Mitochondrial and Nuclear DNA Analyses of Rhipicephalus microplus from Mizoram, Northeast India: Insights into Genetic Diversity and Endosymbiont
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. DNA Isolation and PCR Amplification
2.3. Detection of the Endosymbiont in Tick
2.4. Systematics and Molecular Analyses
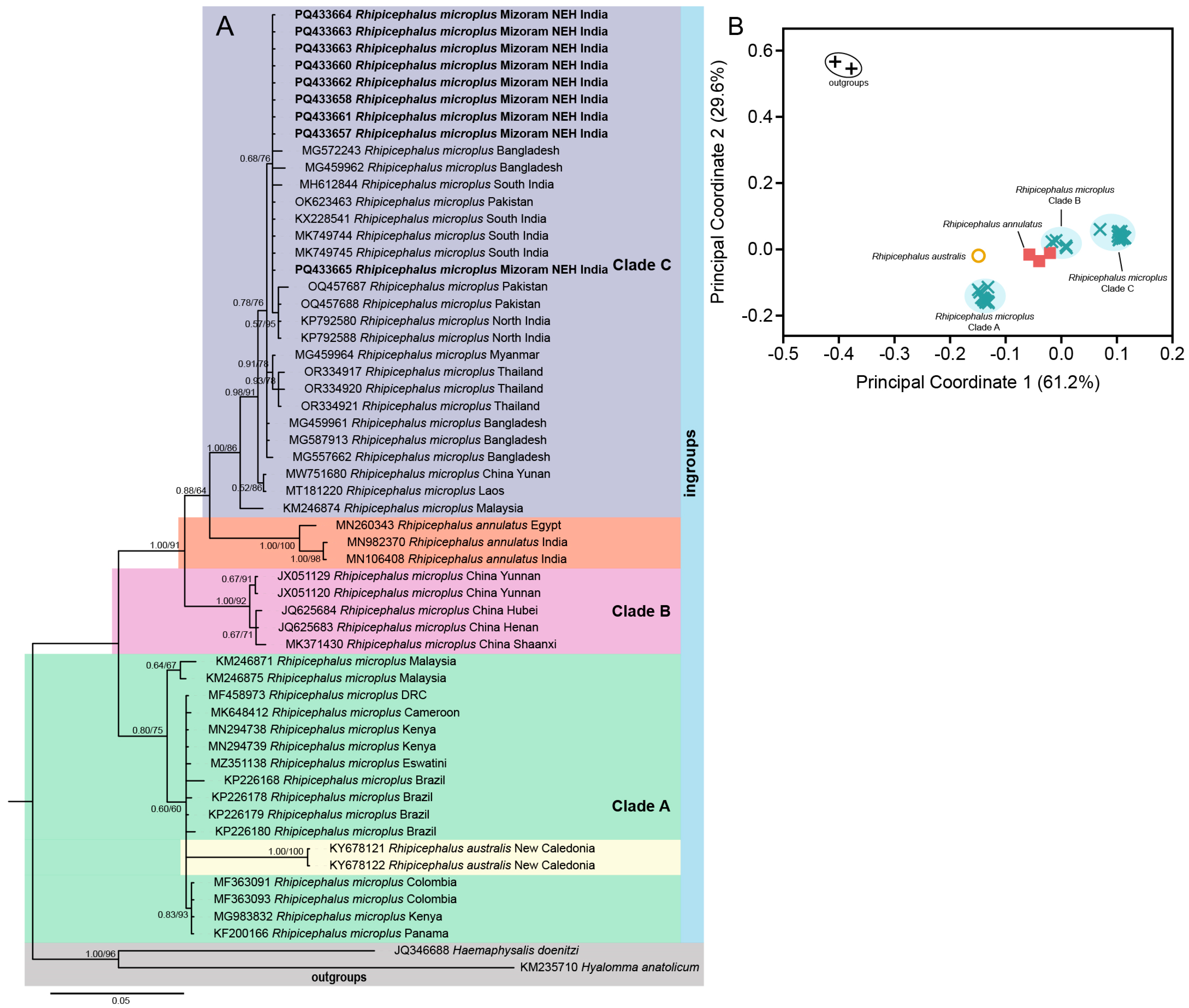
3. Results
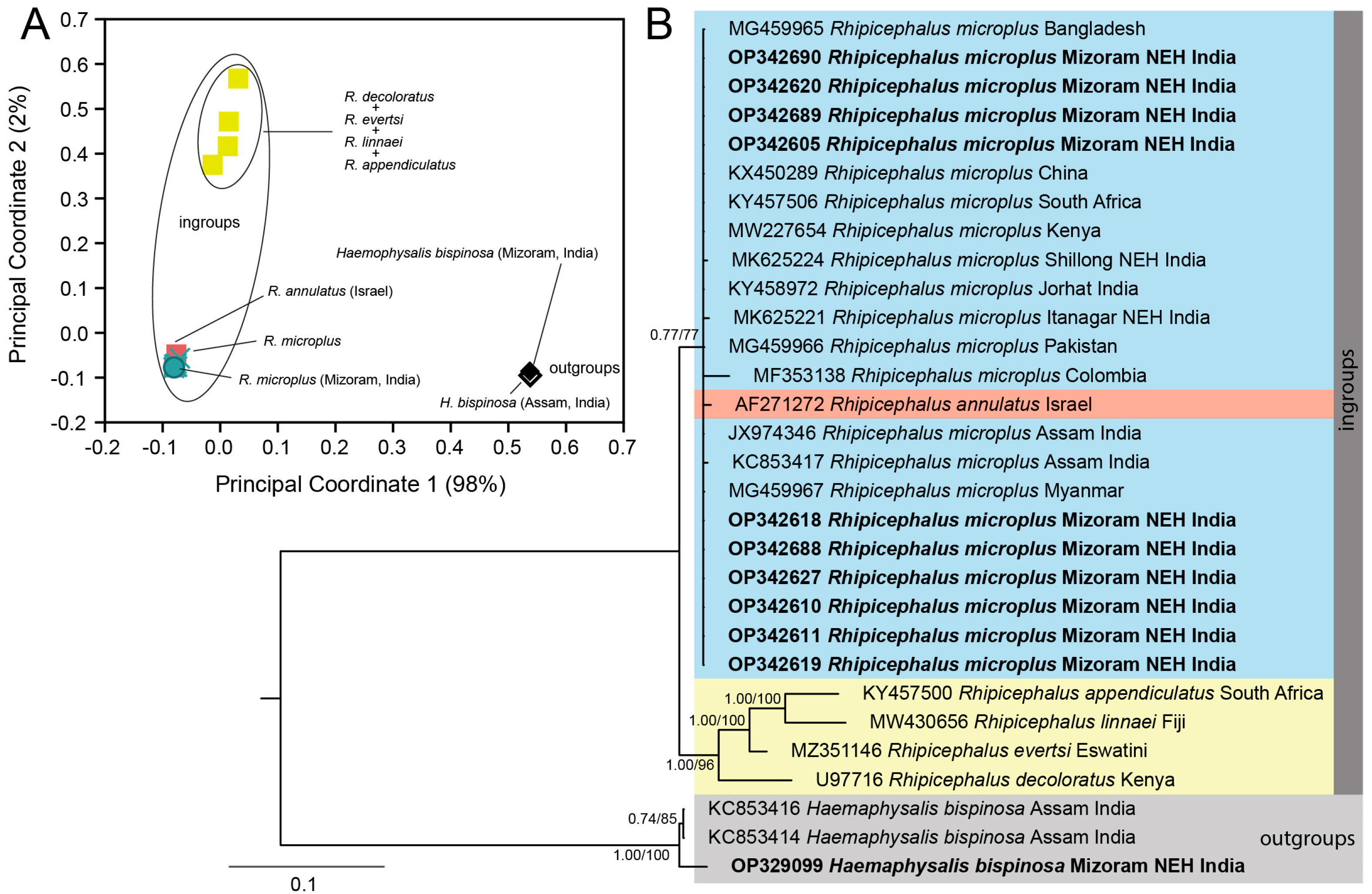
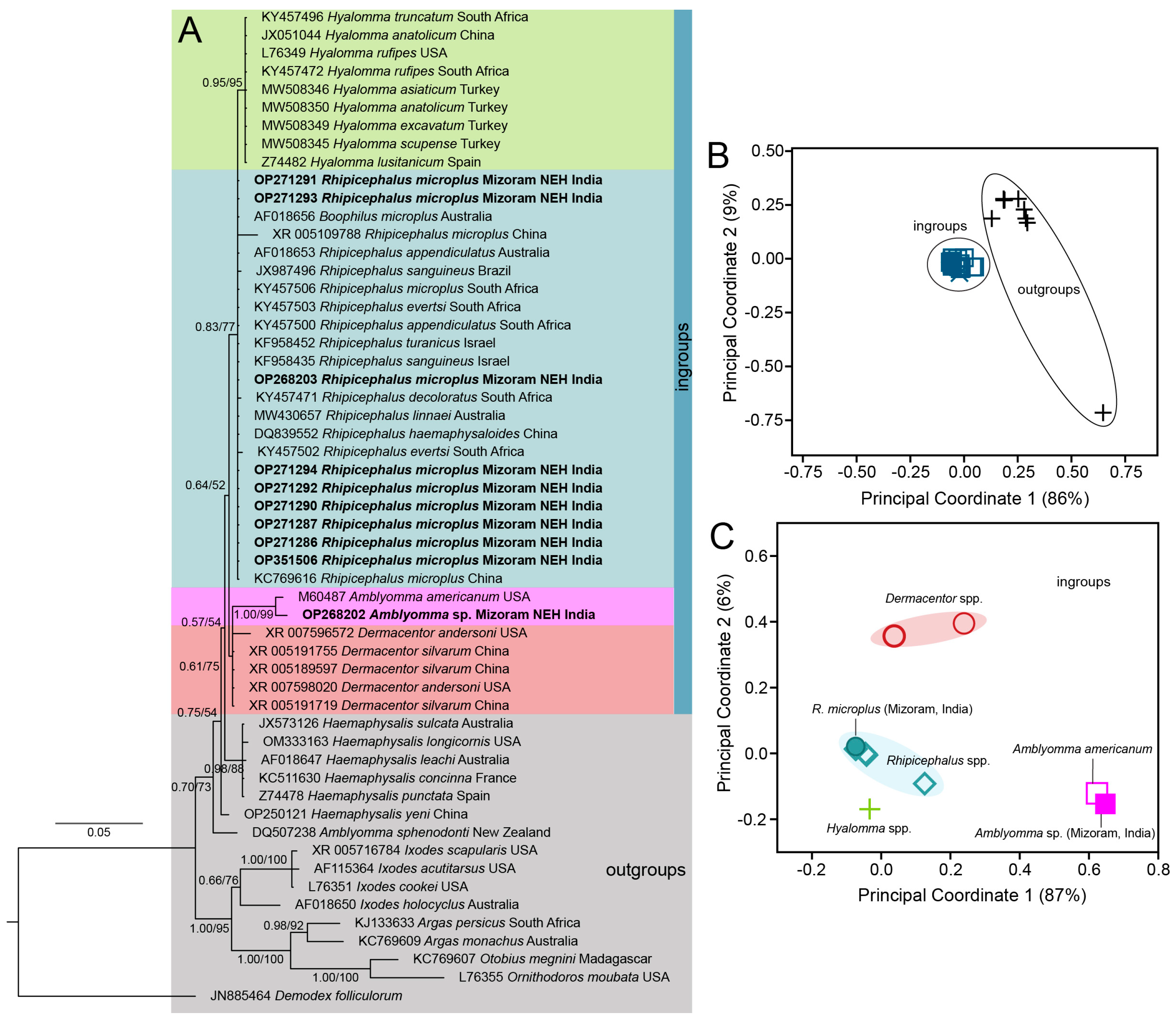
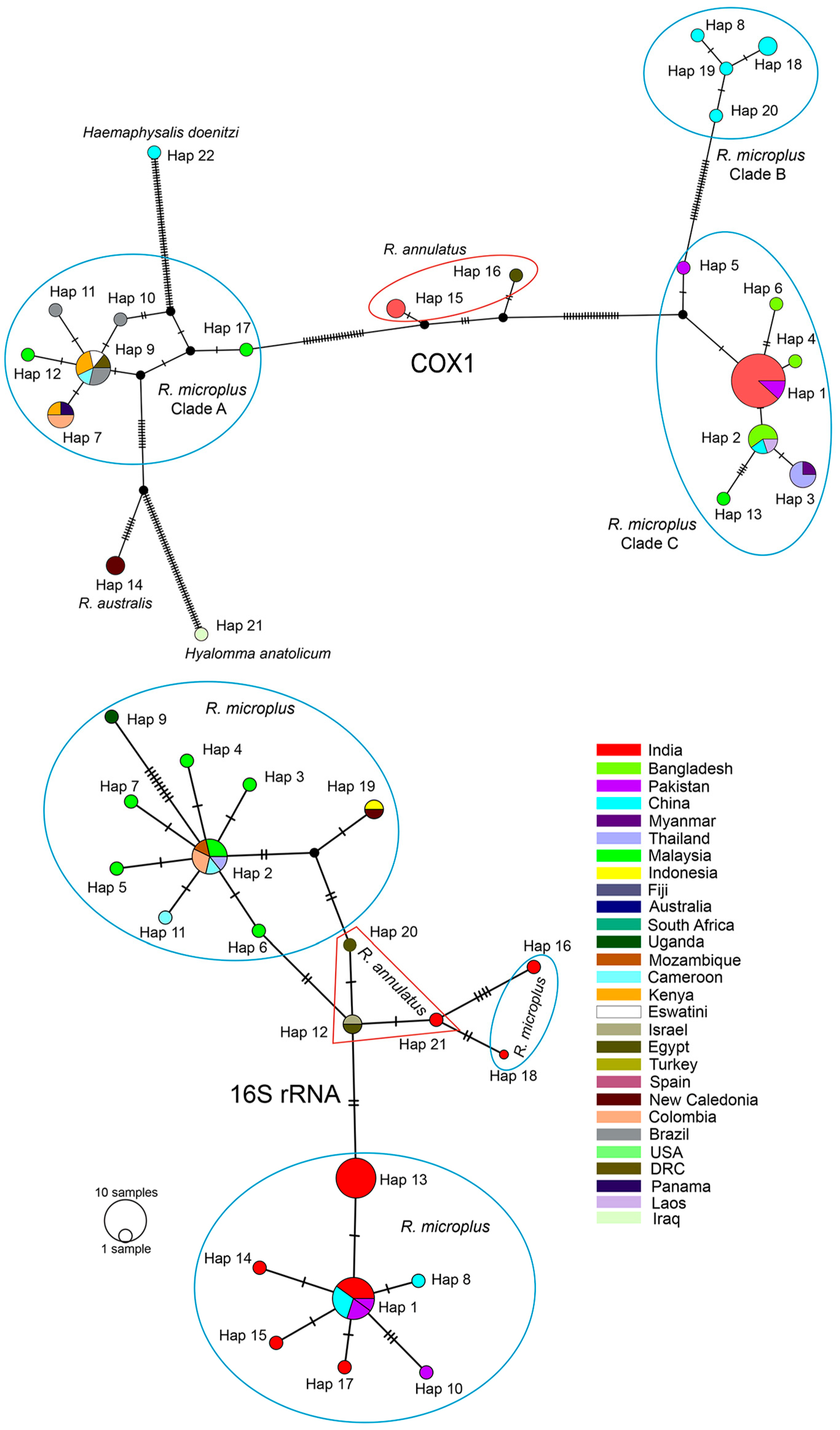
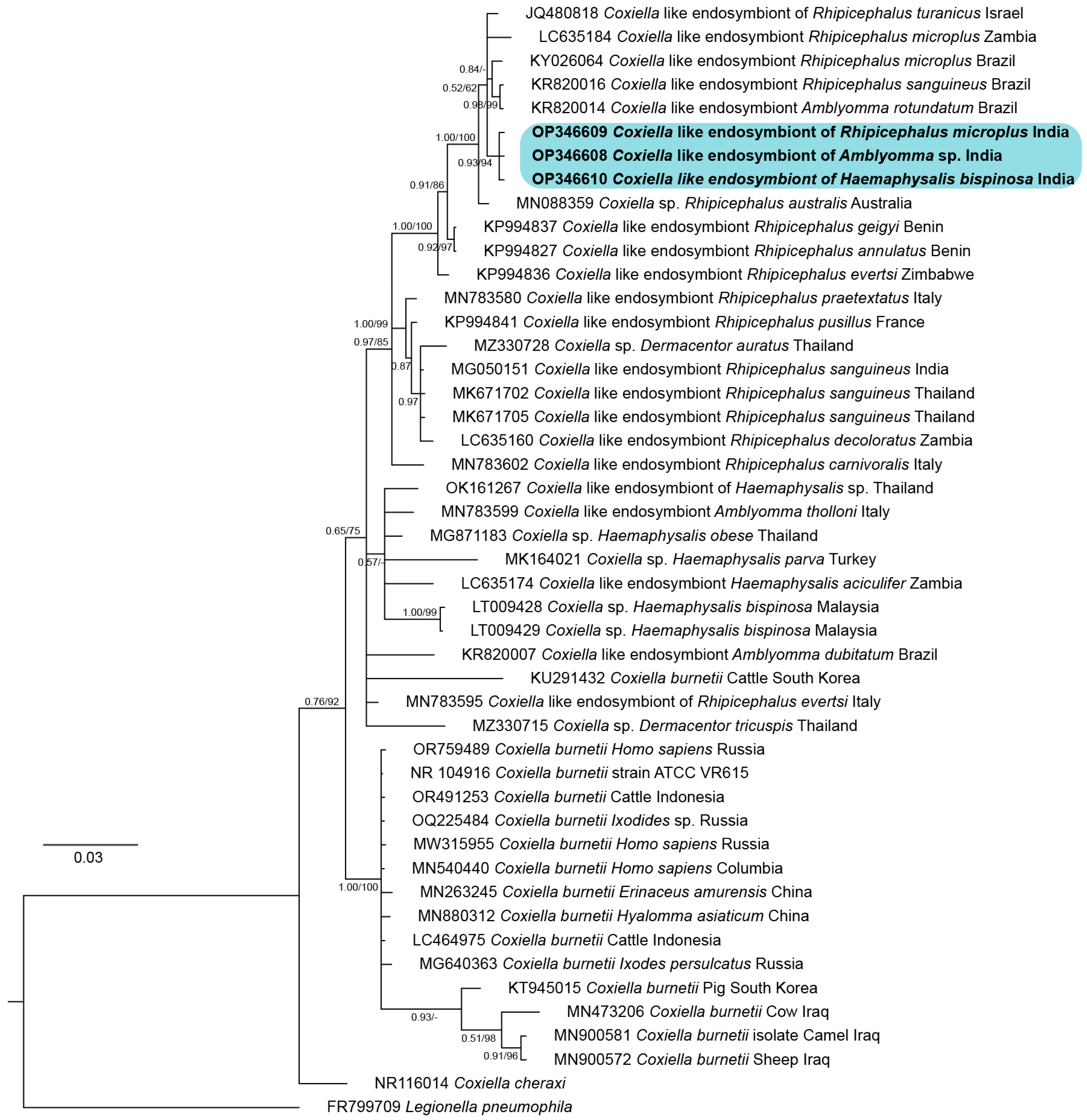
3.1. Systematics and Molecular Phylogeny of R. microplus
3.2. Detection of the Endosymbiont in Tick
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NEH | Northeastern Hill |
| COX1 | Cytochrome C Oxidase sub-unit 1 |
| PCoA | Principal Coordinate Analysis |
| rDNA | Ribosomal Deoxyribonucleic acid |
| TTBDs | Ticks and tick-borne diseases |
| IAEC | Institutional Animal Ethics Committee |
| ITS-2 | Internal Transcribed Spacer-2 |
| BIC | Bayesian Information Criterion |
| UFB | Ultrafast bootstrap |
| CLEs | Coxiella-like endosymbionts |
References
- Johansson, M.; Mysterud, A.; Flykt, A. Livestock owners’ worry and fear of tick-borne diseases. Parasit. Vectors 2020, 13, 331. [Google Scholar] [CrossRef]
- Singh, K.; Kumar, S.; Sharma, A.K.; Jacob, S.S.; RamVerma, M.; Singh, N.K.; Shakya, M.; Sankar, M.; Ghosh, S. Economic impact of predominant ticks and tick-borne diseases on Indian dairy production systems. Exp. Parasitol. 2022, 243, 108408. [Google Scholar] [CrossRef] [PubMed]
- Bishop, R.P.; Githaka, N.W.; Bazarusanga, T.; Bhushan, C.; Biguezoton, A.; Vudriko, P.; Muhanguzi, D.; Tumwebaze, M.; Bosco, T.J.; Shacklock, C.; et al. Control of ticks and tick-borne diseases in Africa through improved diagnosis and utilisation of data on acaricide resistance. Parasit. Vectors 2023, 16, 224. [Google Scholar] [CrossRef] [PubMed]
- Mizoram Government Bans Import of Cattle from Myanmar. Northeast Today. 22 October 2022. Available online: https://www.indiatodayne.in/mizoram/story/mizoram-government-bans-import-cattle-myanmar-461713-2022-10-22 (accessed on 21 October 2022).
- Rajkhowa, T.K.; Mohanarao, J.G.; Gogoi, A.; Hauhnar, L. Indian porcine reproductive and respiratory syndrome virus bears discontinuous deletion of 30 amino acids in nonstructural protein 2. Virusdisease 2016, 27, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Brahma, R.K.; Dixit, V.; Sangwan, A.K.; Doley, R. Identification and characterization of Rhipicephalus (Boophilus) microplus and Haemaphysalis bispinosa ticks (Acari: Ixodidae) of North East India by ITS2 and 16S rDNA sequences and morphological analysis. Exp. Appl. Acarol. 2014, 62, 253–265. [Google Scholar] [CrossRef]
- Bagra, N.; Roy, P.; Elango, A.; Basumatary, M.J.; Mize, D.; Doley, R. Morpho-molecular characterization of tick species prevalent in domesticated and semi-domesticated bovine species in three districts of Arunachal Pradesh. Acta Trop. 2025, 268, 107720. [Google Scholar] [CrossRef]
- Chamuah, J.K.; Bhattacharjee, K.; Sarmah, P.C.; Raina, O.K.; Mukherjee, S.; Rajkhowa, C. Report of Amblyomma testudinarium in mithuns (Bos frontalis) from eastern Mizoram (India). J. Parasit. Dis. 2016, 40, 1217–1220. [Google Scholar] [CrossRef]
- Ghosh, S.; Patra, G.; Borthakur, S.K.; Behera, P.; Tolenkhomba, T.C.; Deka, A.; Khare, R.K.; Biswas, P. Prevalence of haemoprotozoa in cattle of Mizoram, India. Biol. Rhythm Res. 2020, 51, 76–87. [Google Scholar] [CrossRef]
- Muhanguzi, D.; Byaruhanga, J.; Amanyire, W.; Ndekezi, C.; Ochwo, S.; Nkamwesiga, J.; Mwiine, F.N.; Tweyongyere, R.; Fourie, J.; Madder, M.; et al. Invasive cattle ticks in East Africa: Morphological and molecular confirmation of the presence of Rhipicephalus microplus in south-eastern Uganda. Parasit. Vectors 2020, 13, 165. [Google Scholar] [CrossRef]
- Low, V.L.; Tay, S.T.; Kho, K.L.; Koh, F.X.; Tan, T.K.; Lim, Y.A.; Ong, B.L.; Panchadcharam, C.; Norma-Rashid, Y.; Sofian-Azirun, M. Molecular characterisation of the tick Rhipicephalus microplus in Malaysia: New insights into the cryptic diversity and distinct genetic assemblages throughout the world. Parasit. Vectors 2015, 8, 341. [Google Scholar] [CrossRef]
- Tantrawatpan, C.; Vaisusuk, K.; Chatan, W.; Pilap, W.; Suksavate, W.; Andrews, R.H.; Petney, T.N.; Saijuntha, W. Genetic diversity and phylogenetic analyses of ixodid ticks infesting cattle in northeast Thailand: The discovery of Rhipicephalus microplus clade C and the rarely detected R. haemaphysaloides. Exp. Appl. Acarol. 2022, 86, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Barker, S.C.; Barker, D. Ticks of Australasia: 125 species of ticks in and around Australia. Zootaxa 2023, 5253, 1–670. [Google Scholar] [CrossRef] [PubMed]
- Li, L.H.; Zhang, Y.; Wang, J.Z.; Li, X.S.; Yin, S.Q.; Zhu, D.; Xue, J.B.; Li, S.G. High genetic diversity in hard ticks from a China-Myanmar border county. Parasit. Vectors 2018, 11, 469. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Khan, M.A.; Zahid, H.; Yaseen, P.M.; Khan, Q.M.; Nawab, J.; Rehman, Z.; Ateeq, M.; Khan, S.; Ibrahim, M. Seasonal dynamics, record of ticks infesting humans, wild and domestic animals and molecular phylogeny of Rhipicephalus microplus in Khyber Pakhtunkhwa Pakistan. Front. Physiol. 2019, 10, 793. [Google Scholar] [CrossRef] [PubMed]
- Burger, T.D.; Shao, R.; Barker, S.C. Phylogenetic analysis of mitochondrial genome sequences indicates that the cattle tick, Rhipicephalus (Boophilus) microplus, contains a cryptic species. Mol. Phylogenet. Evol. 2014, 76, 241–253. [Google Scholar] [CrossRef]
- Thinnabut, K.; Rodpai, R.; Sanpool, O.; Maleewong, W.; Tangkawanit, U. Genetic diversity of tick (Acari: Ixodidae) populations and molecular detection of Anaplasma and Ehrlichia infesting beef cattle from upper-northeastern Thailand. Infect. Genet. Evol. 2023, 107, 105394. [Google Scholar] [CrossRef]
- Barker, S.C.; Walker, A.R. Ticks of Australia. The species that infest domestic animals and humans. Zootaxa 2014, 3816, 1–144. [Google Scholar] [CrossRef]
- Csordas, B.G.; Garcia, M.V.; Cunha, R.C.; Giachetto, P.F.; Blecha, I.M.; Andreotti, R. New insights from molecular characterization of the tick Rhipicephalus (Boophilus) microplus in Brazil. Rev. Bras. Parasitol. Vet. 2016, 25, 317–326. [Google Scholar] [CrossRef]
- Chitimia, L.; Lin, R.Q.; Cosoroaba, I.; Wu, X.Y.; Song, H.Q.; Yuan, Z.G.; Zhu, X.Q. Genetic characterization of ticks from southwestern Romania by sequences of mitochondrial cox1 and nad5 genes. Exp. Appl. Acarol. 2010, 52, 305–311. [Google Scholar] [CrossRef]
- Lv, J.; Wu, S.; Zhang, Y.; Zhang, T.; Feng, C.; Jia, G.; Lin, X. Development of a DNA barcoding system for the Ixodida (Acari: Ixodida). Mitochondrial DNA 2014, 25, 142–149. [Google Scholar] [CrossRef]
- Ceraul, S.M.; Dreher-Lesnick, S.M.; Mulenga, A.; Rahman, M.S.; Azad, A.F. Functional characterization and novel rickettsiostatic effects of a Kunitz-type serine protease inhibitor from the tick Dermacentor variabilis. Infect. Immun. 2008, 76, 5429–5435. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Gower, J.C. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 1966, 53, 325–338. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Soubrier, J.; Steel, M.; Lee, M.S.; Sarkissian, C.; Guindon, S.; Ho, S.Y.; Cooper, A. The influence of rate heterogeneity among sites on the time dependence of molecular rates. Mol. Biol. Evol. 2012, 29, 3345–3358. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, 293–296. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 110–1116. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Petney, T.N.; Saijuntha, W.; Boulanger, N.; Chitimia-Dobler, L.; Pfeffer, M.; Eamudomkarn, C.; Andrews, R.H.; Ahamad, M.; Putthasorn, N.; Muders, S.V.; et al. Ticks (Argasidae, Ixodidae) and tick-borne diseases of continental Southeast Asia. Zootaxa 2019, 4558, 1–89. [Google Scholar] [CrossRef]
- Kasaija, P.D.; Estrada-Peña, A.; Contreras, M.; Kirunda, H.; de la Fuente, J. Cattle ticks and tick-borne diseases: A review of Uganda’s situation. Ticks Tick Borne Dis. 2021, 12, 101756. [Google Scholar] [CrossRef] [PubMed]
- Kakati, P.; Sarmah, P.C.; Ray, D.; Bhattacharjee, K.; Sharma, R.K.; Barkalita, L.M.; Sarma, D.K.; Baishya, B.C.; Borah, P.; Stanley, B. Emergence of oriental theileriosis in cattle and its transmission through Rhipicephalus (Boophilus) microplus in Assam, India. Vet. World 2015, 8, 1099–1104. [Google Scholar] [CrossRef] [PubMed]
- Martins, K.R.; Garcia, M.V.; Bonatte-Junior, P.; Duarte, P.O.; Csordas, B.G.; Higa, L.O.S.; Zimmermann, N.P.; Barros, J.C.; Andreotti, R. Seasonal fluctuations of Babesia bigemina and Rhipicephalus microplus in Brangus and Nellore cattle reared in the Cerrado biome, Brazil. Parasit. Vectors 2022, 15, 395. [Google Scholar] [CrossRef]
- Amrutha, B.M.; Kumar, K.G.A.; Kurbet, P.S.; Varghese, A.; Deepa, C.K.; Pradeep, R.K.; Nimisha, M.; Asaf, M.; Juliet, S.; Ravindran, R.; et al. Morphological and molecular characterization of Rhipicephalus microplus and Rhipicephalus annulatus from selected states of southern India. Ticks Tick Borne Dis. 2023, 14, 102086. [Google Scholar] [CrossRef]
- Uilenberg, G. Boophilus (Uroboophilus) fallax Minning, 1934, synonyme de Boophilus microplus (Canestrini, 1887) (Ixodidae). Rev. Elev. Méd. Vét. Pays Trop. 1962, 15, 387–398. [Google Scholar] [CrossRef]
- Madder, M.; Thys, E.; Achi, L.; Toure, A.; De Deken, R. Rhipicephalus (Boophilus) microplus: A most successful invasive tick species in West-Africa. Exp. Appl. Acarol. 2011, 53, 139–145. [Google Scholar] [CrossRef]
- Silatsa, B.A.; Kuiate, J.R.; Njiokou, F.; Simo, G.; Feussom, J.K.; Tunrayo, A.; Amzati, G.S.; Bett, B.; Bishop, R.; Githaka, N.; et al. A countrywide molecular survey leads to a seminal identification of the invasive cattle tick Rhipicephalus (Boophilus) microplus in Cameroon, a decade after it was reported in Cote d’Ivoire. Ticks Tick Borne Dis. 2019, 10, 585–593. [Google Scholar] [CrossRef]
- Segura, J.A.; Isaza, J.P.; Botero, L.E.; Alzate, J.F.; Gutierrez, L.A. Assessment of bacterial diversity of Rhipicephalus microplus ticks from two livestock agroecosystems in Antioquia, Colombia. PLoS ONE 2020, 15, e0234005. [Google Scholar] [CrossRef]
- Roy, B.C.; Estrada-Peña, A.; Krucken, J.; Rehman, A.; Nijhof, A.M. Morphological and phylogenetic analyses of Rhipicephalus microplus ticks from Bangladesh, Pakistan and Myanmar. Ticks Tick Borne Dis. 2018, 9, 1069–1079. [Google Scholar] [CrossRef]
- Bakkes, D.K.; Ropiquet, A.; Chitimia-Dobler, L.; Matloa, D.E.; Apanaskevich, D.A.; Horak, I.G.; Mans, B.J.; Matthee, C.A. Adaptive radiation and speciation in Rhipicephalus ticks: A medley of novel hosts, nested predator-prey food webs, off-host periods and dispersal along temperature variation gradients. Mol. Phylogenet. Evol. 2021, 162, 107178. [Google Scholar] [CrossRef]
- Klompen, J.S.; Black, W.C.; Keirans, J.E.; Oliver, J.H. Evolution of ticks. Annu. Rev. Entomol. 1996, 41, 141–161. [Google Scholar] [CrossRef] [PubMed]
- Kozak, K.H.; Wiens, J.J. Accelerated rates of climatic-niche evolution underlie rapid species diversification. Ecol. Lett. 2010, 13, 1378–1389. [Google Scholar] [CrossRef]
- Mangold, A.J.; Bargues, M.D.; Mas-Coma, S. 18S rDNA gene sequences and phylogenetic relationships of European hard-tick species (Acari: Ixodidae). Parasitol. Res. 1998, 84, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Murrell, A.; Campbell, N.J.; Barker, S.C. Phylogenetic analyses of the Rhipicephaline ticks indicate that the genus Rhipicephalus is paraphyletic. Mol. Phylogenet. Evol. 2000, 16, 1–7. [Google Scholar] [CrossRef]
- Murrell, A.; Barker, S.C. Synonymy of Boophilus Curtice, 1891 with Rhipicephalus Koch, 1844 (Acari: Ixodidae). Syst. Parasitol. 2003, 56, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Patra, G.; Ghosh, S.; Priyanka; Efimova, M.A.; Sahara, A.; Al-Awsi, G.R.L.; Polley, S.; Debbarma, A. Molecular detection of Coxiella burnetti and Borrelia burgdorferi in ticks infesting goats in North-Eastern states of India. Int. J. Acarol. 2020, 46, 431–438. [Google Scholar] [CrossRef]


| Target Gene | Oligonucleotide Primer | PCR Amplification Condition | Expected Amplicon Size | References | |
|---|---|---|---|---|---|
| cox1 | Forward: 5′-CTTCAGCCATTTTACCGCGA-3′ Reverse: 5′-CTCCGCCTGAAGGGTCAAA-3′ | Initial denaturation | 94 °C for 5 min | 643 | [19] |
| Cyclical denaturation (35 Cycles) | 94 °C for 1 min | ||||
| Annealing temperature | 56 °C for 30 s | ||||
| Extension | 72 °C for 1 min | ||||
| Final extension | 72 °C for 10 min | ||||
| Cooling temperature | 4 °C | ||||
| 16s rDNA | Forward: 5′-AATTGCTGTAGTATTTTGAC-3′ Reverse: 5′-TCTGAACTCAGATCAAGTAG-3′ | Initial denaturation | 94 °C for 5 min | 455 | [20] |
| Cyclical denaturation | 94 °C for 30 s | ||||
| Annealing temperature | 49 °C for 30 s × 5 cycles | ||||
| 47 °C for 30 s × 5 cycles | |||||
| 45 °C for 30 s × 5 cycles | |||||
| 43 °C for 30 s × 33 cycles | |||||
| Extension | 72 °C for 45 s | ||||
| Final extension | 72 °C for 10 min | ||||
| Cooling temperature | 4 °C | ||||
| ITS-2 | Forward: 5′-CGAGACTTGGTGTGAATTGCA-3′ Reverse: 5′-CCCATACACCACATTTCCCG-3′ | Initial denaturation | 95 °C for 10 min | 1500 (R. m) 1700 (H. b) | [20] |
| Cyclical denaturation (35 Cycles) | 95 °C for 30 s | ||||
| Annealing temperature | 55 °C for 45 s × 33 cycles | ||||
| Extension | 72 °C for 90 s | ||||
| Final extension | 72 °C for 10 min | ||||
| Cooling temperature | 4 °C | ||||
| 18s rDNA | Forward: 5′-CATTAAATCAGTTATGGTTCC-3′ Reverse: 5′-CGCCGCAATACGAATGC-3′ | Initial denaturation | 95 °C for 10 min | 780 | [21] |
| Cyclical denaturation (35 Cycles) | 95 °C for 30 s | ||||
| Annealing temperature | 52 °C for 30 s × 5 cycles | ||||
| 50 °C for 30 s × 5 cycles | |||||
| 48 °C for 30 s × 5 cycles | |||||
| 46 °C for 30 s × 33 cycles | |||||
| Extension | 72 °C for 90 s | ||||
| Final extension | 72 °C for 10 min | ||||
| Cooling temperature | 4 °C | ||||
| Final extension | 72 °C for 10 min | ||||
| Initial denaturation | 4 °C | ||||
| Endosymbiont Primer (16s rRNA) | Forward: 5′-GTTCGGAATTACTGGGCGTA-3′ Reverse: 5′-AATTAAACCGCATGCTCCAC-3′. | Initial denaturation | 94 °C for 5 min | 405 | [22] |
| Cyclical denaturation (35 cycles) | 94 °C for 1 min | ||||
| Annealing temperature | 52 °C for 30 s | ||||
| Extension | 72 °C for 45 s | ||||
| Final extension | 72 °C for 10 min | ||||
| Initial denaturation | 94 °C for 5 min | ||||
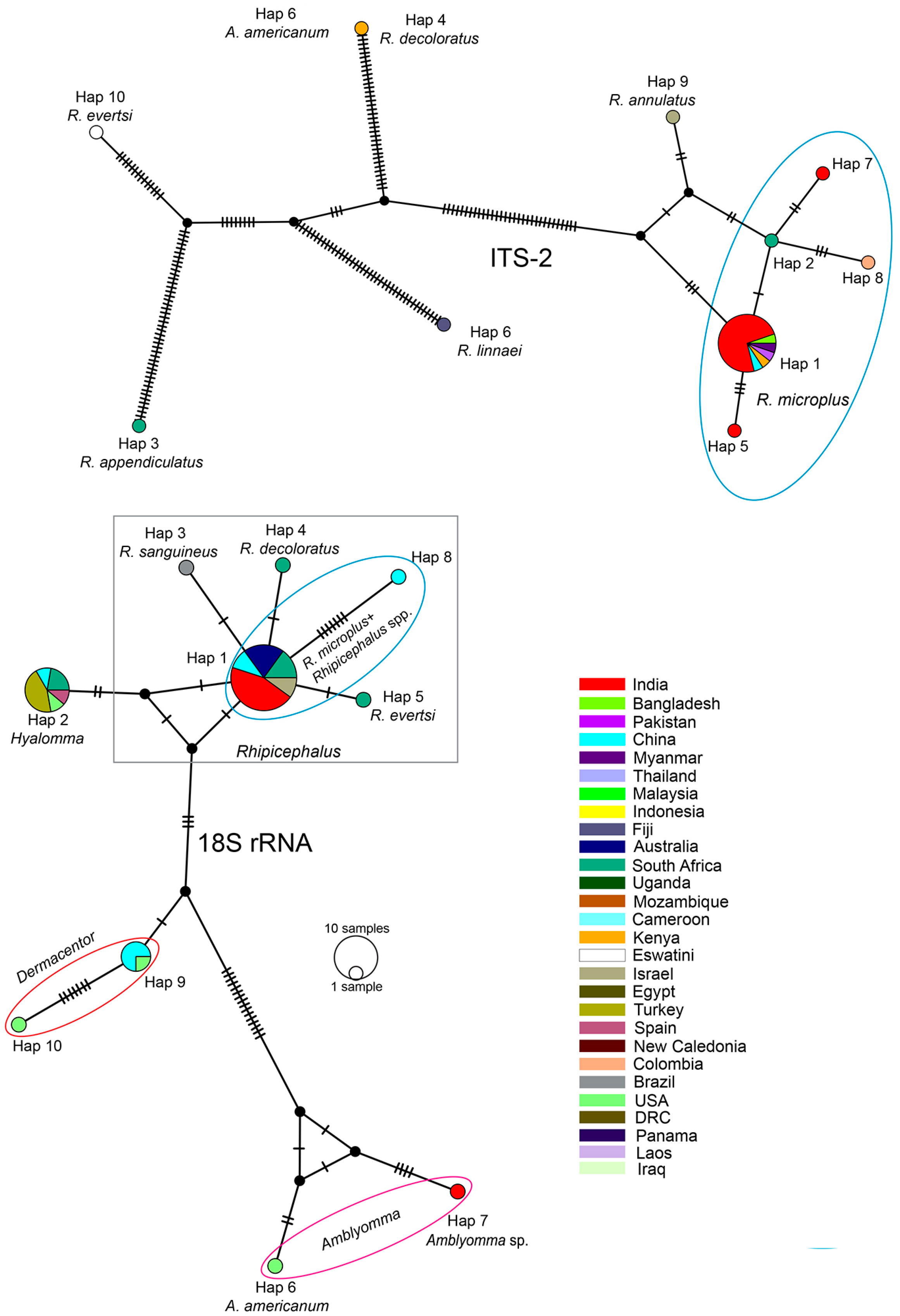
| Sl. No | Genetic Marker | No. of Haplotypes | Haplotype Diversity |
|---|---|---|---|
| 1. | COX1 (among ingroups) | 20 | 0.8774 |
| 2. | COX1 (among R. microplus) | 17 | 0.8531 |
| 3. | 16S rRNA (among ingroup) | 21 | 0.8995 |
| 4. | 16S rRNA (among R. microplus) | 18 | 0.8804 |
| 5. | ITS-2 (among ingroup) | 10 | 0.5476 |
| 6. | ITS-2 (among R. microplus) | 5 | 0.3377 |
| 7. | 18S rRNA (among ingroup) | 10 | 0.7026 |
| 8. | 18S rRNA (among R. microplus) | 2 | 0.1538 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lalawmpuii, K.; Jacob, S.S.; Tolenkhomba, T.C.; Behera, P.; Lalmuanpuia, J.; Lalremsanga, H.T.; Lalrintluanga, K.; Lalchhandama, C.; Biakzuala, L.; Lalrinkima, H. Mitochondrial and Nuclear DNA Analyses of Rhipicephalus microplus from Mizoram, Northeast India: Insights into Genetic Diversity and Endosymbiont. Genes 2025, 16, 1216. https://doi.org/10.3390/genes16101216
Lalawmpuii K, Jacob SS, Tolenkhomba TC, Behera P, Lalmuanpuia J, Lalremsanga HT, Lalrintluanga K, Lalchhandama C, Biakzuala L, Lalrinkima H. Mitochondrial and Nuclear DNA Analyses of Rhipicephalus microplus from Mizoram, Northeast India: Insights into Genetic Diversity and Endosymbiont. Genes. 2025; 16(10):1216. https://doi.org/10.3390/genes16101216
Chicago/Turabian StyleLalawmpuii, Khawlhring, Siju Susan Jacob, Thingujam Chaa Tolenkhomba, Parthasarathi Behera, Joy Lalmuanpuia, Hmar Tlawmte Lalremsanga, Khawlhring Lalrintluanga, Chhakchhuak Lalchhandama, Lal Biakzuala, and Hmar Lalrinkima. 2025. "Mitochondrial and Nuclear DNA Analyses of Rhipicephalus microplus from Mizoram, Northeast India: Insights into Genetic Diversity and Endosymbiont" Genes 16, no. 10: 1216. https://doi.org/10.3390/genes16101216
APA StyleLalawmpuii, K., Jacob, S. S., Tolenkhomba, T. C., Behera, P., Lalmuanpuia, J., Lalremsanga, H. T., Lalrintluanga, K., Lalchhandama, C., Biakzuala, L., & Lalrinkima, H. (2025). Mitochondrial and Nuclear DNA Analyses of Rhipicephalus microplus from Mizoram, Northeast India: Insights into Genetic Diversity and Endosymbiont. Genes, 16(10), 1216. https://doi.org/10.3390/genes16101216






