Abstract
Background/Objectives: GATA transcription factors play pivotal roles in regulating plant growth and development, physiological metabolism, and responses to environmental stress. However, research on GATA genes in sweetpotato remains limited. Methods: In this study, we identified 25 IbGATA genes in sweetpotato (Ipomoea batatas [Lam.] L.) through a genome-wide analysis. These genes were analyzed for their physicochemical properties, chromosomal localization, synteny, phylogenetic relationships, gene structure, promoter cis-elements, protein interaction networks, and expression profiles across various tissues and under drought stress. To elucidate the function of drought-resistant candidate genes, an in situ one-step transformation method was employed. Results: Sweetpotato GATA genes have a complex evolutionary history, including replication events, different selection pressures, and functional diversification. They may be involved in multiple plant stress signaling pathways. Furthermore, functional analysis revealed that IbGATA17 enhances drought tolerance in sweetpotato by promoting proline biosynthesis and reinforcing ROS scavenging capacity. Our findings provide novel insights into the roles of IbGATAs, particularly IbGATA17, in mediating drought-stress responses in sweetpotato. Conclusions: This study provides foundational insights into the GATA gene family in sweetpotato and reveals the pivotal role of IbGATA17 in simulated drought-stress response, providing a potential candidate gene for the development of drought-resistant varieties.
1. Introduction
Drought is considered one of the most detrimental abiotic stresses, with severe ramifications for plant growth and development [1,2]. It also imposes serious constraints on global agricultural production [3,4]. Under drought conditions, plants experience water deficit, oxidative stress, and metabolic disruption, which can result in reduced yields and compromised quality [5,6]. Sweetpotato (Ipomoea batata (L.) Lam.) is a globally vital crop and a crucial food source. it is rich in carbohydrates, carotenoids, vitamins, and minerals [7,8]. It plays a pivotal role in ensuring food security and combating hunger [9,10,11,12]. The crop’s remarkable adaptability and resilience to diverse planting environments, including marginal lands with poor soil conditions, contribute to its importance as a climate-resilient crop [13]. However, drought stress significantly limits sweetpotato storage root formation and storage root development, posing a serious threat to sustainable agriculture in arid and semi-arid regions [14]. Sweetpotato is a hexaploid crop with highly complex genome, complex genetic patterns, and high degree of inbreeding both within and between species [15]. Genetic engineering represents a significant method of enhancing the drought tolerance of sweetpotato. At present, some drought-tolerance-related genes have been cloned from sweetpotato, and their overexpression can enhance drought tolerance in transgenic sweetpotato plants [16,17,18,19,20,21,22].
The GATA transcription factors (TFs) are a class of type IV zinc finger DNA-binding proteins that regulate the transcription of downstream genes by binding to the W-GATA-R motif in promoter regions [23]. GATA family genes have been identified in multiple plant species, such as Arabidopsis thaliana [24], Triticum aestivum [25], Capsicum annuum and Solanum tuberosum [26,27], as well as Setaria italica [28]. In plants, GATA TFs are known to modulate diverse physiological processes, including light signaling, hormonal responses, and tolerance to abiotic stress [23]. In A. thaliana, GATA TFs (GNC, GNL, and B-GATA) have been shown to regulate physiological processes, including chlorophyll synthesis, flowering time, and cold resistance, and they also contribute to the balance of phototropic and gravitropic growth responses [29,30]. In rice, the GATA TFs (GATA6, GATA8, and GATA16) have been shown to exert a pivotal regulatory influence on grain size, the number of tillers, and the response to cold stress and reactive oxygen species (ROS) [31,32,33,34,35]. At present, there are few reports on GATA genes in sweetpotato. Overexpression of IbGATA24 enhances the hormone signaling pathway and ROS scavenging, thereby improving the drought resistance of A. thaliana [22]. Wang et al. identified the GATA gene family in sweetpotato, however, no functional analysis of these genes was performed [36]. The molecular mechanisms through which GATA family genes confer drought tolerance in plants are not yet fully elucidated.
In this study, we conducted a comprehensive genome-wide analysis of the GATA gene family in sweetpotato, leading to the identification of 25 IbGATA genes. Their phylogenetic relationships, gene structures, promoter cis-acting elements, and expression patterns in response to drought stress were systematically characterized. Notably, IbGATA17 exhibited significant upregulation under drought stress. Functional analysis revealed that its overexpression enhances drought tolerance by facilitating proline biosynthesis and scavenging ROS. This provides novel insights into the molecular basis of drought tolerance in sweetpotato.
2. Materials and Methods
2.1. Plant Materials
The sweetpotato drought-resistant line ‘Xu55-2′ was utilized in the analysis of gene expression and cloning of IbGATA17, and the cultivar ‘Shangshu 19′ was employed for the genetic transformation and the characterization of gene functions. The sweetpotato plants were grown in a substrate made of peat and vermiculite in a 1:1 ratio. The greenhouse cultivation took place at 25 ± 3 °C, which is the natural light environment of the China Agricultural University in Beijing, China.
2.2. Identification of IbGATAs
We obtained the whole-genome sequences of sweetpotato from the Ipomoea Genome Hub (https://sweetpotao.com/: accessed on 4 May 2025) and the protein sequences of A. thaliana GATA transcription factors from TAIR (https://www.arabidopsis.org/: accessed on 4 May 2025). Utilizing these A. thaliana GATAs, a search was conducted for potential GATA genes in the sweetpotato genome, employing a BlastP search with an E-value cutoff of no more than 1 × 10−5.
To further refine our search, we retrieved the Hidden Markov Model (HMM) profile for the zinc finger domain (PF00320) from the Pfam database (now hosted by InterPro) and conducted an HMM search using TBtools-II v2.326. We merged the results from both BlastP and HMM searches and validated all candidate GATA genes using the CD-Search tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi: accessed on 4 May 2025) to confirm the presence of conserved domains. Finally, the remaining GATA genes were systematically numbered based on their chromosomal positions.
2.3. Protein Property Prediction and Chromosome Distribution of IbGATAs
In order to assess the physical and chemical characteristics of IbGATA proteins, such as their molecular mass, isoelectric point, instability index, and water solubility, the ExPASy ProtParam tool (https://web.expasy.org/protparam/: accessed on 4 May 2025) was employed. In addition, Cell-PLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/: accessed on 4 May 2025) was employed to predict their subcellular locations. In order to ascertain the chromosomal locations of IbGATA genes, genomic data from the Sweetpotato Genomics Resource (http://sweetpotato.uga.edu/index.shtml: accessed on 4 May 2025) was utilized, and the results were visualized using TBtools-II v2.326 to create an in-depth chromosomal distribution map.
2.4. Phylogenetic Analysis of GATAs
The amino acid sequences of the GATAs transcription factors from A. thaliana (At) and sweetpotato (Ipomoea batatas, Ib) were aligned using the ClustalW tool in MEGA 11.0 software [37]. The same software was used to build a phylogenetic tree following the alignment, employing the Neighbor-Joining method with a bootstrap test of 1000 [38]. The phylogenetic trees were visualized using the Interactive Tree of Life (ITOL) platform (https://itol.embl.de/index.shtml: accessed on 15 May 2025).
2.5. Collinearity Analysis of IbGATAs
The collinear gene pairs were predicted by a one-step MCScanX of TBtools-II v2.326 software and the resulting data were visualized using TBtools-II v2.326 [39].
2.6. Conserved Structural Domains and Gene Structure Analysis of IbGATAs
Ten distinct motifs within the protein sequences were pinpointed through the tried-and-true discovery process with the Multiple Em for Motif Elicitation tool, also known as MEME (https://web.mit.edu/meme/current/share/doc/overview.html: accessed on 15 May 2025). To dissect the exon–intron framework, we utilized the TBtools-II v2.326 software package in tandem with the GSDS2.0 website (https://gsds.gao-lab.org/: accessed on 15 May 2025). Our examination was grounded in the meticulous GFF annotation files.
2.7. Identification of Cis-Acting Elements in the IbGATAs Promoter
TBtools was used to extract 2000 bp upstream of the CDS of the IbGATAs gene, and then the PlantCARE tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/: accessed on 17 May 2025) was used to analyze the cis-acting elements in the promoter region. TBtools-II v2.326 software and Python v3.7 were used to analyze the homeostatic elements in the promoter region [40]. Visualization was performed using TBtools-II v2.326 software and the Python v3.7 package Seaborn.
2.8. Prediction of IbGATAs Secondary Dimensional Structures
The secondary structure of the IbGATAs protein was predicted using NetSurfP-3.0 [41].
2.9. Protein Interaction Network of IbGATAs
The protein interaction network of the IbGATAs was analyzed on the STRING website (https://cn.string-db.org/: accessed on 23 May 2025) based on the AtGATAs proteins of A. thaliana [42].
2.10. Transcriptome and Expression Analysis
The RNA-seq data for IbGATAs in different sweetpotato tissues were obtained from the NCBI SRA repository (PRJCA000640) [43]. Additionally, the RNA-seq data for Xu 55-2 under PEG6000 treatment were obtained from related research [44]. Sweetpotato stem explants grown in vitro were cultured on Murashige and Skoog (MS) solid medium under 13 h of cold white light and 11 h of darkness. The 4-week-old plants were then treated with 20% polyethylene glycol (PEG) 6000 for 24 h. The sweetpotato IbACTIN (Genbank AY905538) gene served as an internal control and quantified gene expression by the comparative CT method. The expression of GATA genes (IbGATA4, IbGATA8, IbGATA12, IbGATA16, and IbGATA17) in sweetpotato was confirmed by quantitative reverse-transcription qRT-PCR on a 7500 Real-Time PCR instrument (Applied Biosystems, Foster City, CA, USA). The Premier 5 used for qRT-PCR primer design is listed in Supplementary Table S1. PCR cycle conditions were 95 °C for 5 min as the first denaturing step, followed by 40 cycles at 95 °C for 10 s, 60 °C for 30 min, and a gradual increase in temperature from 60 to 95 °C during the dissociation stage to monitor the specificity of each primer pair.
2.11. Transcriptional-Activation Assay
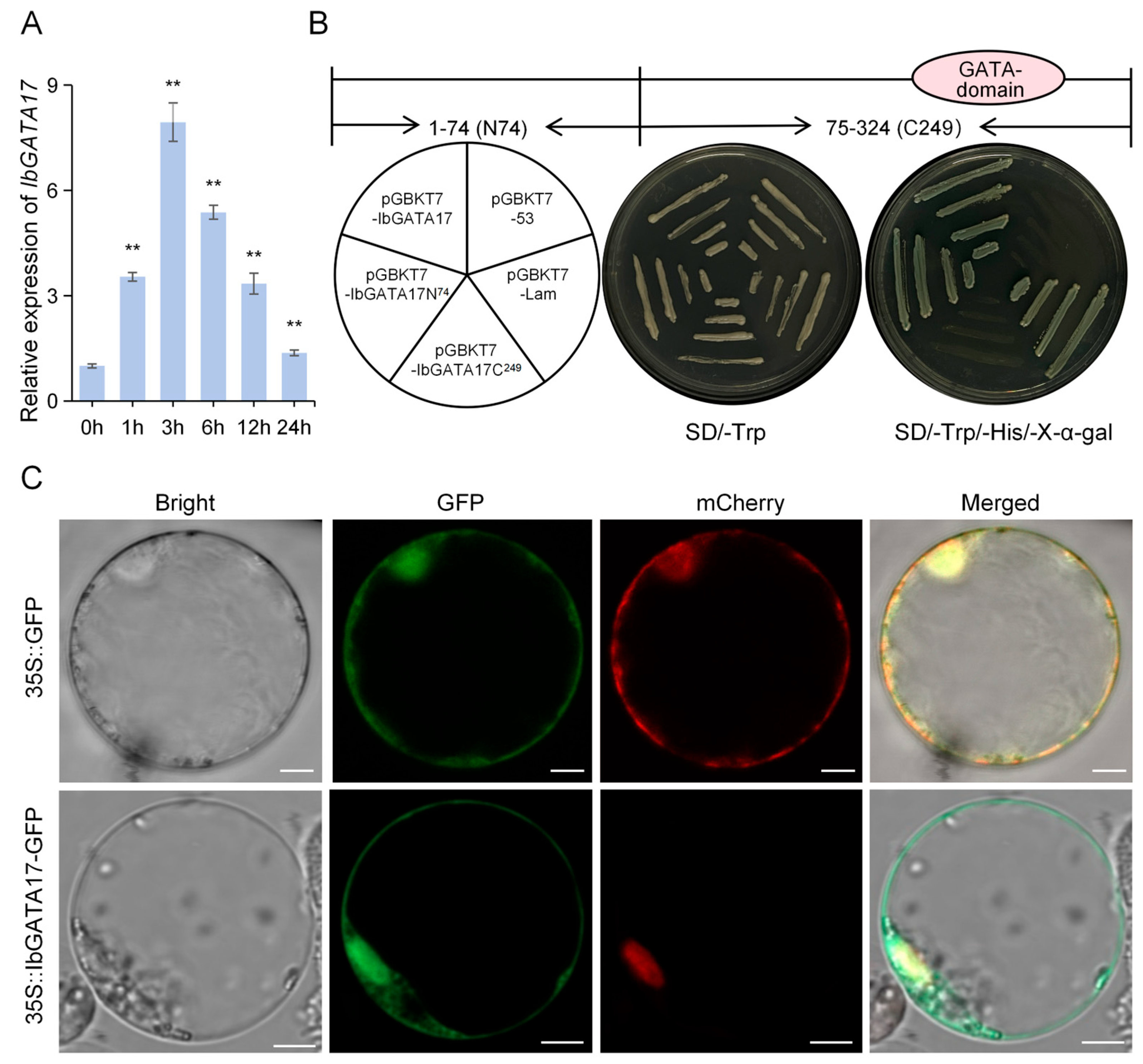
The coding sequence (CDS) of IbGATA17, as well as fragments encoding amino acids 1–74 and 75–324, were inserted into the pGBKT7 vector. The positive control was pGBKT7-53 while the negative control was pGBKT7-Lam. These constructs were transformed into the yeast strain Y2H Gold. The transformed yeast was then streaked onto SD/-Trp and SD/-Trp/-His/X-α-Gal plates and incubated at 30 °C for 2–3 days to observe growth [45].
2.12. Subcellular Localization of IbGATA17
The CDS of IbGATA17, excluding the stop codon, were inserted into pCAMBIA1300-35S-GFP. The construct and the control were both transfected into rice protoplasts using a polyethylene glycol–calcium-mediated method. This was followed by an 18 h incubation period to allow for transient expression. The isolation of the protoplasts and the transfection of the vectors were performed according to the method described by Yoo et al. [46]. The green and red fluorescent protein signals (GFP and RFP, respectively) were then collected using confocal laser scanning microscopy (LSM880; Zeiss, Oberkochen, Germany).
2.13. Production of Transgenic Plants
The CDS of IbGATA17 was separately inserted into the pCAMBIA1300-GFP vector. A pair of forward and reverse highly specific fragments of the gene IbGATA17 were inserted into the plant RNAi vector pFGC5941. The pCAMBIA1300-IbGATA17-GFP and pFGC5941-IbGATA17 constructs were then introduced into Agrobacterium strain K599. The production of the IbGATA17-OE and IbGATA17-Ri plants was achieved through the utilization of Shangshu 19 cuttings employing one-step Agrobacterium-rhizogenes-mediated transformation. After PCR analysis of the transgenic sweetpotato storage roots, we obtained 5 overexpression and 5 RNAi lines for further study [47].
2.14. Drought Tolerance Assays
The WT, IbGATA17-OE, and IbGATA17-Ri plants were cultivated in flowerpots with a substrate consisting of a 1:1 mixture of peat soil and vermiculite. The PEG6000 stress treatment was performed according to the method of Zhang et al. [17], with minor modifications. Select plants with consistent growth were irrigated with Hoagland solution containing 20% PEG6000 every 4 h for a period of 48 h.
2.15. Measurement of Abiotic Stress Tolerance Indices
Proline, malondialdehyde (MDA), and H2O2 contents were determined using assay kits (Cominbio, Suzhou, China) following the manufacturer’s instructions using leaves of transgenic sweetpotato.
2.16. Statistical Analysis
All data were analyzed using a one-way ANOVA, followed by a post hoc Tukey’s test, in SPSS 27.0. Data are presented as means ± standard deviation (SD). These statistical methods were employed to analyze the gene expression and resistance-related indicators of drought tolerance. Three biological replicates were performed for each experiment.
3. Results
3.1. Characteristics of GATA Genes in Sweetpotato
The HMM profile PF00320 (GATA DNA-binding domain) from Pfam database was first employed to screen the sweetpotato proteome. Next, the GATAs protein sequence in A. thaliana was utilized as control and the BLASTp search was conducted on protein sequence databases of sweetpotato. After removing redundant and incomplete sequences, a total of 25 members of the GATA gene family were identified in the entire sweetpotato genome. The genes were separately named IbGATA1–IbGATA25 according to their position on the chromosome. The 25 identified GATA proteins exhibited considerable diversity in their physicochemical properties. The amino acid lengths of these peptides ranged from 142 to 370 residues, the molecular weights varied from 14.87 to 94.10 kDa, and the aliphatic indices ranged from 37.05 to 73.85. Theoretical isoelectric points of the 25 GATA proteins ranged from 5.38 to 10.33. The stability of all GATA proteins was predicted, with instability indices exceeding 40. The median hydrophilicity for the 25 GATA proteins fell below zero, signifying their hydrophilic nature. Subcellular localization predictions indicated that the sweetpotato’s GATA are predominantly located within the nucleus or chloroplasts. (Table 1)

Table 1.
Information on the characterization of the identified GATA proteins.
3.2. Chromosome Mapping of GATA Family Genes in Sweetpotato
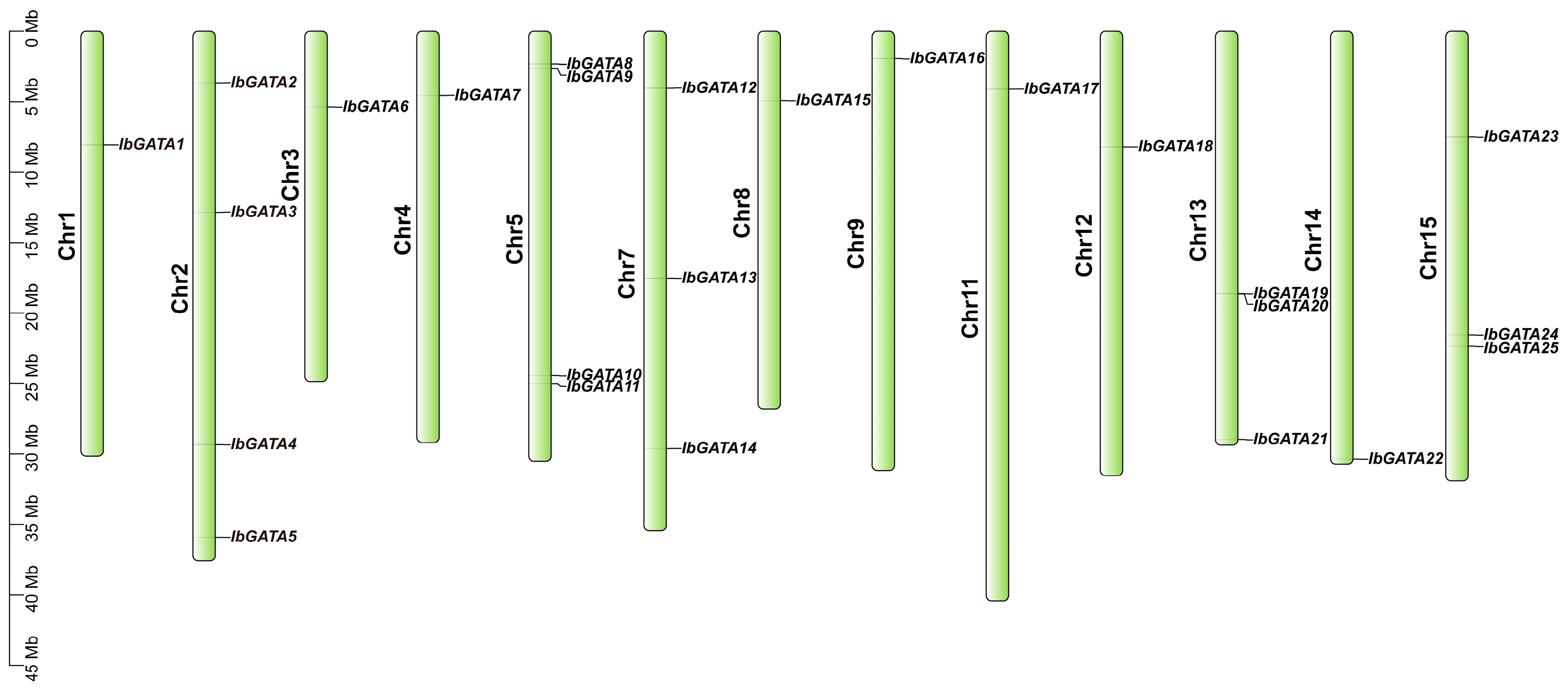
The distribution of GATA genes on sweetpotato chromosomes was determined based on the results of the chromosome location analysis of the 25 GATA family genes in sweetpotato (Figure 1). Chromosomal mapping revealed an uneven distribution of the 25 IbGATA genes across 13 out of the 15 sweetpotato chromosomes. Chromosomes 2 and 5 contained the largest number of IbGATAs (4 genes), followed by chromosomes 7, 13, and 15 (3 genes each). It is noteworthy that no GATA genes were identified on chromosomes 6 and 10.
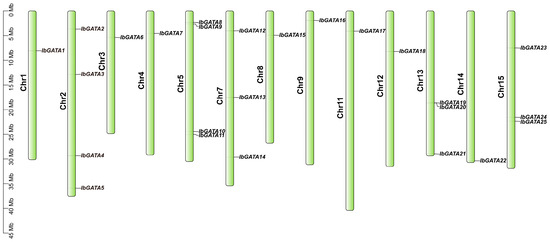
Figure 1.
Chromosomal distribution of the GATA gene family in sweetpotato.
The vertical bars in the figure represent chromosomes, with chromosome numbers labeled on the left. GATA gene names are shown on the right, and their precise positions are marked by black ticks along the chromosomes. Scale bars indicate physical distances (Mbp).
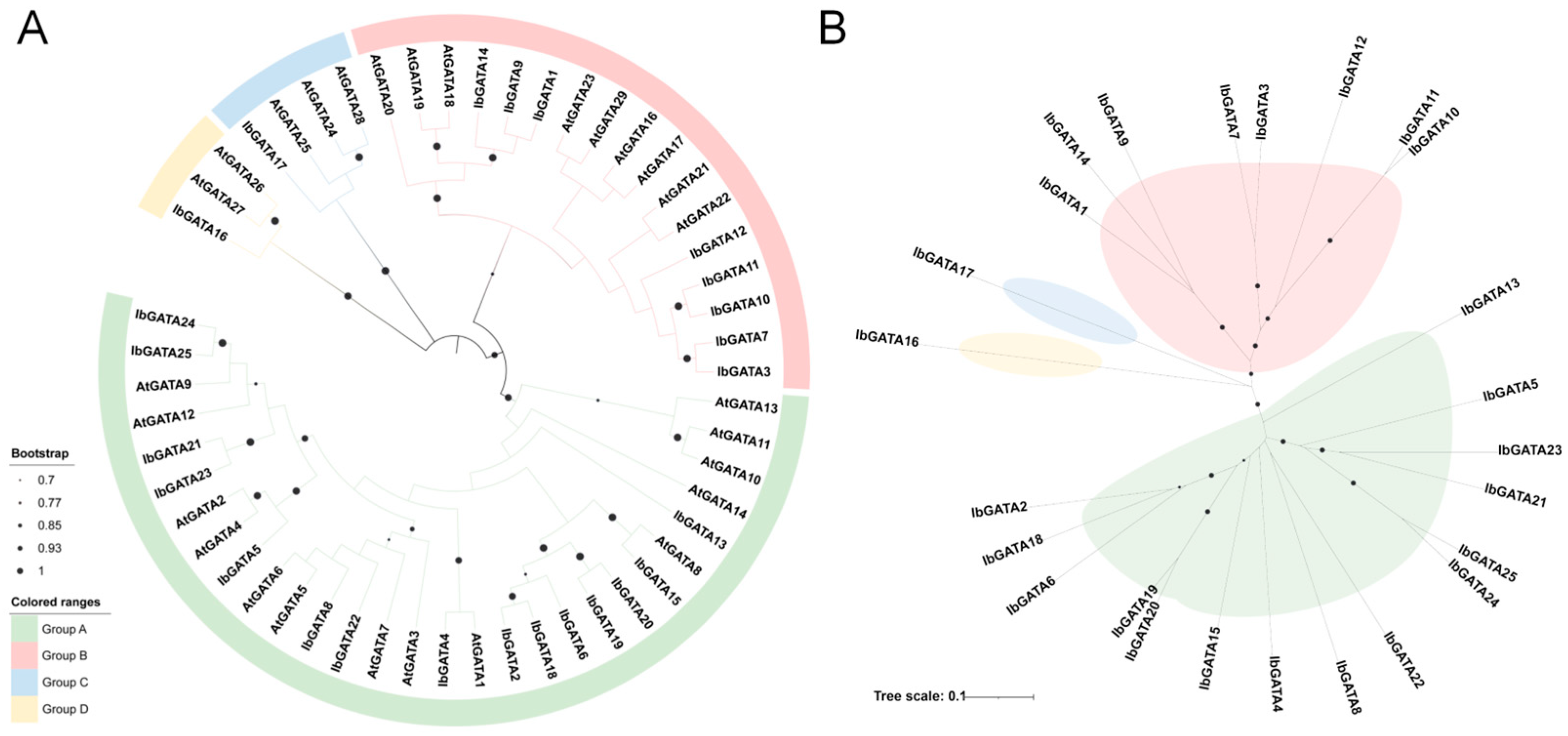
3.3. Cluster Analysis of GATA Family Genes in Sweetpotato
In order to achieve complete clarification of the genetic relationship and biological function of sweetpotato GATA family genes, a phylogenetic tree was constructed by clustering the identified GATA family members by multiple sequence alignment (Figure 2). The classification results of the A. thaliana GATAs revealed the presence of four distinct subgroups, designated Groups A, B, C, and D, according to the established nomenclature. The distribution of GATAs across the phylogenetic tree was found to be uneven. The largest number of sweetpotato GATAs was found in Group A (fifteen), followed by Group B with eight. Groups C and D contained one each. A comparison of GATAs within analogous subgroups reveals a striking similarity in their structural and functional characteristics. Consequently, the biological function of the GATA gene in sweetpotato can be deduced from the results of analogous genes in A. thaliana. Phylogenetic analysis of the GATA gene family revealed distinct evolutionary patterns, with Group A genes (GATA2, GATA18, and GATA6) forming a monophyletic cluster suggestive of common ancestry through gene duplication, potentially followed by functional conservation or divergence. Notably, GATA16 and GATA17 exhibited elongated branches indicative of accelerated evolution, possibly due to relaxed purifying selection or positive selection for novel functions, while Group B genes (GATA10 and GATA11) showed shorter branches consistent with strong functional constraints or recent duplication events. These findings collectively demonstrate the complex evolutionary history of sweetpotato GATA genes, involving duplication events, differential selection pressures, and functional diversification.
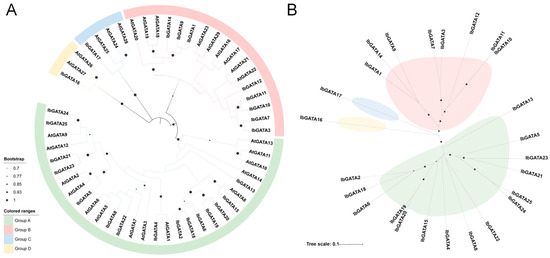
Figure 2.
Phylogenetic analysis of IbGATA proteins. (A) Phylogenetic tree of IbGATAs without evolutionary rates. (B) Phylogenetic tree of IbGATAs with evolutionary rates. The length of branches represents the evolutionary distance.
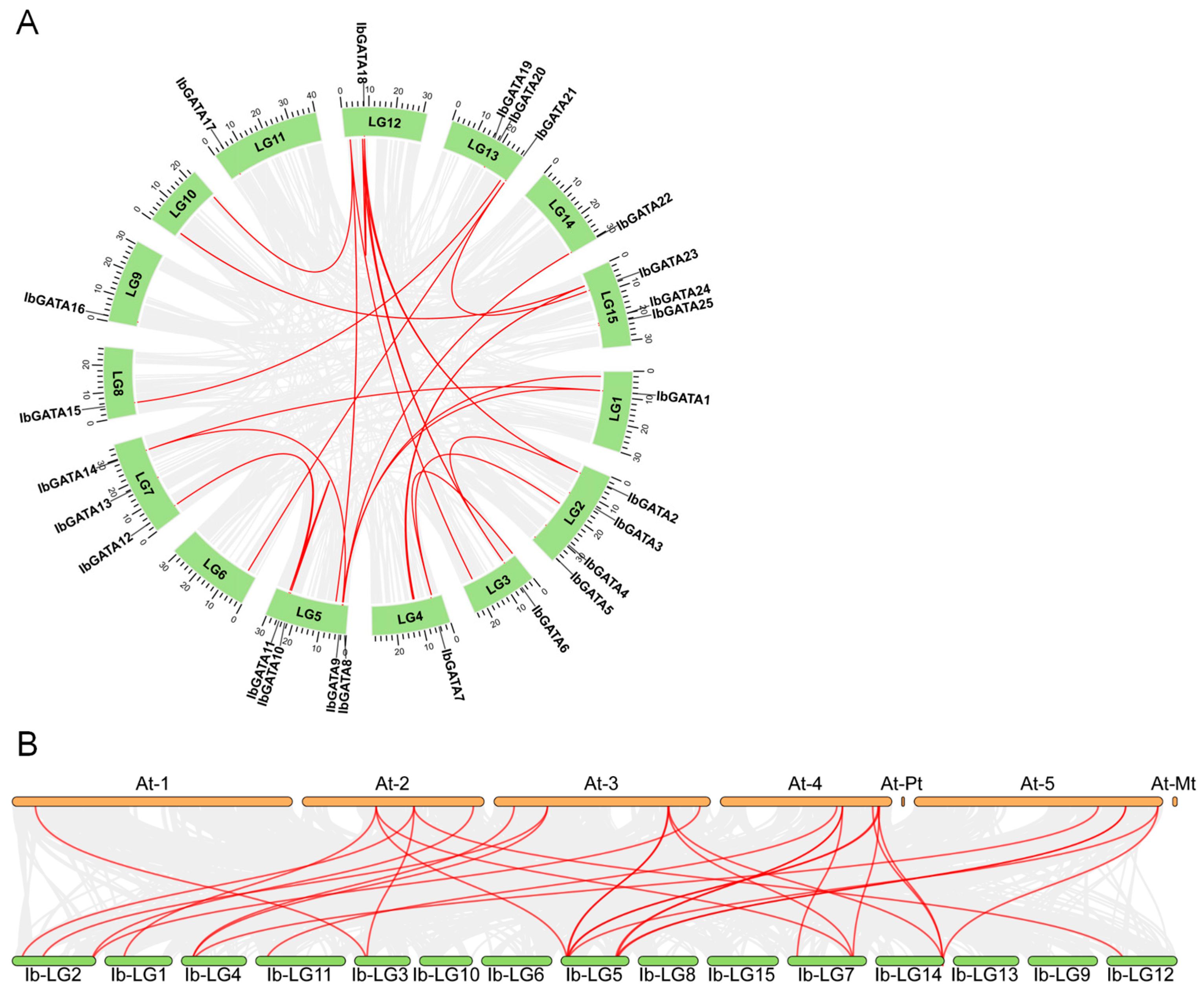
3.4. Evolutionary Dynamics of the IbGATAs Gene Family in Sweetpotato
To investigate the evolutionary mechanisms of the IbGATAs gene family in sweetpotato, a genome-wide collinearity analysis was performed. The results identified 13 collinear IbGATAs genes (Figure 3A). Notably, five genes (IbGATA1, IbGATA9, IbGATA11, IbGATA14, and IbGATA18) showed evidence of at least two duplication events, suggesting recurrent gene duplication during evolution. These duplications may have arisen through whole-genome duplication, segmental duplication, or tandem repeats, potentially driving functional diversification. The repeated duplication of these genes implies their important roles in sweetpotato evolution and development, possibly through mechanisms like neofunctionalization or subfunctionalization.
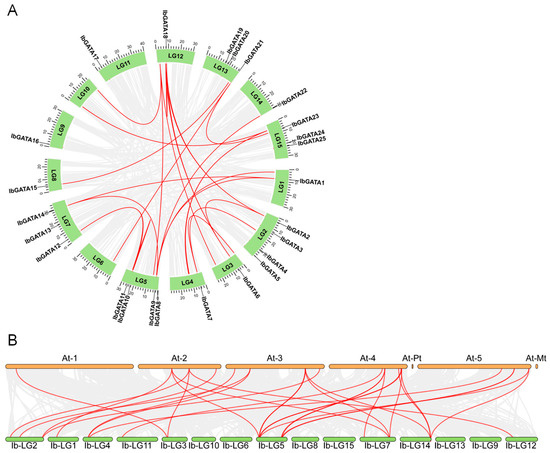
Figure 3.
Collinearity analysis. (A) Syntenic relationships of GATA gene family in sweetpotato. The red lines represent syntenic GATA genes. (B) Interspecies collinearity of sweetpotato and A. thaliana. The red lines represent syntenic GATA genes.
To elucidate the evolutionary mechanisms of the IbGATA gene family, we performed a comparative collinearity analysis between sweetpotato and A. thaliana (Figure 3B). Our analysis revealed 29 collinear relationships between 15 sweetpotato IbGATA genes (IbGATA1, IbGATA2, IbGATA3, IbGATA5, IbGATA6, IbGATA7, IbGATA8, IbGATA9, IbGATA10, IbGATA11, IbGATA12, IbGATA14, IbGATA17, IbGATA18, and IbGATA22) and 15 A. thaliana GATA genes (AtGATA2, AtGATA3, AtGATA4, AtGATA5, AtGATA6, AtGATA7, AtGATA10, AtGATA13, AtGATA16, AtGATA17, AtGATA18, AtGATA19, AtGATA21, AtGATA22, and AtGATA25).
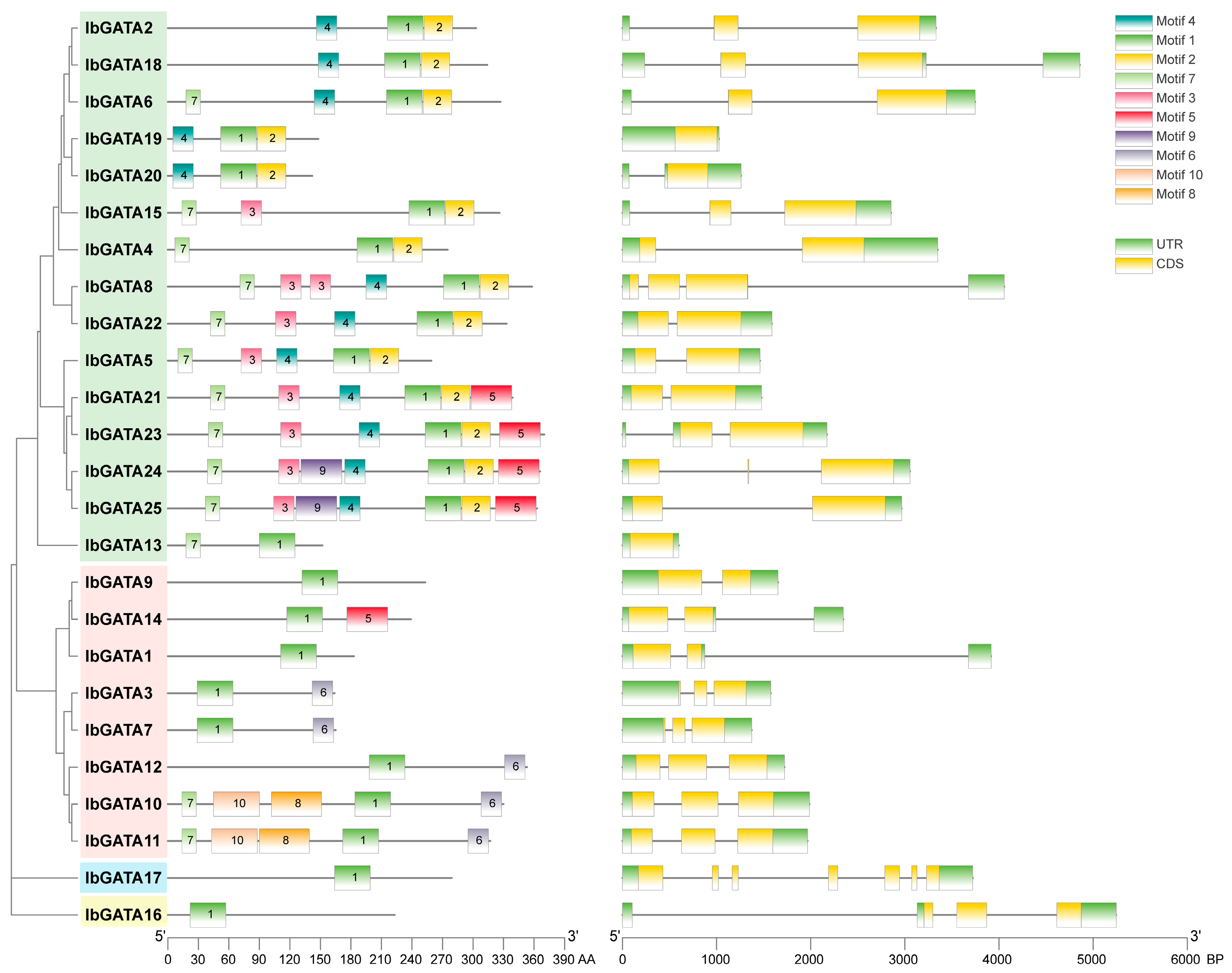
3.5. Motifs of IbGATAs and Exon–Intron Analysis of IbGATAs Genes
The structural architecture of a gene, including its regulatory elements, exon–intron organization and splicing patterns, and the three-dimensional conformation of the protein it encodes, are critical determinants of protein functionality. These factors influence stability, interaction capacity, and subcellular localization [48]. The structure of genes and protein conformation are key determinants of protein function. In order to further understand the GATA protein, motif and gene structure analyses were performed. Sequence motif analysis showed that there were one to ten different motifs in the IbGATAs protein family and the distributions of motifs in different groups showed high degrees of similarity (Figure 4). All of the groups contained Motif 1, most genes in group A contained Motif 4 and Motif 7, most genes in group B contained Motif 6, and genes in groups C and D contained only motif 1. Among them, IbGATA24 and IbGATA25 have the most with seven motifs, while IbGATA1, IbGATA9, IbGATA16, and IbGATA17 have only one motif. This suggests that Motif 1 is the most important motif for GATAs protein function and that different groups contain different conserved motifs which may be related to their different functions. These results also further illustrate the accuracy of phylogenetic analysis.
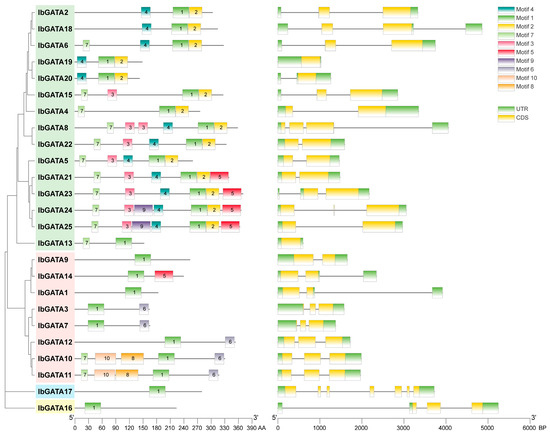
Figure 4.
Domains and gene structure of GATAs proteins in sweetpotato.
The exon–intron structures and motif compositions of sweetpotato GATA family members were subjected to visualization. An analysis of the exon–intron structures of IbGATAs genes was conducted, revealing a range of exon numbers from one to seven, and a range of intron numbers from one to six. Additionally, all GATA genes have UTR structures (Figure 4). Overall, there is significant variation in gene structures among different categories. Proteins with close phylogenetic relationships exhibit similar gene structures, such as nine out of fifteen members of group A of GATA genes having two exons and one intron. In group B, five out of eight genes have three exons and two introns, while the remaining two members, which are located on a separate branch of the phylogenetic tree, possess two exons and one intron. The IbGATA17 gene contains the most exons and introns, with seven and six. It indicates that the gene structure of this gene family is relatively stable.
3.6. Cis-Acting Elements of IbGATA Gene Promoters
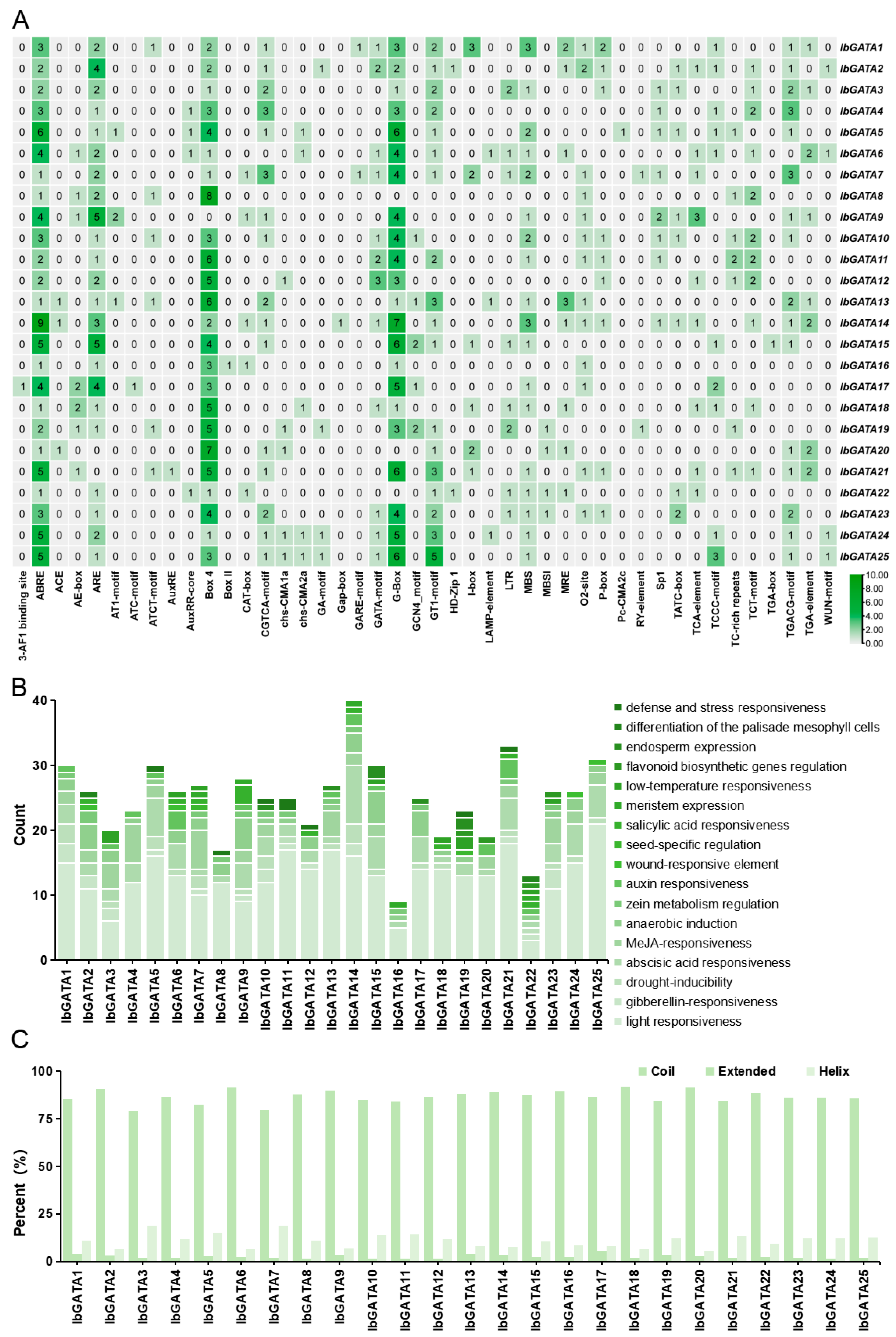
The regulation of gene expression in hormone signals and abiotic stress responses is typically associated with factors related to the spatial distribution of relevant elements in gene promoters [2]. To elucidate the regulatory mechanisms of IbGATA family genes in sweetpotato growth, development, and stress responses, we employed PlantCARE for the comprehensive analysis of cis-acting elements within the 2000 base pair promoter regions of the 25 IbGATA genes (Figure 5A). The results showed that promoters of GATAs contained a variety of light-responsive elements such as G-box (TACGAT) and Box 4 (ATTAAT) and resistance-related elements include abscisic acid (ABA)-responsive element ABRE (ACGTG), antioxidant-response element ARE (AAACCA), and defense-response element WUN-motif (TAATTACTC).
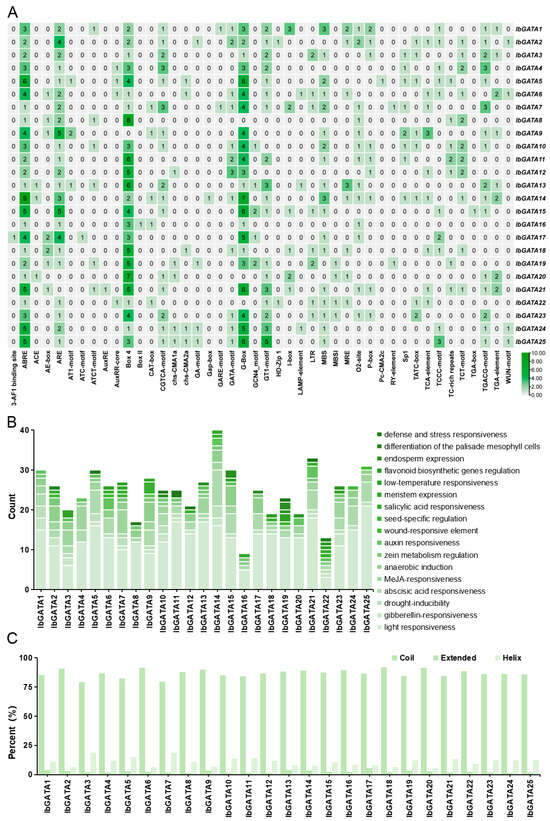
Figure 5.
Analysis of GATA gene promoters. (A) Distribution of cis-acting elements in the promoters of sweetpotato GATA genes. The degree of green color represents the number of cis-acting elements upstream of the GATAs. (B) Functional categorization and number of cis- acting elements in the promoters of sweetpotato GATA genes. (C) Secondary structure analysis of the GATA protein.
Subsequently, the number of elements present in specific biological pathways was counted. The results indicated that light responsiveness was the most widely regulated feature. Following these are resistance-related elements, including abscisic acid (ABA) responsiveness, methyl jasmonate (MeJA) responsiveness, and anaerobic induction. IbGATA14 exhibits the highest number of ABA-responsiveness elements, with a total of nine, and IbGATA7 has the highest number of MeJA-responsiveness elements, with a total of six (Figure 5B). This suggests that IbGATAs may be involved in plant adversity stress pathways.
3.7. Significant Variations Are Evident in the Secondary Structures of GATA Proteins in Sweetpotato
In order to provide further elucidation on the structural characteristics of the GATA protein in sweetpotato, an analysis was conducted on its secondary structure. The results demonstrated that the proportion of ‘coil’ exceeded 79% for all proteins; this was the most prevalent secondary structure. The extended structures were found to be minimal, ranging from 1 to 6%. The proportion of helical structures was found to be intermediate, ranging from 6 to 19%, thus occupying a position between coil and extended structures (Figure 5C).
3.8. Protein Interaction Network of IbGATAs in Sweetpotato
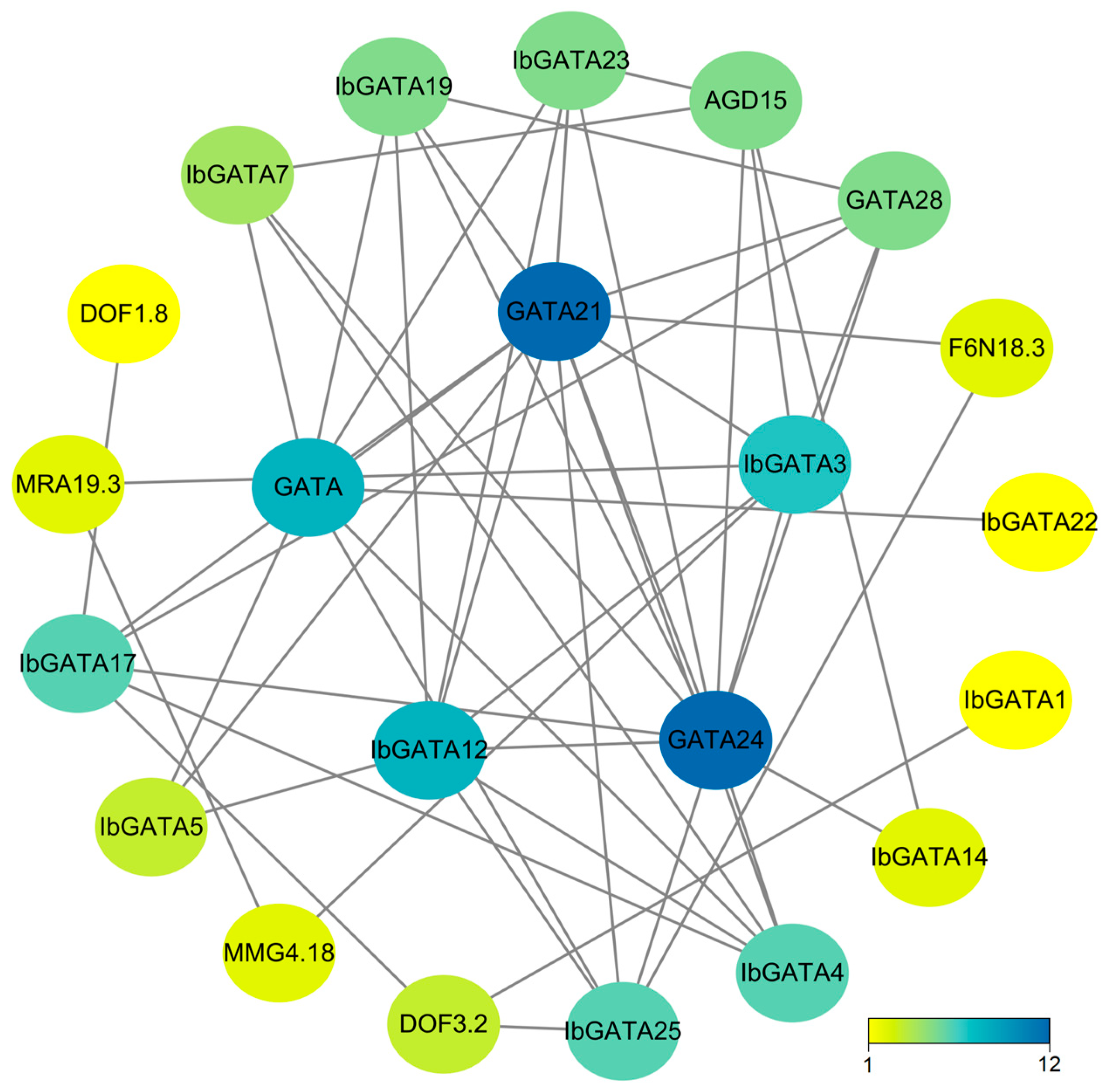
To delve deeper into the functions of the 25 IbGATA genes in sweetpotato, we utilized STRING 12.0 to compare IbGATAs with A. thaliana and identify homologous genes, thereby predicting the protein interaction network. The analysis of this network reveals that core proteins function in diverse aspects of plant biology. As indicated by protein–protein interaction (PPI) analysis, nine interactions were identified among the 25 sweetpotato GATA proteins (Figure 6). It can also interact with a variety of transcription factors. Among the proteins under consideration, IbGATA21 and IbGATA24 exhibit the most complex interaction networks. It has been established that there are a total of 12 interacting genes. By analyzing the protein interaction network, we can speculate that GATA proteins may play key roles in plant responses to abiotic and biotic stress. These findings provide a valuable basis for future functional studies of sweetpotato GATA genes.
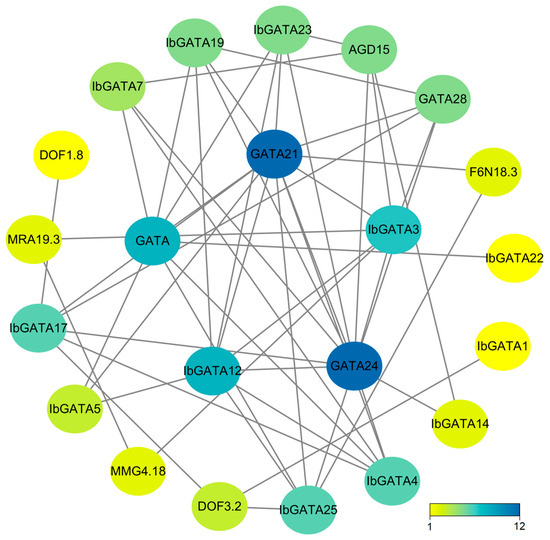
Figure 6.
The protein interaction network of IbGATAs. The nodes in the figure represent proteins, with the gray lines denoting the interactions between them. The color of the nodes is indicative of the number of interacting proteins.
3.9. Tissue-Specific Expression Analysis of IbGATAs
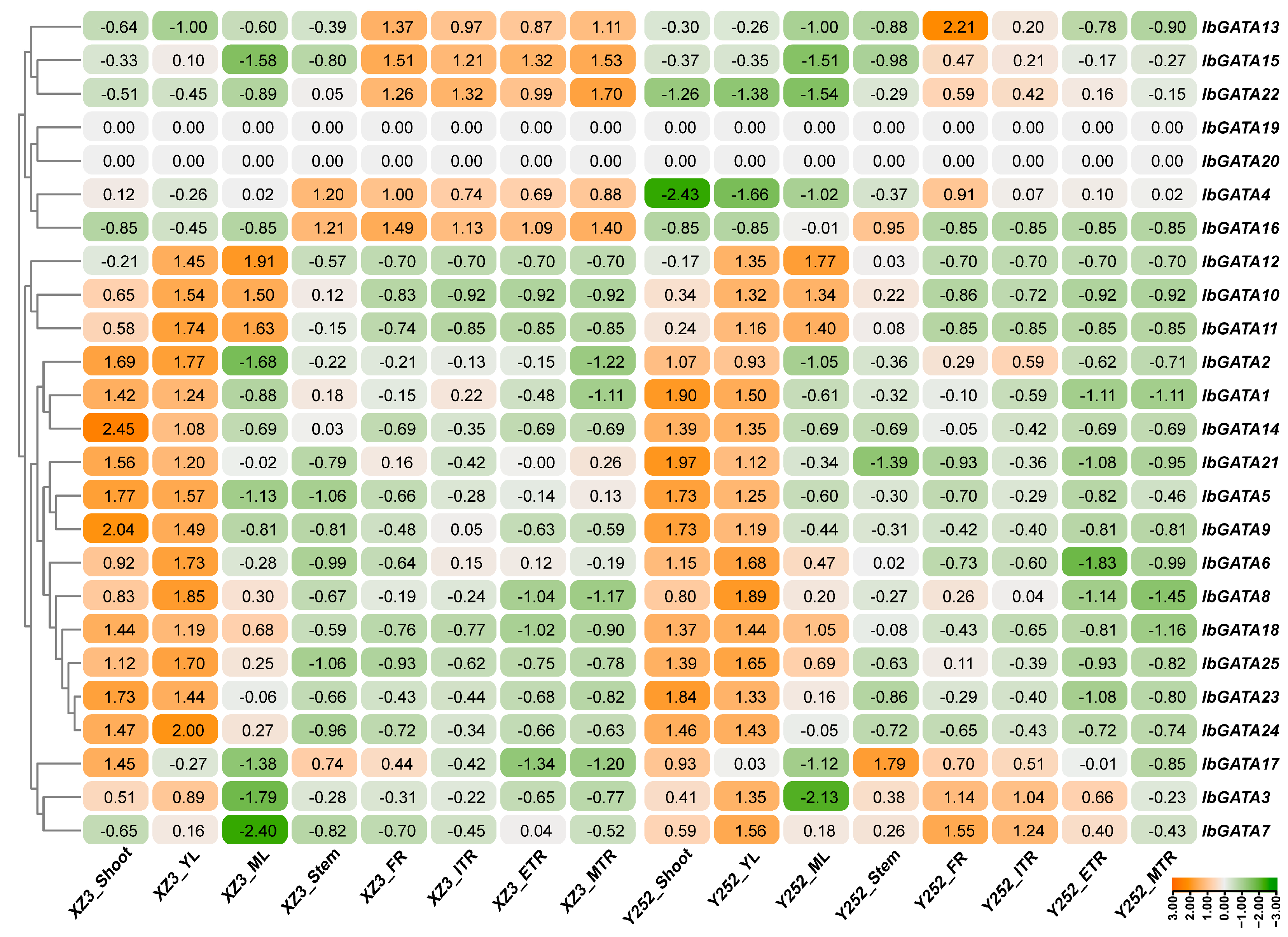
In order to explore the biological function of IbGATAs in sweetpotato, the expression of IbGATAs was detected in eight representative sweetpotato tissues (i.e., fibrous root (FR), initial tuberous root (ITR), expanding tuberous root (ETR), mature tuberous root (MTR), stem, shoot, young leaf (YL), and mature leaf (ML)) using RNA-seq data of sweetpotato varieties Xuzi3 and Yan252. IbGATAs expression were observed in different sweetpotato tissues, showing tissue-specific expression in a few of the IbGATAs such as IbGATA13, 15, and 22 (highly expressed in the FR, ITR, ETR, and MTR tissues), and IbGATA10, 11, and 12 (highly expressed in YL and ML tissues) (Figure 7). Most importantly, these findings suggest that IbGATAs have a different pattern of expression and play a role in different tissues during the growth and development of sweetpotato.
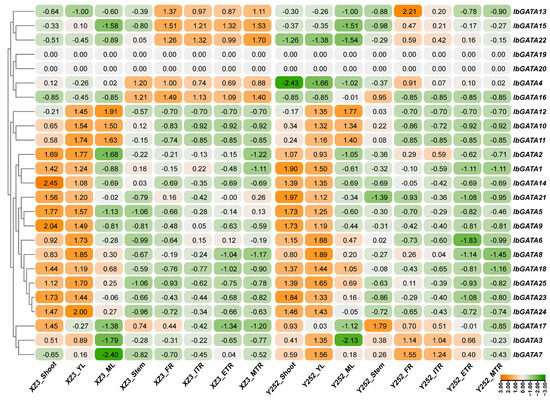
Figure 7.
Expression levels of IbGATAs in different tissue from RNA-seq data. The log2 (FPKM + 1) values are displayed within the confines of the designated boxes. Higher transcript levels are shown in orange (0 to 3) and lower transcript levels are shown in green (−3 to 0).
3.10. The Analysis of Induction of Abiotic Stress Gene Expression
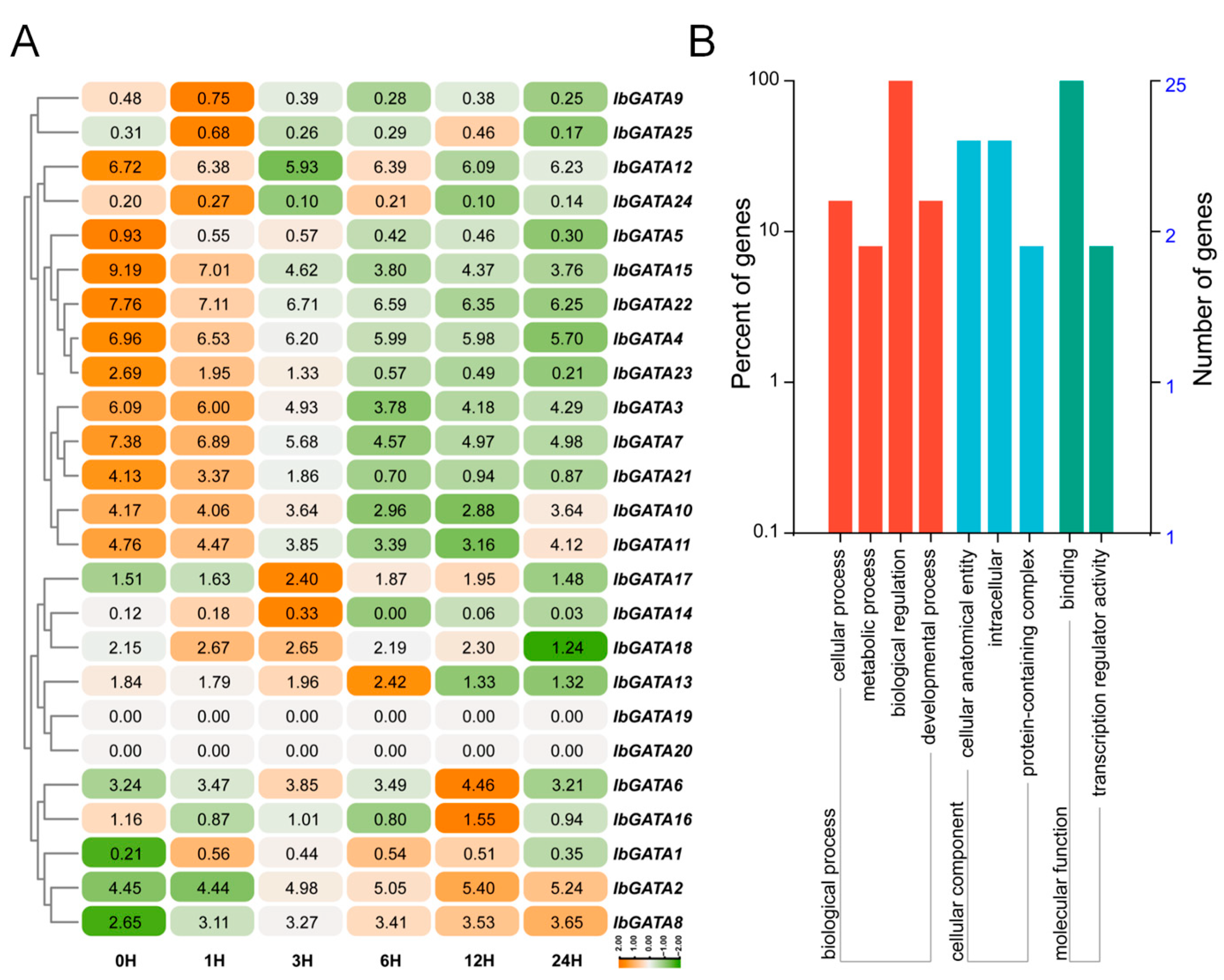
The growth and development of plants are limited by abiotic stress. To investigate the responses of GATA family genes to abiotic stress, we analyzed RNA-seq data from the sweetpotato line Xu55-2 subjected to 30% PEG treatment (Figure 8A). Transcriptome analysis revealed differential expression among IbGATA genes, with significantly more upregulated than downregulated genes. Specifically, IbGATA6, IbGATA13, IbGATA14, IbGATA16, and IbGATA17 were markedly upregulated under PEG treatment, whereas IbGATA4, IbGATA7, IbGATA10, IbGATA15, and IbGATA22 were downregulated. These genes were involved in cellular progress, metabolic progress, biological progress and developmental progress, etc. (Figure 8B). Subsequently, an analysis of the gene expression was conducted by means of qRT-PCR. The results obtained were consistent with the trend observed in the transcriptome (Figure 9A and Figure S1). These results suggest that IbGATA genes play a role in sweetpotato’s drought tolerance, potentially regulating this adaptive process.
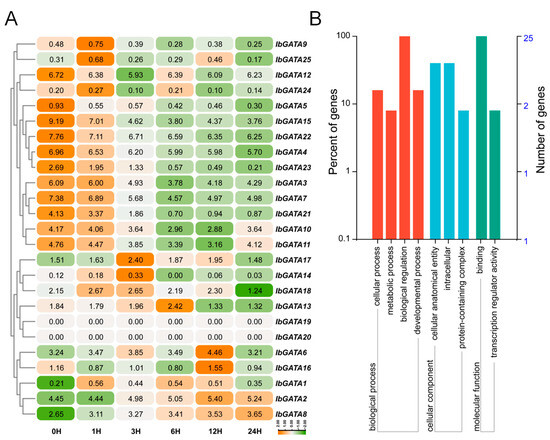
Figure 8.
Expression analysis of IbGATAs under 30% PEG6000 treatment in sweetpotato. (A) Heat map of the expression. The log2 (FPKM + 1) values are shown in boxes. Higher transcript levels are shown in orange (0 to 2) and lower transcript levels are shown in green (−2 to 0). (B) GO-enrichment analysis of IbGATA genes differentially expressed under 30% PEG treatment. Biological process, molecular function and cellular component categories are shown.
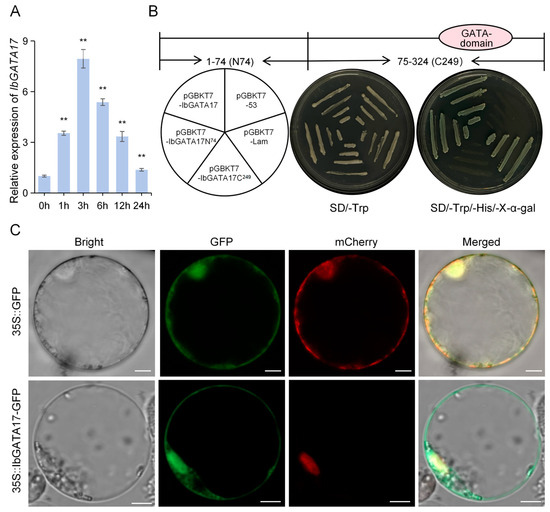
Figure 9.
Expression analysis. Transcriptional-activation analysis and subcellular localization of IbGATA17. (A) Expression analysis of IbGATA17 in sweetpotato upon PEG6000 over a 24 h period. Data are shown as mean ± SD (n = 3). ** indicate significant differences from that of WT at p < 0.01, based on Student’s t-test. The sweetpotato IbACTIN gene was used as a reference. (B) Transcriptional-activation assay of IbGATA17. Fusion proteins of the GAL4 DNA-binding domain and different portions of IbPIF1 were expressed in yeast strain Y2H and examined on SD/-Trp and SD/-Trp/-His/X-a-gal selection media. (C) Subcellular localization of IbGATA17. Rice protoplasts cells were transformed with the fusion construct (IbGATA17-GFP) and the nuclear marker NLS-mCherry. Bars = 5 μm.
3.11. IbGATA17 Acts as a Pivotal Modulator in Drought-Stress Responses
The IbGATA17 gene demonstrated a significant increase in expression in response to drought treatment, suggesting a potential role in enhancing plant resilience against abiotic stresses. The relative transcript levels of IbGATA17 were quantified via qRT-PCR in sweetpotato under drought treatment. Under drought treatments, the expression levels of IbGATA17 were upregulated nearly 7.94-fold (at 3 h) in sweetpotato (Figure 9A).
The IbGATA17 gene demonstrated a significant increase in expression in response to drought treatment, suggesting a potential role in enhancing plant resilience against abiotic stresses. The relative transcript levels of IbGATA17 were quantified via qRT-PCR in sweetpotato under drought treatment. Under drought treatments, the expression levels of IbGATA17 were upregulated nearly 7.94-fold (at 3 h) in sweetpotato (Figure 9A).In order to identify potential regulators of IbGATA17, the gene was cloned. The protein is composed of 279 amino acids (aa) and is predicted to have a molecular weight of 30.55 kD. This is encoded by an 837-base-pair open reading frame (ORF). In order to investigate the transcription-activation activity of IbGATA17, three fragments encoding the full-length protein, the N-terminal 74aa residue (IbGATA17N74), and the C-terminal 249aa residue (IbGATA17C249) into the GAL4 pGBKT7 vector were constructed. Subsequent to this, the resulting fusion constructs were then transformed into a Y2H Gold yeast strain (Figure 9B). The experimental results demonstrated that yeast transformants carrying BD-IbGATA17 and BD-IbGATA17N74 exhibited robust growth on SD/-Trp/-His/X-α-Gal medium, accompanied by the manifestation of blue fluorescence. In vivo plate assays confirmed that IbGATA17 possesses transcription-activation activity, with its transcription-activation activity located in the N-terminal region of 74 aa residues. The present findings demonstrate that IbGATA17 functions as a transcription activator (Figure 9B). In order to ascertain the subcellular localization of IbGATA17, the open reading frame of IbGATA17 was fused with GFP under the control of the 35S promoter to construct the IbGATA17-GFP fusion protein. The transient expression of this fusion protein in rice protoplasts was achieved via polyethylene glycol–calcium-mediated transformation. Confocal microscopy revealed that green fluorescence emitted by IbGATA17-GFP was localized to the nucleus and cell membrane (Figure 9C).
3.12. IbGATA17 Enhances Drought Tolerance of Sweetpotato
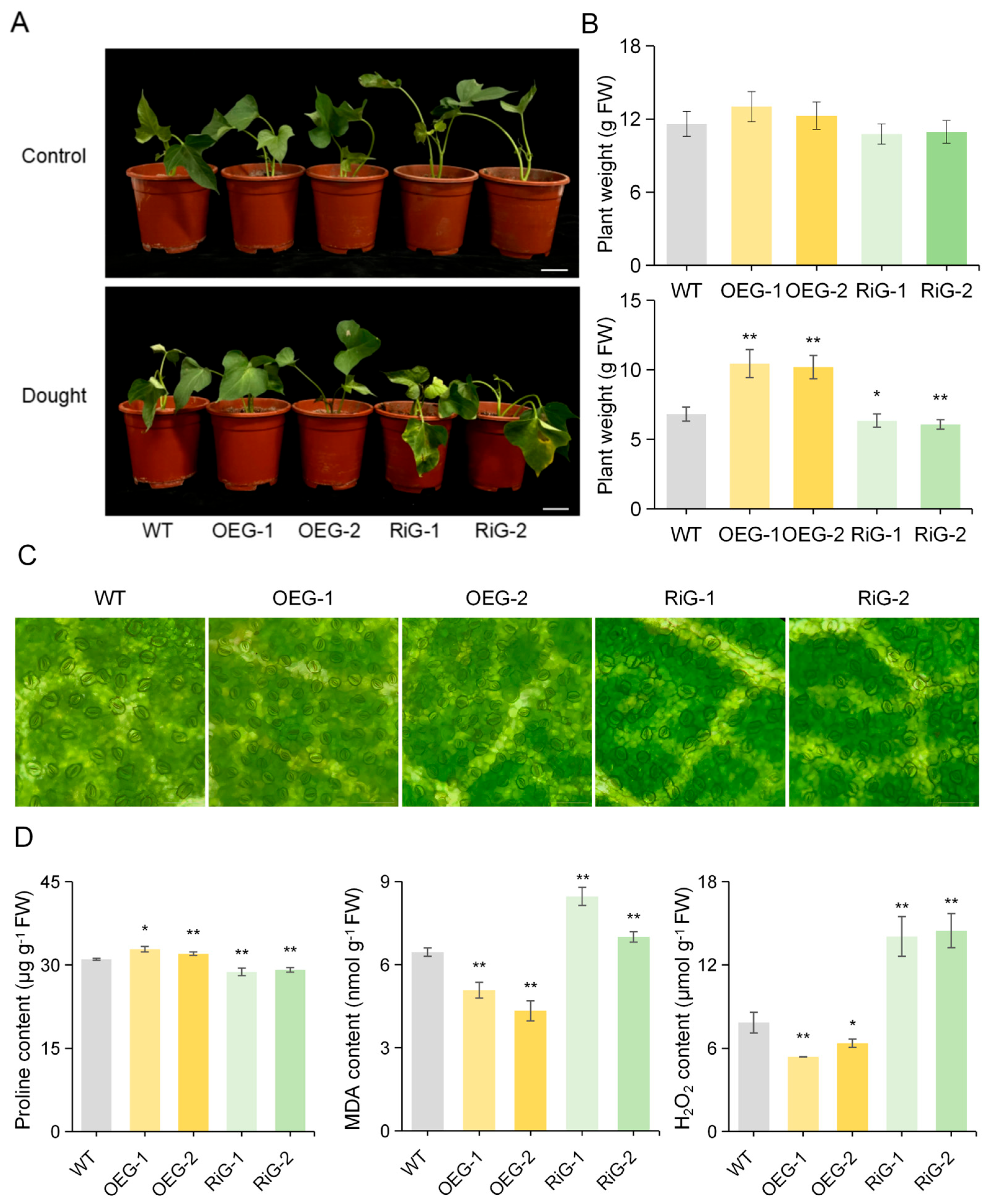
The objective of this study was to ascertain whether IbGATA17 exerts an influence on the drought response of sweetpotato. To this end, we generated overexpressing (OE) lines (designated as OEG-1 and OEG-2) and resistance introgression (Ri) lines (designated as RiG-1 and RiG-2) via Agrobacterium-rhizogenes-mediated transformation for the purpose of conducting drought tolerance assays (Figure S2). In the control condition, no discernible morphological differences were observed between the transgenic and WT plants. After 48 h of PEG treatment, the IbGATA17-OE plants exhibited better growth, whereas their IbGATA17-Ri lines became wilt earlier than that of the WT plants (Figure 10A,B). We observed the stomatal openings of both transgenic and wild-type plants. We found that IbGATA17-OE promoted stomatal contraction, while IbGATA17-Ri exhibited the opposite effect (Figure 10C and Figure S3). A significantly higher level of proline content was observed in IbGATA17-OE than in WT plants, while lower levels were observed in IbGATA17-Ri (Figure 10D). Furthermore, lower levels of H2O2 and MDA accumulation were found in IbGATA17-OE, while higher levels were found in IbGATA17-Ri. These results reveal that the overexpression of IbGATA17 activates the sweetpotato’s ROS scavenging system.
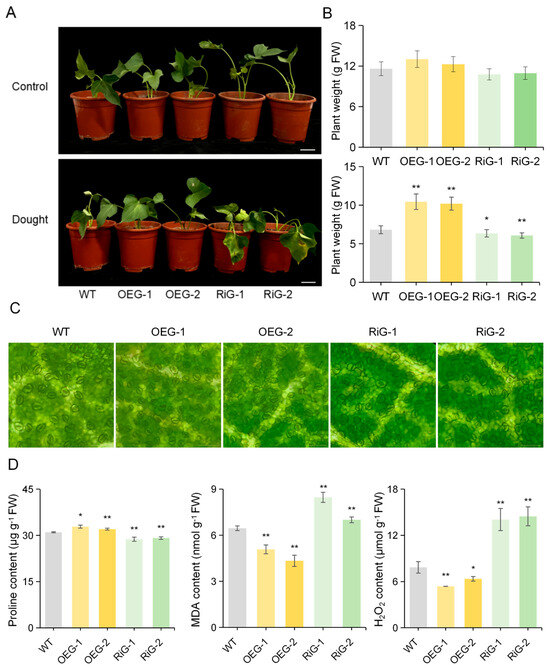
Figure 10.
Overexpression of IbGATA17 enhanced drought tolerance in sweetpotato. (A) Responses of IbGATA17 transgenic and WT sweetpotato plants. Scale bars = 5 cm. (B) Plant weight grown in a flowerpot under normal conditions (control) or subjected to PEG stress for 48 h. Data are shown as mean ± SD (n = 3). * and ** indicate significant differences from that of WT at p < 0.05 and p < 0.01, based on Student’s t-test. (C) Stomatal aperture of the leaves of transgenic and WT plants. Scale bars = 100 μm. (D) Proline content, MDA content and H2O2 content in leaves of IbGATA17 transgenic and WT plants. Data are shown as mean ± SD (n = 3). * and ** indicate significant differences compared to the WT at p < 0.05 and p < 0.01, based on Student’s t-test.
4. Discussion
GATA TFs are type IV zinc finger DNA-binding proteins which play a crucial role in diverse physiological processes of plant growth, development, and responses to abiotic stresses. Research has demonstrated that the GATA family genes are widely distributed throughout plants species, including 28 GATA genes in Oryza sativa [23], 30 in Lycopersicon esculentum [49], 49 in Solanum tuberosum [50], 64 in Glycine max [51], and 37 in Zea mays [52]. Nevertheless, the intricate workings and the core regulatory systems of IbGATAs remain largely unknown. Consequently, a comprehensive identification and functional study of the GATA gene family at the genome-wide level is imperative to enhance our understanding of their functions and the molecular intricacies underlying sweetpotato development.
In this study, a total of 25 IbGATAs were identified in the sweetpotato (I. batatas) genome. Chromosomal localization showed an uneven distribution of these genes, with chromosomes 2 and 5 harboring the highest numbers (Figure 1). Phylogenetic analysis classified these genes into four subgroups (A, B, C, and D), with subgroup A containing the largest number of genes (Figure 2). This classification is consistent with findings in A. thaliana [24] and Zea mays [52]. The distribution suggests functional diversification within the family, which is supported by the presence of conserved motifs, such as Motif 1, that appear to be critical for GATA protein function. Gene structure analysis revealed variability in exon–intron organization; notably, IbGATA17 exhibited the most complex structure (seven exons and six introns). This structural diversity may reflect adaptive evolution, enabling GATA genes to acquire specialized roles in the sweetpotato.
Synteny analysis provides key insights into the mechanisms of whole-genome duplication, chromosomal rearrangement, and the functional divergence of gene families [53]. In this study, we found that there are thirteen collinear genes, including five (IbGATA1, IbGATA9, IbGATA11, IbGATA14, and IbGATA18) that have undergone duplication in the sweetpotato genome (Figure 3A). These results suggest that collinear genes have played a key role in the expansion and functional diversification of the GATA family in sweetpotato, potentially contributing to its adaptation to environmental stresses.
Cis-acting elements are defined as transcription factor binding sites that regulate the precise initiation and efficiency of gene transcription [54]. In tomato, SlHY5 suppresses the expression of SlGATA17 by binding to the GATA-box in its promoter, thereby reducing the salt tolerance of tomato [55]. Similarly, BdGATA13 influences primary root development in in Arabidopsis under gibberellic acid (GA) treatment [56]. In this study, we identified a multitude of cis-acting elements associated with abiotic stress, hormone signaling, and stress responses in the promoter regions of IbGATAs. These results suggest that IbGATAs are regulated by complex networks that integrate environmental and developmental cues (Figure 5).
Gene expression patterns are related to gene function to some extent. In wheat, TaGATAs showed significantly different expression patterns across different tissues and under various abiotic stress conditions [25]. Similarly, in potato, StGATA genes showed tissue-specific and stress-responsive expression patterns, with StGATA12 overexpression enhancing tolerance to salinity and osmotic stresses [50]. In our study, IbGATAs were differentially expressed in different tissues (Figure 7) and IbGATA6, IbGATA13, IbGATA14, IbGATA16, and IbGATA17 were significantly upregulated after PEG treatment (Figure 8A). These results suggest that different IbGATA genes may play different roles in different tissues and participate in drought responses to improve drought tolerance in sweetpotato.
The GATA family genes play a crucial role in various physiological processes during plant growth and development [29,30,31,32,33,34,57], and responses to abiotic stresses such as cold, hot, salt, and drought [35,50,58,59]. At present, there are few reports on the drought-resistance function of the GATA gene. Guo et al. cloned the gene BdGATA13 in Brachypodium distachyon and overexpressed it in Arabidopsis to enhance drought tolerance [56]. Guo et al. reported that SlGATA17, which is localized in the nucleus, functions to regulate drought tolerance in tomato through the phenylpropanoid biosynthetic pathway [49]. In this study, we found that the IbGATA17 was significantly upregulated under drought stress, with its expression increasing nearly 8-fold after 3 h of PEG treatment. This strong induction implicates IbGATA17 as a key player in sweetpotato’s drought response (Figure 9A). Subcellular localization analysis showed that IbGATA17 was localized in both the nucleus and the cell membrane (Figure 9C). Under simulated drought conditions, IbGATA17 overexpression enhanced growth vigor in transgenic sweetpotato plants compared to WT, whereas IbGATA17 RNAi lines exhibited the opposite phenotype (Figure 10A-C). Further analysis revealed that IbGATA17 likely enhances drought tolerance in transgenic sweetpotato by modulating stomatal regulation, proline biosynthesis, and reactive oxygen species (ROS) scavenging (Figure 10D). These results suggest that the regulatory mechanisms of GATA17 genes in response to drought stress may differ across plant species.
Sweetpotato is a vital crop for food security, particularly in regions prone to drought. We used whole-genome identification, combined with transcriptome data analysis and qRT-PCR experiments to explore the potential drought tolerance genes in sweetpotato. At present, no GATA17 drought tolerance reports have been reported in sweetpotato. Our identification of IbGATA17 as a drought-responsive gene creates new opportunities for the genetic improvement of sweetpotato. Targeting IbGATA17 or its regulatory network could enable the development of cultivars with enhanced drought tolerance, thereby contributing to sustainable agriculture in the face of climate change. While this study establishes a fundamental understanding of IbGATA genes in sweetpotatoes, further research is required to elucidate the precise mechanisms through which IbGATA17 and other family members mediate stress responses. Functional studies, such as CRISPR-based gene editing or detailed protein interaction analyses, could deepen our understanding of their roles. Furthermore, investigating the interactions between IbGATAs and other stress-responsive pathways would be valuable.
In summary, we present genome-wide results for sweetpotato GATA genes, alongside analyses of their expression patterns under drought conditions. Additionally, we performed a functional analysis of IbGATA17 and demonstrated that it can improve the drought tolerance of sweetpotato. This information could contribute to future research investigating the IbGATAs gene family, especially IbGATA17 in relation to drought tolerance.
5. Conclusions
In this study, we used whole-genome identification and analysis techniques to elucidate the pivotal functions of the GATA gene family in the evolution of sweetpotato, the intricacies of their regulatory mechanisms, and their capacity for drought tolerance. The research identified genes that may be involved in the regulation of drought stress, with the IbGATA17 gene demonstrating enhanced drought tolerance in transgenic sweetpotato plants under simulated drought conditions. These findings provide a basis for the identification of candidate genes that may be useful in the development of drought-resistant sweetpotato varieties, and they also serve as markers that can be utilized in genome-assisted crop improvement.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/genes16101237/s1. Table S1: Sequences of the primers used in this study; Figure S1: Analysis of the gene expressions associated with PEG6000 over a 24 h period; Figure S2: Production of transgenic sweetpotato plants overexpressing and interfering with the IbGATA17 gene; Figure S3: Stomatal aperture of the leaves.
Author Contributions
Conceptualization, H.Z. (Hong Zhai) and Y.Y.; methodology, Y.Y. and R.L.; software, Y.Y. and R.L.; validation, H.Z. (Hong Zhai) and Y.Y.; formal analysis, Y.Y.; investigation, Y.Y. and R.L.; data curation, Y.Y. and R.L.; writing—original draft preparation, Y.Y.; writing—review and editing, H.Z. (Hong Zhai) and Y.Y.; visualization, Y.Y. and R.L.; supervision, H.Z. (Hong Zhai), Q.L., S.H., S.G., and N.Z.; project administration, H.Z. (Hong Zhai), Q.L., S.H., and H.Z. (Huan Zhang).; funding acquisition, H.Z. (Hong Zhai). All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the earmarked fund for CARS-10-Sweetpotato, the National Natural Science Foundation of China (32472120), and the 2115 Talent Development Program of China Agricultural University.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data in this study are available in this article or in the Supplementary Materials.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Farooq, M.; Hussain, M.; Wahid, A.; Siddique, K.H.M. Drought Stress in Plants: An Overview. In Plant Responses to Drought Stress; Aroca, R., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–33. [Google Scholar]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef]
- Dai, A. Increasing drought under global warming in observations and models. Nature Clim. Change 2013, 3, 52–58. [Google Scholar] [CrossRef]
- Chaves, M.M.; Maroco, J.P.; Pereira, J.S. Understanding plant responses to drought—from genes to the whole plant. Funct. Plant Biol. 2003, 30, 239–264. [Google Scholar] [CrossRef]
- Lamaoui, M.; Jemo, M.; Datla, R.; Bekkaoui, F. Heat and Drought Stresses in Crops and Approaches for Their Mitigation. Front. Chem. 2018, 6, 26. [Google Scholar] [CrossRef]
- Bovell-Benjamin, A.C. Sweet potato: A review of its past, present, and future role in human nutrition. Adv. Food Nutr. Res. 2007, 52, 1–59. [Google Scholar]
- Alam, M.K. A comprehensive review of sweet potato (Ipomoea batatas [L.] Lam): Revisiting the associated health benefits. Trends Food Sci. Tech. 2021, 115, 512–529. [Google Scholar] [CrossRef]
- Low, J.W.; Mwanga, R.O.; Andrade, M.; Carey, E.; Ball, A.-M. Tackling vitamin A deficiency with biofortified sweetpotato in sub-Saharan Africa. Glob. Food Sec. 2017, 14, 23–30. [Google Scholar] [CrossRef]
- Sohindji, F.S.; Quenum, F.J.-B.; Fassinou-Hotegni, N.V.; Adékounlé Oke, A.; Adje, C.O.A.; Achigan-Dako, E.G. Crossing possibility for breeding promising orange-fleshed sweetpotato genotypes in Benin. Czech J. Genet. Plant Breed. 2023, 59, 253–262. [Google Scholar] [CrossRef]
- Laveriano-Santos, E.P.; López-Yerena, A.; Jaime-Rodríguez, C.; González-Coria, J.; Lamuela-Raventós, R.M.; Vallverdú-Queralt, A.; Romanyà, J.; Pérez, M. Sweet Potato Is Not Simply an Abundant Food Crop: A Comprehensive Review of Its Phytochemical Constituents, Biological Activities, and the Effects of Processing. Antioxidants 2022, 11, 1648. [Google Scholar] [CrossRef]
- Lebot, V. Tropical Root and Tuber Crops: Cassava, Sweet Potato, Yams and Aroids, 2nd ed.; CABI: Boston, MA, USA, 2020. [Google Scholar]
- Andrade, M.I.; Naico, A.; Ricardo, J.; Eyzaguirre, R.; Makunde, G.S.; Ortiz, R.; Grüneberg, W.J. Genotype × environment interaction and selection for drought adaptation in sweetpotato (Ipomoea batatas [L.] Lam.) in Mozambique. Euphytica 2016, 209, 261–280. [Google Scholar] [CrossRef]
- Hartemink, A.E.; Johnston, M.; O’Sullivan, J.N.; Poloma, S. Nitrogen use efficiency of taro and sweet potato in the humid lowlands of Papua New Guinea. Agric. Ecosyst. Environ. 2020, 79, 271–280. [Google Scholar] [CrossRef]
- Dufresne, F.; Stift, M.; Vergilino, R.; Mable, B.K. Recent progress and challenges in population genetics of polyploid organisms: An overview of current state-of-the-art molecular and statistical tools. Mol. Ecol. 2014, 23, 40–69. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhao, H.; Xue, L.; Nie, N.; Zhang, H.; Zhao, N.; He, S.; Liu, Q.; Gao, S.; Zhai, H. IbMYC2 Contributes to Salt and Drought Stress Tolerance via Modulating Anthocyanin Accumulation and ROS-Scavenging System in Sweet Potato. Int. J. Mol. Sci. 2024, 25, 2096. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, X.; Zhi, Y.; Li, X.; Zhang, Q.; Niu, J.; Wang, J.; Zhai, H.; Zhao, N.; Li, J.; et al. A non-tandem CCCH-type zinc-finger protein, IbC3H18, functions as a nuclear transcriptional activator and enhances abiotic stress tolerance in sweet potato. New Phytol. 2019, 223, 1918–1936. [Google Scholar] [CrossRef]
- Sun, S.; Li, X.; Gao, S.; Nie, N.; Zhang, H.; Yang, Y.; He, S.; Liu, Q.; Zhai, H. A Novel WRKY Transcription Factor from Ipomoea trifida, ItfWRKY70, Confers Drought Tolerance in Sweet Potato. Int. J. Mol. Sci. 2020, 23, 686. [Google Scholar] [CrossRef]
- Xue, L.; Wei, Z.; Zhai, H.; Xing, S.; Wang, Y.; He, S.; Gao, S.; Zhao, N.; Zhang, H.; Liu, Q. The IbPYL8-IbbHLH66-IbbHLH118 complex mediates the abscisic acid-dependent drought response in sweet potato. New Phytol. 2022, 236, 2151–2171. [Google Scholar] [CrossRef]
- Meng, X.; Liu, S.; Zhang, C.; He, J.; Ma, D.; Wang, X.; Dong, T.; Guo, F.; Cai, J.; Long, T.; et al. The unique sweet potato NAC transcription factor IbNAC3 modulates combined salt and drought stresses. Plant Physiol. 2023, 191, 747–771. [Google Scholar] [CrossRef]
- Zhu, H.; Yang, X.; Wang, X.; Li, Q.; Guo, J.; Ma, T.; Zhao, C.; Tang, Y.; Qiao, L.; Wang, J.; et al. The sweetpotato β-amylase gene IbBAM1.1 enhances drought and salt stress resistance by regulating ROS homeostasis and osmotic balance. Plant Physiol. Biochem. 2021, 168, 167–176. [Google Scholar] [CrossRef]
- Zhu, H.; Zhai, H.; He, S.; Zhang, H.; Gao, S.; Liu, Q. A novel sweetpotato GATA transcription factor, IbGATA24, interacting with IbCOP9-5a positively regulates drought and salt tolerance. Environ. Exp. Bot. 2022, 194, 104735. [Google Scholar] [CrossRef]
- Reyes, J.C.; Muro-Pastor, M.I.; Florencio, F.J. The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 2004, 134, 1718–1732. [Google Scholar] [CrossRef]
- Kim, M.; Xi, H.; Park, J. Genome-wide comparative analyses of GATA transcription factors among 19 Arabidopsis ecotype genomes: Intraspecific characteristics of GATA transcription factors. PLoS ONE 2021, 16, e0252181. [Google Scholar] [CrossRef]
- Feng, X.; Yu, Q.; Zeng, J.; He, X.; Liu, W. Genome-wide identification and characterization of GATA family genes in wheat. BMC Plant Biol. 2022, 22, 372. [Google Scholar] [CrossRef]
- Yu, C.; Li, N.; Yin, Y.; Wang, F.; Gao, S.; Jiao, C.; Yao, M. Genome-wide identification and function characterization of GATA transcription factors during development and in response to abiotic stresses and hormone treatments in pepper. J. Appl. Genet. 2021, 62, 265–280. [Google Scholar] [CrossRef]
- Khatun, M.S.; Islam, M.S.U.; Shing, P.; Zohra, F.T.; Rashid, S.B.; Rahman, S.M.; Sarkar, M.A.R. Genome-wide identification and characterization of FORMIN gene family in potato (Solanum tuberosum L.) and their expression profiles in response to drought stress condition. PLoS ONE 2024, 19, e0309353. [Google Scholar] [CrossRef] [PubMed]
- Lai, D.; Yao, X.; Yan, J.; Gao, A.; Yang, H.; Xiang, D.; Ruan, J.; Fan, Y.; Cheng, J. Genome-wide identification, phylogenetic and expression pattern analysis of GATA family genes in foxtail millet (Setaria italica). BMC Genom. 2022, 23, 549. [Google Scholar] [CrossRef] [PubMed]
- Richter, R.; Behringer, C.; Zourelidou, M.; Schwechheimer, C. Convergence of auxin and gibberellin signaling on the regulation of the GATA transcription factors GNC and GNL in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2013, 110, 13192–13197. [Google Scholar] [CrossRef] [PubMed]
- Sala, J.; Mosesso, N.; Isono, E.; Schwechheimer, C. Arabidopsis thaliana B-GATA factors repress starch synthesis and gravitropic growth responses. New Phytol. 2023, 239, 979–991. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Zhang, Y.; Zhang, L.L.; He, J.X.; Xue, H.W.; Wang, J.W.; Lin, W.H. The transcription factor OsGATA6 regulates rice heading date and grain number per panicle. J. Exp. Bot. 2022, 73, 6133–6149. [Google Scholar] [CrossRef]
- Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. The Saltol QTL-localized transcription factor OsGATA8 plays an important role in stress tolerance and seed development in Arabidopsis and rice. J. Exp. Bot. 2020, 71, 684–698. [Google Scholar] [CrossRef]
- Wu, W.; Dong, X.; Chen, G.; Lin, Z.; Chi, W.; Tang, W.; Yu, J.; Wang, S.; Jiang, X.; Liu, X.; et al. The elite haplotype OsGATA8-H coordinates nitrogen uptake and productive tiller formation in rice. Nat. Genet. 2024, 56, 1516–1526. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Kim, Y.; Shim, Y.; Cho, S.H.; Yang, T.J.; Song, Y.H.; Kang, K.; Paek, N.C. Rice OsGATA16 is a positive regulator for chlorophyll biosynthesis and chloroplast development. Plant J. 2024, 117, 599–615. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wu, T.; Li, Z.; Huang, K.; Kim, N.E.; Ma, Z.; Kwon, S.W.; Jiang, W.; Du, X. OsGATA16, a GATA Transcription Factor, Confers Cold Tolerance by Repressing OsWRKY45-1 at the Seedling Stage in Rice. Rice 2021, 14, 42. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lan, M.; Xiao, M.; Peng, Y.; Pan, H.; Deng, J.; Wu, W. Genome-wide identification of GATA family genes in sweet potato (Ipomoea batatas L.) and their expression patterns under abiotic stress. Front. Genet. 2025, 16, 1635749. [Google Scholar] [CrossRef]
- Hung, J.H.; Weng, Z. Sequence Alignment and Homology Search with BLAST and ClustalW. Cold Spring Harb. Protoc. 2016, 2016, 10. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant. 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Høie, M.H.; Kiehl, E.N.; Petersen, B.; Nielsen, M.; Winther, O.; Nielsen, H.; Hallgren, J.; Marcatili, P. NetSurfP-3.0: Accurate and fast prediction of protein structural features by protein language models and deep learning. Nucleic Acids Res. 2022, 50, W510–W515. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Sun, H.; Mei, J.; Zhao, W.; Hou, W.; Zhang, Y.; Xu, T.; Wu, S.; Zhang, L. Phylogenetic Analysis of the SQUAMOSA Promoter-Binding Protein-Like Genes in Four Ipomoea Species and Expression Profiling of the IbSPLs During Storage Root Development in Sweet Potato (Ipomoea batatas). Front. Plant Sci. 2022, 12, 801061. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhou, Y.Y.; Zhai, H.; He, S.Z.; Zhao, N.; Liu, Q.C. Transcriptome profiling reveals insights into the molecular mechanism of drought tolerance in sweetpotato. Int. J. Mol. Sci. 2023, 24, 14398. [Google Scholar] [CrossRef]
- Nie, N.; Yang, Y.; Huo, J.; Wang, F.; Liu, R.; Sun, S.; Hu, Y.; Chen, Y.; Wu, W.; Liu, Q.; et al. IbPIF1 confers stem nematode resistance by regulating secondary metabolites in sweet potato. Plant Biotechnol. J. 2025, 23, 4650–4664. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Zhang, W.; Zuo, Z.; Zhu, Y.; Feng, Y.; Wang, Y.; Zhao, H.; Zhao, N.; Zhang, H.; He, S.; Liu, Q.; et al. Fast track to obtain heritable transgenic sweet potato inspired by its evolutionary history as a naturally transgenic plant. Plant Biotechnol. J. 2023, 21, 671–673. [Google Scholar] [CrossRef]
- Dunker, A.K.; Silman, I.; Uversky, V.N.; Sussman, J.L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008, 18, 756–764. [Google Scholar] [CrossRef]
- Zhao, T.; Wu, T.; Pei, T.; Wang, Z.; Yang, H.; Jiang, J.; Zhang, H.; Chen, X.; Li, J.; Xu, X. Overexpression of SlGATA17 Promotes Drought Tolerance in Transgenic Tomato Plants by Enhancing Activation of the Phenylpropanoid Biosynthetic Pathway. Front. Plant Sci. 2021, 12, 634888. [Google Scholar] [CrossRef]
- Zhu, X.; Duan, H.; Zhang, N.; Majeed, Y.; Jin, H.; Li, W.; Chen, Z.; Chen, S.; Tang, J.; Zhang, Y.; et al. Genome-Wide Identification of GATA Family Genes in Potato and Characterization of StGATA12 in Response to Salinity and Osmotic Stress. Int. J. Mol. Sci. 2024, 25, 12423. [Google Scholar] [CrossRef]
- Zhang, C.; Hou, Y.; Hao, Q.; Chen, H.; Chen, L.; Yuan, S.; Shan, Z.; Zhang, X.; Yang, Z.; Qiu, D.; et al. Genome-wide survey of the soybean GATA transcription factor gene family and expression analysis under low nitrogen stress. PLoS ONE 2015, 10, e0125174. [Google Scholar] [CrossRef]
- Zhao, F.; Li, X.; Chen, Z.; Guo, C. Pan-Genome-Wide Investigation and Expression Analysis of GATA Gene Family in Maize. Plants 2025, 14, 1693. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef]
- Haberle, V.; Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 2018, 19, 621–637. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, X.; Zhang, D.; Li, Y.; Wang, Q.; Ma, F.; Xu, X.; Zhang, X.; Hu, T. SlGATA17, A tomato GATA protein, interacts with SlHY5 to modulate salinity tolerance and germination. Environ. Exp. Bot. 2023, 206, 105191. [Google Scholar] [CrossRef]
- Guo, J.; Bai, X.; Dai, K.; Yuan, X.; Guo, P.; Zhou, M.; Shi, W.; Hao, C. Identification of GATA Transcription Factors in Brachypodium distachyon and Functional Characterization of BdGATA13 in Drought Tolerance and Response to Gibberellins. Front. Plant Sci. 2021, 12, 763665. [Google Scholar] [CrossRef]
- An, Y.; Zhou, Y.; Han, X.; Shen, C.; Wang, S.; Liu, C.; Yin, W.; Xia, X. The GATA transcription factor GNC plays an important role in photosynthesis and growth in poplar. J. Exp. Bot. 2020, 71, 1969–1984. [Google Scholar] [CrossRef]
- Du, X.; Lu, Y.; Sun, H.; Duan, W.; Hu, Y.; Yan, Y. Genome-Wide Analysis of Wheat GATA Transcription Factor Genes Reveals Their Molecular Evolutionary Characteristics and Involvement in Salt and Drought Tolerance. Int. J. Mol. Sci. 2022, 24, 27. [Google Scholar] [CrossRef]
- Zhu, X.; Duan, H.; Jin, H.; Chen, S.; Chen, Z.; Shao, S.; Tang, J.; Zhang, Y. Heat responsive gene StGATA2 functions in plant growth, photosynthesis and antioxidant defense under heat stress conditions. Front. Plant Sci. 2023, 14, 1227526. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).