Insights into Familial Hypospadias Identifying Rare Variants and Their Potential Role in Urethral Development. Genes. 2025; 16(11):1340. https://doi.org/10.3390/genes16111340
Chicago/Turabian StyleAl-Shafai, Kholoud N., Seem Arar, Asma Jamil, Amina Azzah, Maraeh Mancha, Luis R. Saraiva, and Tariq Abbas. 2025. "Genetic Insights into Familial Hypospadias Identifying Rare Variants and Their Potential Role in Urethral Development" Genes 16, no. 11: 1340. https://doi.org/10.3390/genes16111340
APA StyleAl-Shafai, K. N., Arar, S., Jamil, A., Azzah, A., Mancha, M., Saraiva, L. R., & Abbas, T. (2025). Genetic Insights into Familial Hypospadias Identifying Rare Variants and Their Potential Role in Urethral Development. Genes, 16(11), 1340. https://doi.org/10.3390/genes16111340






