A Phylogenetic Perspective on the Evolutionary Patterns of the Animal Interleukin-10 Signaling System
Abstract
1. Introduction
2. Materials and Methods
2.1. Construction of a Phylogenetic Tree
2.2. Preparation of Genome and Annotation Files
2.3. Identification of the IL-10 and IL-10R Families
2.4. Conserved Motif Identification
2.5. Gene Family Clustering and Classification
2.6. Analysis of the IL-10 and IL-10R Family Variants
2.7. dN/dS Analysis
3. Results
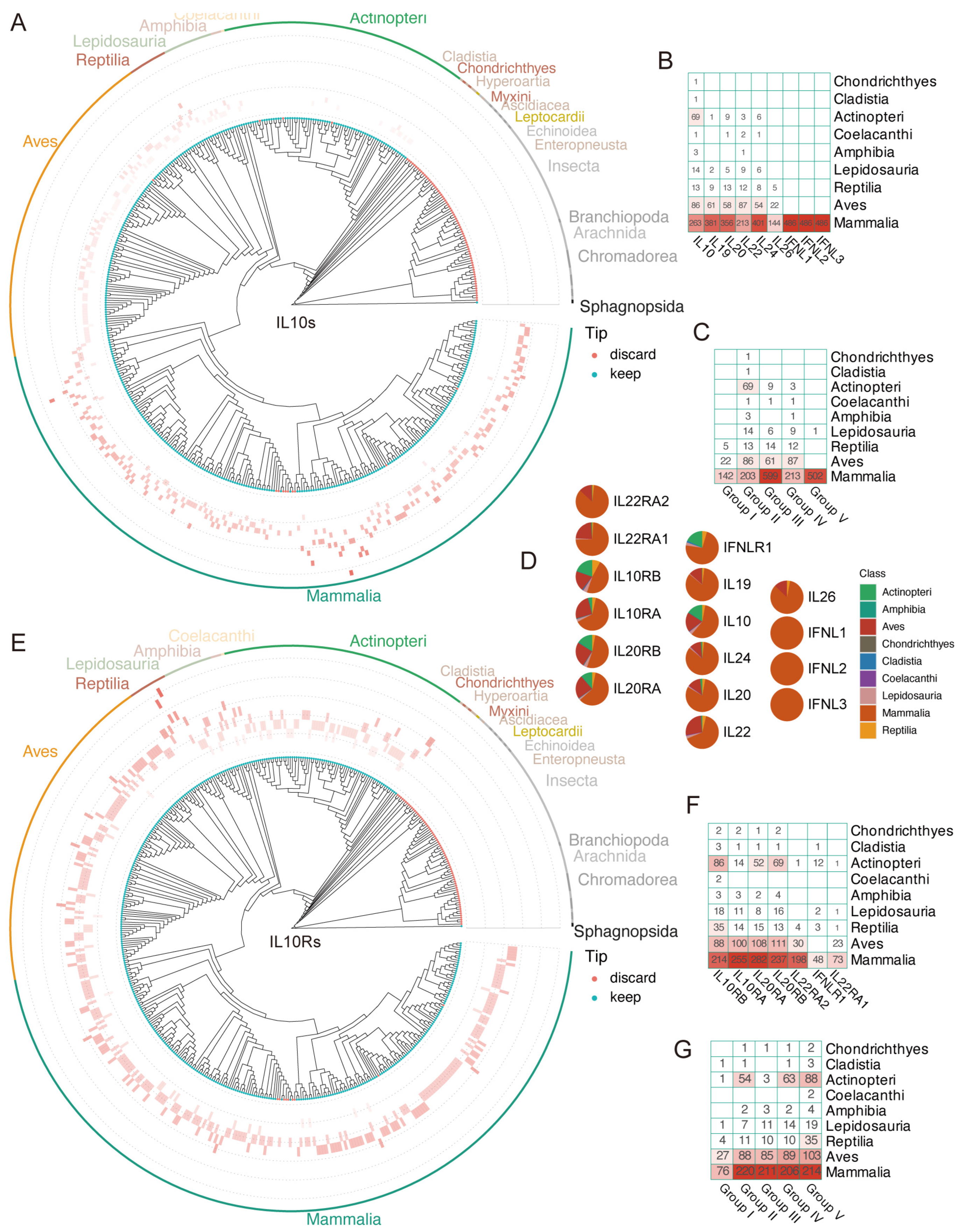
3.1. Identification and Distribution of IL-10s and IL-10Rs in the Animal Class
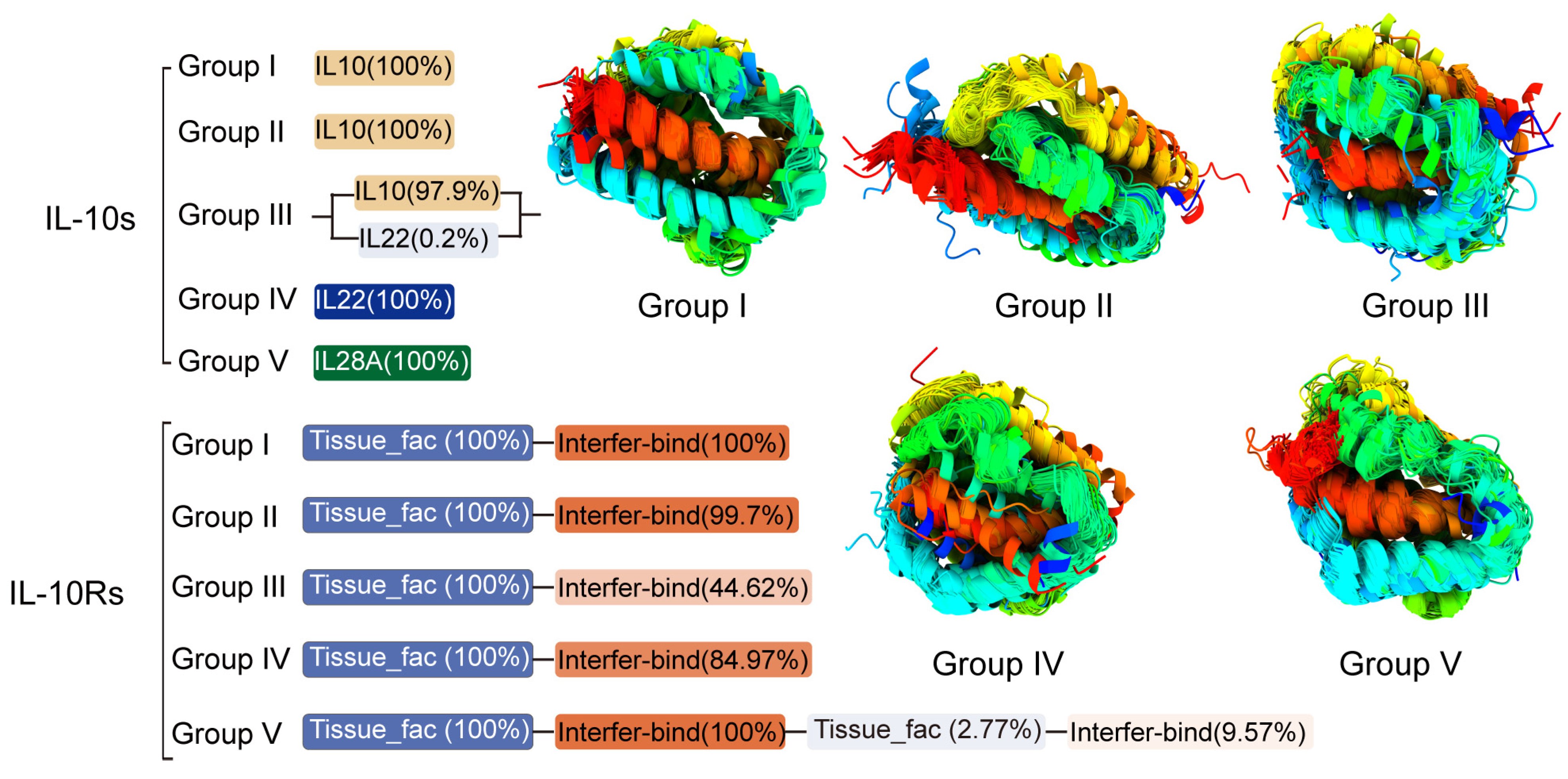
3.2. Conserved Motif, Domain, and 3D Structure of IL-10s and IL-10Rs Across Several Groups
3.3. The Evolution Pattern of IL-10s and IL-10Rs
4. Discussion
4.1. The Ancient Origin of IL-10s and IL-10-Rs
4.2. IL-10s and IL-10-Rs Structure and Cross-Species Characteristics
4.3. The Association of IL-10s and IL-10Rs with Diseases
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wei, S.H.-Y.; Ming-Lum, A.; Liu, Y.; Wallach, D.; Ong, C.J.; Chung, S.W.; Moore, K.W.; Mui, A.L.-F. Proteasome-Mediated Proteolysis of the Interleukin-10 Receptor Is Important for Signal Downregulation. J. Interferon Cytokine Res. 2006, 26, 281–290. [Google Scholar] [CrossRef]
- Ouyang, W.; O’Garra, A. IL-10 Family Cytokines IL-10 and IL-22: From Basic Science to Clinical Translation. Immunity 2019, 50, 871–891. [Google Scholar] [CrossRef]
- Wang, X.; Wong, K.; Ouyang, W.; Rutz, S. Targeting IL-10 Family Cytokines for the Treatment of Human Diseases. Cold Spring Harb. Perspect. Biol. 2019, 11, a028548. [Google Scholar] [CrossRef]
- Sabat, R. IL-10 Family of Cytokines. Cytokine Growth Factor Rev. 2010, 21, 315–324. [Google Scholar] [CrossRef]
- Ouyang, W.; Rutz, S.; Crellin, N.K.; Valdez, P.A.; Hymowitz, S.G. Regulation and Functions of the IL-10 Family of Cytokines in Inflammation and Disease. Annu. Rev. Immunol. 2011, 29, 71–109. [Google Scholar] [CrossRef] [PubMed]
- Fickenscher, H.; Hör, S.; Küpers, H.; Knappe, A.; Wittmann, S.; Sticht, H. The Interleukin-10 Family of Cytokines. Trends Immunol. 2002, 23, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Renauld, J.-C. Class II Cytokine Receptors and Their Ligands: Key Antiviral and Inflammatory Modulators. Nat. Rev. Immunol. 2003, 3, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, R.P.; Sheikh, F.; Kotenko, S.V.; Dickensheets, H. The Expanded Family of Class II Cytokines That Share the IL-10 Receptor-2 (IL-10R2) Chain. J. Leukoc. Biol. 2004, 76, 314–321. [Google Scholar] [CrossRef]
- Monte, M.M.; Wang, T.; Collet, B.; Zou, J.; Secombes, C.J. Molecular Characterisation of Four Class 2 Cytokine Receptor Family Members in Rainbow Trout, Oncorhynchus Mykiss. Dev. Comp. Immunol. 2015, 48, 43–54. [Google Scholar] [CrossRef]
- Kotenko, S.V. The Family of IL-10-Related Cytokines and Their Receptors: Related, but to What Extent? Cytokine Growth Factor Rev. 2002, 13, 223–240. [Google Scholar] [CrossRef]
- Parrish-Novak, J.; Xu, W.; Brender, T.; Yao, L.; Jones, C.; West, J.; Brandt, C.; Jelinek, L.; Madden, K.; McKernan, P.A.; et al. Interleukins 19, 20, and 24 Signal through Two Distinct Receptor Complexes. Differences in Receptor-Ligand Interactions Mediate Unique Biological Functions. J. Biol. Chem. 2002, 277, 47517–47523. [Google Scholar] [CrossRef]
- Moore, K.W.; de Waal Malefyt, R.; Coffman, R.L.; O’Garra, A. Interleukin-10 and the Interleukin-10 Receptor. Annu. Rev. Immunol. 2001, 19, 683–765. [Google Scholar] [CrossRef]
- Walter, M.R. Structure of Interleukin-10/Interleukin-10R1 Complex: A Paradigm for Class 2 Cytokine Activation. Immunol. Res. 2002, 26, 303–308. [Google Scholar] [CrossRef]
- Zdanov, A. Structural Features of the Interleukin-10 Family of Cytokines. Curr. Pharm. Des. 2004, 10, 3873–3884. [Google Scholar] [CrossRef] [PubMed]
- Brocker, C.; Thompson, D.; Matsumoto, A.; Nebert, D.W.; Vasiliou, V. Evolutionary Divergence and Functions of the Human Interleukin (IL) Gene Family. Hum. Genom. 2010, 5, 30. [Google Scholar] [CrossRef]
- Trivella, D.B.B.; Ferreira-Júnior, J.R.; Dumoutier, L.; Renauld, J.-C.; Polikarpov, I. Structure and Function of Interleukin-22 and Other Members of the Interleukin-10 Family. Cell. Mol. Life Sci. CMLS 2010, 67, 2909–2935. [Google Scholar] [CrossRef] [PubMed]
- Nagalakshmi, M.L.; Murphy, E.; McClanahan, T.; de Waal Malefyt, R. Expression Patterns of IL-10 Ligand and Receptor Gene Families Provide Leads for Biological Characterization. Int. Immunopharmacol. 2004, 4, 577–592. [Google Scholar] [CrossRef]
- Kuntzel, T.; Spenlé, C.; Pham-Van, L.D.; Birmpili, D.; Riou, A.; Loeuillet, A.; Charmarke-Askar, I.; Bagnard, D. Implication of the Transmembrane Domain in the Interleukin 10 Receptor Platform Oligomerisation. Cells 2023, 12, 1361. [Google Scholar] [CrossRef] [PubMed]
- Xuan, L.; Zhang, N.; Wang, X.; Zhang, L.; Bachert, C. IL-10 Family Cytokines in Chronic Rhinosinusitis with Nasal Polyps: From Experiments to the Clinic. Front. Immunol. 2022, 13, 947983. [Google Scholar] [CrossRef]
- Porro, C.; Cianciulli, A.; Panaro, M.A. The Regulatory Role of IL-10 in Neurodegenerative Diseases. Biomolecules 2020, 10, 1017. [Google Scholar] [CrossRef]
- Zhu, H.; Hu, S.; Li, Y.; Sun, Y.; Xiong, X.; Hu, X.; Chen, J.; Qiu, S. Interleukins and Ischemic Stroke. Front. Immunol. 2022, 13, 828447. [Google Scholar] [CrossRef]
- Wegenka, U.M. IL-20: Biological Functions Mediated through Two Types of Receptor Complexes. Cytokine Growth Factor Rev. 2010, 21, 353–363. [Google Scholar] [CrossRef]
- Chiang, H.-Y.; Lu, H.-H.; Sudhakar, J.N.; Chen, Y.-W.; Shih, N.-S.; Weng, Y.-T.; Shui, J.-W. IL-22 Initiates an IL-18-Dependent Epithelial Response Circuit to Enforce Intestinal Host Defence. Nat. Commun. 2022, 13, 874. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Sun, X.; Wang, Y.; Luan, H.; Zhang, R.; Hu, F.; Sun, X.; Li, X.; Guo, J. Role of IL-24 in NK Cell Activation and Its Clinical Implication in Systemic Lupus Erythematosus. Clin. Rheumatol. 2021, 40, 2707–2715. [Google Scholar] [CrossRef] [PubMed]
- Scala, E.; Di Caprio, R.; Cacciapuoti, S.; Caiazzo, G.; Fusco, A.; Tortorella, E.; Fabbrocini, G.; Balato, A. A New T Helper 17 Cytokine in Hidradenitis Suppurativa: Antimicrobial and Proinflammatory Role of Interleukin-26. Br. J. Dermatol. 2019, 181, 1038–1045. [Google Scholar] [CrossRef]
- Diegelmann, J.; Beigel, F.; Zitzmann, K.; Kaul, A.; Göke, B.; Auernhammer, C.J.; Bartenschlager, R.; Diepolder, H.M.; Brand, S. Comparative Analysis of the Lambda-Interferons IL-28A and IL-29 Regarding Their Transcriptome and Their Antiviral Properties against Hepatitis C Virus. PLoS ONE 2010, 5, e15200. [Google Scholar] [CrossRef] [PubMed]
- Nagata, K.; Nishiyama, C. IL-10 in Mast Cell-Mediated Immune Responses: Anti-Inflammatory and Proinflammatory Roles. Int. J. Mol. Sci. 2021, 22, 4972. [Google Scholar] [CrossRef]
- Zdanov, A. Structural Analysis of Cytokines Comprising the IL-10 Family. Cytokine Growth Factor Rev. 2010, 21, 325–330. [Google Scholar] [CrossRef]
- Hazlett, L.D.; Jiang, X.; McClellan, S.A. IL-10 Function, Regulation, and in Bacterial Keratitis. J. Ocul. Pharmacol. Ther. Off. J. Assoc. Ocul. Pharmacol. Ther. 2014, 30, 373–380. [Google Scholar] [CrossRef]
- Asadullah, K.; Sterry, W.; Volk, H.D. Interleukin-10 Therapy--Review of a New Approach. Pharmacol. Rev. 2003, 55, 241–269. [Google Scholar] [CrossRef]
- Hofmann, S.R.; Rösen-Wolff, A.; Tsokos, G.C.; Hedrich, C.M. Biological Properties and Regulation of IL-10 Related Cytokines and Their Contribution to Autoimmune Disease and Tissue Injury. Clin. Immunol. 2012, 143, 116–127. [Google Scholar] [CrossRef]
- Wang, J.; Chen, Z.; Li, M.; Zhang, Z.; Song, Y.; Xu, W.; Wang, L.; Chen, S. Genome-Wide Identification, Immune Response Profile and Functional Characterization of IL-10 from Spotted Knifejaw (Oplegnathus Punctatus) during Host Defense against Bacterial and Viral Infection. Fish Shellfish Immunol. 2022, 124, 513–524. [Google Scholar] [CrossRef]
- Liu, Y.; Wenren, M.; Cheng, W.; Zhou, X.; Xu, D.; Chi, C.; Lü, Z.; Liu, H. Identification, Functional Characterization and Immune Response Profiles of Interleukin-10 in Nibea Albiflora. Fish Shellfish Immunol. 2024, 151, 109654. [Google Scholar] [CrossRef]
- Wang, Q.; Zhao, Z.; Sun, R.; Shi, Z.; Zhang, Y.; Wang, B.; Zhang, X.; Ji, W. Bioinformatics Characteristics and Expression Analysis of IL-8 and IL-10 in Largemouth Bass (Micropterus salmoides) upon Nocardia Seriolae Infection. Fish Shellfish Immunol. 2024, 148, 109465. [Google Scholar] [CrossRef]
- Chakkalakkal, G.J.; Gopakumar, S.T.; Sharma, S.R.K.; Raveendranathan, D.N.; Jagannivasan, A.; Nair, A.V.; Ramachandran, V.; Achamveetil, G. Molecular Features and Expression Kinetics of Interleukin-10 Gene from the Marine Teleost, Snubnose Pompano (Trachinotus blochii). Mol. Biol. Rep. 2024, 52, 79. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Wang, C.; Zhang, W.; Wu, Y.; Wang, D.; Zhao, S.; Zhu, M. Pleiotropic Architectures of Porcine Immune and Growth Trait Pairs Revealed by a Self-Product-Based Transcriptome Method. Anim. Genet. 2023, 54, 123–131. [Google Scholar] [CrossRef]
- Pacheco, A.; Conington, J.; Corripio-Miyar, Y.; Frew, D.; Banos, G.; McNeilly, T.N. Genetic Profile of Adaptive Immune Traits and Relationships with Parasite Resistance and Productivity in Scottish Blackface Sheep. Anim. Int. J. Anim. Biosci. 2024, 18, 101061. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Liu, Y.; Xu, N.; Ai, X.; Yang, Y. Molecular Characterization and Expression Analysis of IL-10 and IL-6 in Channel Catfish (Ictalurus punctatus). Pathogens 2023, 12, 886. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, M.; O’Garra, A. The Regulation of IL-10 Production by Immune Cells. Nat. Rev. Immunol. 2010, 10, 170–181. [Google Scholar] [CrossRef]
- Lin, Z.; Tian, Y.; Chai, C.; Fu, M.; Wu, Q.; Tan, L.; Li, L.; Guan, X.; Wang, Z.; Zhao, J.; et al. The Association of Immune-Related Genes and the Potential Role of IL10 with Biliary Atresia. Pediatr. Res. 2023, 94, 1659–1666. [Google Scholar] [CrossRef]
- Arjmand, B.; Rahim, F. The Probable Protective Effect of Photobiomodulation on the Immunologic Factor’s mRNA Expression Level in the Lung: An Extended COVID-19 Preclinical and Clinical Meta-Analysis. Clin. Pathol. 2023, 16, 2632010X221127683. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Wang, M.; Liu, X.-S. The Evolving Landscape of IL-10, IL-22 and IL-26 in Pleurisy Especially in Tuberculous Pleurisy. Respir. Res. 2024, 25, 275. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, A.; Banos, G.; Lambe, N.; McLaren, A.; McNeilly, T.N.; Conington, J. Genome-Wide Association Studies of Parasite Resistance, Productivity and Immunology Traits in Scottish Blackface Sheep. Anim. Int. J. Anim. Biosci. 2024, 18, 101069. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Zhang, C.; Rabiee, M.; Sayyari, E.; Mirarab, S. ASTRAL-III: Polynomial Time Species Tree Reconstruction from Partially Resolved Gene Trees. BMC Bioinf. 2018, 19, 153. [Google Scholar] [CrossRef]
- Dainat, J.; Hereñú, D.; Pucholt, P. AGAT: Another Gff Analysis Toolkit to Handle Annotations in Any GTF/GFF Format. Zenodo 2020, 431. Available online: https://github.com/NBISweden/AGAT (accessed on 1 October 2025).
- Pertea, G.; Pertea, M. GFF Utilities: GffRead and GffCompare. F1000Research 2020, 9, 304. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Almeida, L.V.; Reis-Cunha, J.L.; Bartholomeu, D.C. Dgfr: An R Package to Assess Sequence Diversity of Gene Families. BMC Bioinform. 2024, 25, 207. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.-Y. Ggtree: An r Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Xu, S.; Dai, Z.; Guo, P.; Fu, X.; Liu, S.; Zhou, L.; Tang, W.; Feng, T.; Chen, M.; Zhan, L.; et al. ggtreeExtra: Compact Visualization of Richly Annotated Phylogenetic Data. Mol. Biol. Evol. 2021, 38, 4039–4042. [Google Scholar] [CrossRef]
- Wilkins, D. Gggenes: Draw Gene Arrow Maps in “Ggplot2”. 2023. Available online: https://wilkox.org/gggenes/ (accessed on 1 October 2025).
- Bodenhofer, U.; Bonatesta, E.; Horejš-Kainrath, C.; Hochreiter, S. Msa: An R Package for Multiple Sequence Alignment. Bioinformatics 2015, 31, 3997–3999. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinf. 2004, 5, 113. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Lin, Z.; Akin, H.; Rao, R.; Hie, B.; Zhu, Z.; Lu, W.; Smetanin, N.; Verkuil, R.; Kabeli, O.; Shmueli, Y.; et al. Evolutionary-Scale Prediction of Atomic-Level Protein Structure with a Language Model. Science 2023, 379, 1123–1130. [Google Scholar] [CrossRef]
- Zhang, C.; Shine, M.; Pyle, A.M.; Zhang, Y. US-Align: Universal Structure Alignments of Proteins, Nucleic Acids, and Macromolecular Complexes. Nat. Methods 2022, 19, 1109–1115. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for Structure Building and Analysis. Protein Sci. Publ. Protein Soc. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Kosakovsky Pond, S.L.; Frost, S.D.W. Not so Different after All: A Comparison of Methods for Detecting Amino Acid Sites under Selection. Mol. Biol. Evol. 2005, 22, 1208–1222. [Google Scholar] [CrossRef]
- Logsdon, N.J.; Jones, B.C.; Allman, J.C.; Izotova, L.; Schwartz, B.; Pestka, S.; Walter, M.R. The IL-10R2 Binding Hot Spot on IL-22 Is Located on the N-terminal Helix and Is Dependent on N-Linked Glycosylation. J. Mol. Biol. 2004, 342, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Al-Abbasi, F.A.; Mohammed, K.; Sadath, S.; Banaganapalli, B.; Nasser, K.; Shaik, N.A. Computational Protein Phenotype Characterization of IL10RA Mutations Causative to Early Onset Inflammatory Bowel Disease (IBD). Front. Genet. 2018, 9, 146. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Fischer, K.P.; Tyrrell, D.L.; Gutfreund, K.S. cDNA Cloning, Genomic Structure, Molecular Characterization and mRNA Expression Analysis of the Pekin Duck Interleukin-10 Receptor 1. Int. J. Immunogenet. 2012, 39, 55–67. [Google Scholar] [CrossRef]
- Preimel, D.; Sticht, H. Molecular Modeling of the Interleukin-19 Receptor Complex. Novel Aspects of Receptor Recognition in the Interleukin-10 Cytokine Family. J. Mol. Model. 2004, 10, 290–296. [Google Scholar] [CrossRef]
- Ding, Y.; Qin, L.; Kotenko, S.V.; Pestka, S.; Bromberg, J.S. A Single Amino Acid Determines the Immunostimulatory Activity of Interleukin 10. J. Exp. Med. 2000, 191, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Dumoutier, L.; Lejeune, D.; Hor, S.; Fickenscher, H.; Renauld, J.-C. Cloning of a New Type II Cytokine Receptor Activating Signal Transducer and Activator of Transcription (STAT)1, STAT2 and STAT3. Biochem. J. 2003, 370, 391–396. [Google Scholar] [CrossRef]
- Xu, D.; Xie, M.; Yang, L. Molecular Characterization and Expression Analysis of IL-10 and IL-20L Genes in Swamp Eel (Monopterus Albus) against Aeromonas Veronii Infection. Aquac. Rep. 2022, 24, 101164. [Google Scholar] [CrossRef]
- Wu, S.; Duan, C.; Kong, L.; Tu, X.; Wang, L.; Guo, Z.; Ye, J. Interleukin-10 (IL-10) Participates in Host Defense against Bacterial Pathogens and Promotes IgM Antibody Production in Nile Tilapia (Oreochromis Niloticus). Aquaculture 2021, 531, 735829. [Google Scholar] [CrossRef]
- Piazzon, M.C.; Lutfalla, G.; Forlenza, M. IL10, a Tale of an Evolutionarily Conserved Cytokine across Vertebrates. Crit. Rev. Immunol. 2016, 36, 99–129. [Google Scholar] [CrossRef]
- Zhang, H.; Kuchroo, V. Epigenetic and Transcriptional Mechanisms for the Regulation of IL-10. Semin. Immunol. 2019, 44, 101324. [Google Scholar] [CrossRef]
- Kwon, G.; Kang, K. Transcriptional Regulation of IL10 Gene in Human Macrophages. J. Immunol. 2020, 204, 152.21. [Google Scholar] [CrossRef]
- Zheng, Z.; Huang, G.; Gao, T.; Huang, T.; Zou, M.; Zou, Y.; Duan, S. Epigenetic Changes Associated with Interleukin-10. Front. Immunol. 2020, 11, 1105. [Google Scholar] [CrossRef]
- Bedke, T.; Muscate, F.; Soukou, S.; Gagliani, N.; Huber, S. IL-10-Producing T Cells and Their Dual Functions. Semin. Immunol. 2019, 44, 101335. [Google Scholar] [CrossRef]
- Lu, M.-C.; Yu, C.-L.; Chen, H.-C.; Yu, H.-C.; Huang, H.-B.; Lai, N.-S. Increased miR-223 Expression in T Cells from Patients with Rheumatoid Arthritis Leads to Decreased Insulin-like Growth Factor-1-Mediated Interleukin-10 Production. Clin. Exp. Immunol. 2014, 177, 641–651. [Google Scholar] [CrossRef]
- Wang, S.; Li, Z.; Chen, Q.; Wang, L.; Zheng, J.; Lin, Z.; Li, W. NF-κB-Induced MicroRNA-211 Inhibits Interleukin-10 in Macrophages of Rats with Lipopolysaccharide-Induced Acute Respiratory Distress Syndrome. Cell Physiol. Biochem. 2018, 45, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Moradi, N.; Fadaei, R.; Ahmadi, R.; Kazemian, E.; Fallah, S. Lower Expression of miR-10a in Coronary Artery Disease and Its Association with pro/Anti-Inflammatory Cytokines. Clin. Lab. 2018, 64, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Huang, Y.Y.; Jin, Y.; Li, C.R.; Yang, J. Effect of miR-21 on the expression of interleukin-10 in B cell of patients with henoch-schonlein purpura. Zhonghua Er Ke Za Zhi Chin. J. Pediatr. 2018, 56, 939–944. [Google Scholar] [CrossRef]
- Alashkar Alhamwe, B.; Gao, Z.; Alhamdan, F.; Harb, H.; Pichene, M.; Garnier, A.; El Andari, J.; Kaufmann, A.; Graumann, P.L.; Kesper, D.; et al. Intranasal Administration of Acinetobacter Lwoffii in a Murine Model of Asthma Induces IL-6-Mediated Protection Associated with Cecal Microbiota Changes. Allergy 2023, 78, 1245–1257. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, N.; Zhang, X.; Qin, M.; Dong, Y.; Jin, L. MiR-410 down-Regulates the Expression of Interleukin-10 by Targeting STAT3 in the Pathogenesis of Systemic Lupus Erythematosus. Cell Physiol. Biochem. 2016, 39, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Sun, P.; Zhang, C.; Li, Z.; Zhou, W. MiR-98 Suppresses the Effects of Tumor-Associated Macrophages on Promoting Migration and Invasion of Hepatocellular Carcinoma Cells by Regulating IL-10. Biochimie 2018, 150, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Minshawi, F.; Lanvermann, S.; McKenzie, E.; Jeffery, R.; Couper, K.; Papoutsopoulou, S.; Roers, A.; Muller, W. The Generation of an Engineered Interleukin-10 Protein with Improved Stability and Biological Function. Front. Immunol. 2020, 11, 1794. [Google Scholar] [CrossRef]
- Chang, C.-C.; Yang, C.-H.; Chuang, C.-H.; Jiang, S.-J.; Hwang, Y.-M.; Liou, J.-W.; Hsu, H.-J. A Peptide Derived from Interleukin-10 Exhibits Potential Anticancer Activity and Can Facilitate Cell Targeting of Gold Nanoparticles Loaded with Anticancer Therapeutics. Commun. Chem. 2023, 6, 278. [Google Scholar] [CrossRef] [PubMed]


| Member | Expressing Cells | Receptors | Signal Activation | Biological Functions |
|---|---|---|---|---|
| IL-10 | T lymphocytes, monocytes, and B cells | IL-10R1/IL-10R2 | STAT1, STAT3, STAT5 | Immunosuppressive, anti-inflammatory [15,19,20] |
| IL-19 | monocytes | IL-20R1/IL-20R2 | STAT3, STAT5 | Immune regulation [15,19,21] |
| IL-20 | Skin keratinocytes | IL-20R1/IL-20R2 | STAT3, STAT5 | Skin development and inflammation, hematopoiesis [15,19,22] |
| IL-22 | Activated T cells and NKT cells | IL-22R/IL-20R2, IL-22R/IL-10R2 | STAT1, STAT3, STAT5 | Acute phase reaction, innate immunity [15,19,23] |
| IL-24 | Melanocytes, Th2 cells, and monocytes | IL-20R1/IL-20R2, IL-22R/IL-20R2 | STAT3, STAT5 | Apoptosis, epidermal function, and the inflammatory cascade [15,19,24] |
| IL-26 | memory Th1 cells, Th17 cells and monocytes | IL-20R1/IL-10R2 | STAT1, STAT3 | Mucosal and skin immunity [15,19,25] |
| IL-28; IL-29 | monocytes | IL-28R/IL-10R2 | STAT1, STAT2 | Antiviral immunity [15,19,26] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, L.; Zhou, Z.; Wang, W.; Li, D.; Hao, T.; Chen, Y. A Phylogenetic Perspective on the Evolutionary Patterns of the Animal Interleukin-10 Signaling System. Genes 2025, 16, 1243. https://doi.org/10.3390/genes16111243
Tang L, Zhou Z, Wang W, Li D, Hao T, Chen Y. A Phylogenetic Perspective on the Evolutionary Patterns of the Animal Interleukin-10 Signaling System. Genes. 2025; 16(11):1243. https://doi.org/10.3390/genes16111243
Chicago/Turabian StyleTang, Liu, Zeyu Zhou, Weibin Wang, Dawei Li, Tingting Hao, and Yue Chen. 2025. "A Phylogenetic Perspective on the Evolutionary Patterns of the Animal Interleukin-10 Signaling System" Genes 16, no. 11: 1243. https://doi.org/10.3390/genes16111243
APA StyleTang, L., Zhou, Z., Wang, W., Li, D., Hao, T., & Chen, Y. (2025). A Phylogenetic Perspective on the Evolutionary Patterns of the Animal Interleukin-10 Signaling System. Genes, 16(11), 1243. https://doi.org/10.3390/genes16111243






