Beyond the Curtains: Identification of the Genetic Cause of Foetal Developmental Abnormalities Through the Application of Molecular Autopsy
Abstract
1. Introduction
2. Materials and Methods
2.1. Recruitment of Cases and Inclusion and Exclusion Criteria
2.2. Diagnostic Investigations Before Termination of Pregnancy
2.3. Diagnostic Investigations After Termination of Pregnancy
2.4. DNA Extraction and Quality Control
2.5. SNP-Array Analysis
2.6. WES Analysis and Data Interpretation
2.7. Minigene Assays
2.8. Cell Culture and Transfection
2.9. RNA Extraction and Transcript Analysis
3. Results
3.1. Demographic and Pregnancies Data
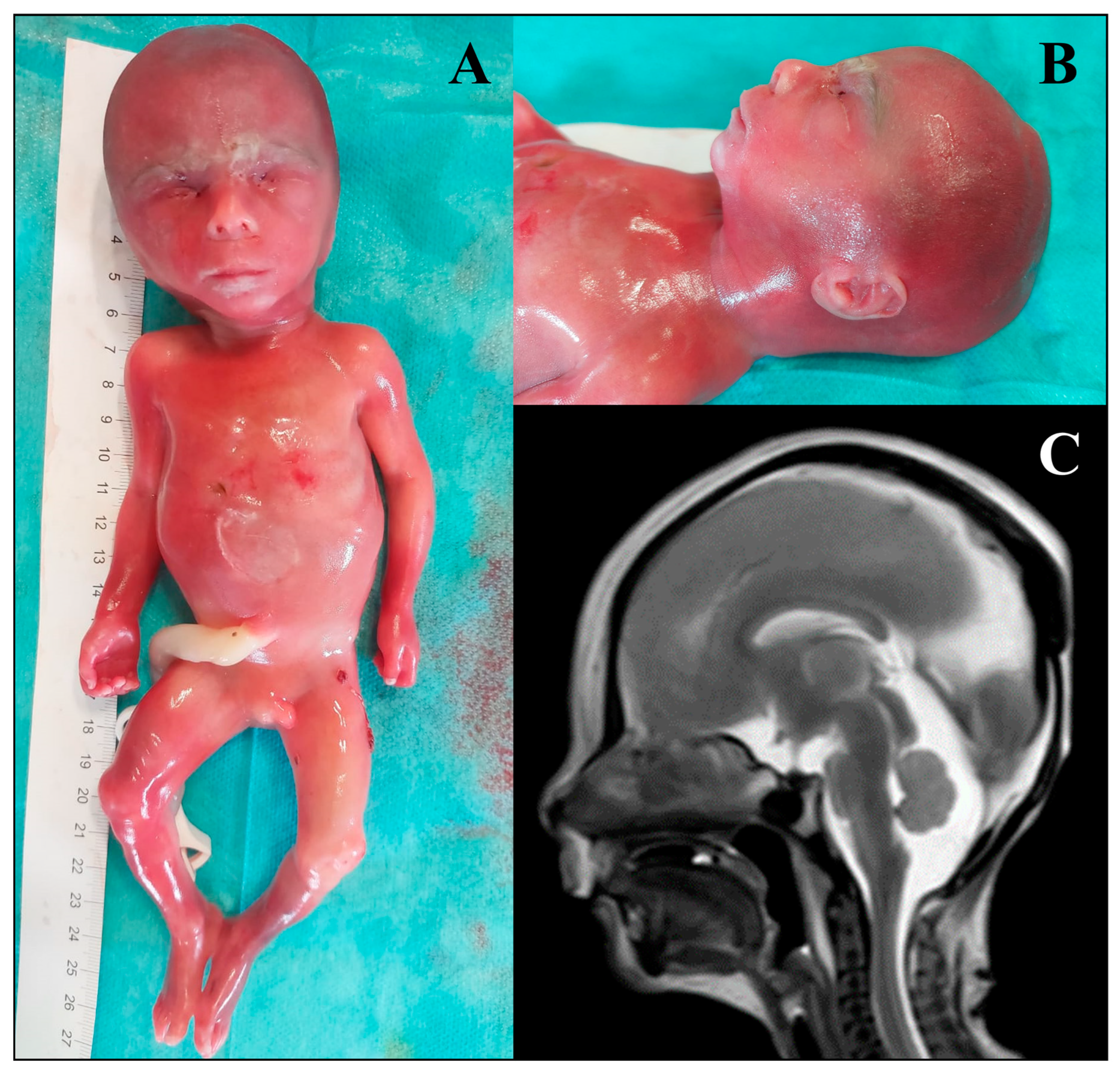
3.2. Phenotypic Classification of Foetuses
3.3. Molecular Autopsy Results
| Section A: Variants Identified in Solved Cases | ||||||||||
| Foetal ID | Diagnostic Group | Phenotypic Group | Gene | Associated Disease and Inheritance Pattern | Variant | Frequency | Zygosity | Inheritance | ACMG/AMP Classification | Ref. |
| 1 | Single major malformation | CNS anomalies | TUBA1A | Lissencephaly 3 (MIM: # 611603) AD | NM_006009.3:c.521C>T p.(Ala174Val) | NA | HET | De novo | Likely Pathogenic (PS2, PM1, PM2_Supporting, PM5) | [44] |
| 2 | Single major malformation | CNS anomalies | TUBA1A | Lissencephaly 3 (MIM: # 611603) AD | NM_006009.3:c.190C>T p.(Arg64Trp) | 0.00003965 | HET | De novo | Pathogenic (PS2; PS3; PM1; PM2_supporting; PM5; PP3_moderate) | [19] |
| 3 | Single major malformation | CNS anomalies | ZIC2 | Holoprosencephaly 5 (MIM: # 609637) AD | NM_007129.5:c.215delG p.(Gly72Ala fs*146) | NA | HET | De novo | Pathogenic (PVS1; PS2; PM2_supporting) | NA |
| 4 | Single major malformation | CNS anomalies | ASPM | Microcephaly 5, primary, autosomal recessive (MIM: # 608716) AR | NM_018136.5:c.7551T>A p.(Tyr2517*) | 0.000002481 | COMP HET | Maternal | Pathogenic (PVS1; PM2_supporting; PM3) | NA |
| NM_018136.5:c.9926delG p.(Ser3309Ile fs*31) | 6.198 × 10−7 | Paternal | Pathogenic (PVS1; PM2_supporting; PM3) | NA | ||||||
| 5 | Single major malformation | CNS anomalies | FOXG1 | Rett syndrome, congenital variant (MIM: # 613454) AD | NM_005249.5:c.256delC p.(Gln86Arg fs*106) | NA | HET | De novo | Pathogenic (PVS1; PS2_very strong; PM2_supporting) | [45] |
| 6 | Single major malformation | Skeletal anomalies | COL1A2 | Osteogenesis imperfecta, type II (MIM: # 166210) AD | NM_000089.3:c.3125G>T p.(Gly1042Val) | NA | HET | Assumed de novo | Likely Pathogenic (PM1; PM2_supporting; PM5; PM6; PP3) | NA |
| 7 | Single major malformation | Skeletal anomalies | EBP | Chondrodysplasia punctata, X-linked dominant (MIM: # 302960) XLD | NM_006579.2:c.364G>A p.(Glu122Lys) | NA | HET | De novo | Pathogenic (PS2; PM1; PM2_supporting; PP3_strong) | [46] |
| 8 | Single major malformation | Skeletal anomalies | TRIP11 | Achondrogenesis, type IA (MIM: # 200600) AR | NM_004239.3:c.673C>T p.(Arg225*) | 0.000008070 | HET | Maternal | Likely Pathogenic (PVS1; PM2_supporting) | [47] |
| 9 | Single major malformation | Urogenital anomalies | GREB1L | Renal hypodysplasia/ aplasia 3 (MIM: # 617805) AD | NM_001142966.3:c.3007dupA p.(Thr1003Asn fs*14) | NA | HET | Assumed paternal | Likely Pathogenic (PVS1; PM2_supporting) | NA |
| 10 | Single major malformation | Urogenital anomalies | GREB1L | Renal hypodysplasia/ aplasia 3 (MIM: # 617805) AD | NM_001142966.3:c.3007dupA p.(Thr1003Asn fs*14) | NA | HET | Assumed paternal | Likely Pathogenic (PVS1; PM2_supporting) | NA |
| 11 | Single major malformation | Fluid accumulation | CTSA | Galactosialidosis (MIM: # 256540) AR | NM_000308.4:c.990dupC p.(Cys331Leu fs*56) | 0.000003099 | COMP HET | Maternal | Likely Pathogenic (PVS1; PM2_supporting) | [48] |
| NM_000308.4:c.753_755 del p.(Asn251del) | NA | Paternal | Variant of Uncertain Significance (PM2_supporting; PM3; PM4) | NA | ||||||
| 12 | Multiple malformations | Multisystemic condition | ARID1B | Coffin-Siris syndrome 1 (MIM: # 135900) AD | NM_001374820.1:c.4359G>A p.(Pro1453Pro) | 6.258 × 10−7 | HET | De novo | Likely Pathogenic (PS2; PM2_supporting; PP3) | [49] |
| 13 | Multiple malformations | Multisystemic condition | ARID1B | Coffin-Siris syndrome 1 (MIM: # 135900) AD | NM_001374820.1:c.1293_1314 del p.(Gly434Metfs*11) | NA | HET | De novo | Pathogenic (PVS1; PS2; PS4_moderate; PM2_supporting) | NA |
| 14 | Multiple malformations | Multisystemic condition | MECOM | Radioulnar synostosis with amegakaryocytic thrombocytopenia 2 (MIM: # 616738) AD | NM_001105077.3:c.2005C>T p.(Arg669*) | NA | HET | De novo | Pathogenic (PVS1; PS2; PM2_supporting) | NA |
| 15 | Multiple malformations | Multisystemic condition | SLC26A2 | Achondrogenesis Ib (MIM: # 600972) AR | NM_000112.4:c.1336A>T p.(Lys446*) | 0.000002478 | HOM | Maternal and paternal | Pathogenic (PVS1; PM2_supporting; PM3_supporting) | NA |
| 16 | Multiple malformations | Multisystemic condition | ARX | Lissencephaly, X-linked 2 (MIM: # 300215) XL | NM_139058.3:c.206delA p.(Lys69fs*99) | NA | HET | De novo | Pathogenic (PVS1; PS2; PM2_supporting) | NA |
| 17 | Multiple malformations | Multisystemic condition | FGFR2 | Apert syndrome (MIM: # 101200) AD | NM_000141.5:c.755C>G p.(Ser252Trp) | 0.00004030 | HET | De novo | Pathogenic (PS2; PS3; PM1; PM2_supporting; PM5_moderate) | [50] |
| 18 | Multiple malformations | Multisystemic condition | NAA10 | Ogden syndrome (MIM: # 300855) XLD | NM_003491.4:c.92A>G p.(Tyr31Cys) | NA | HET | De novo | Likely Pathogenic (PS2; PS4_moderate; PM1_supporting; PM2_supporting) | [51] |
| 19 | Multiple malformations | Multisystemic condition | RPS19 | Diamond-Blackfan anemia 1 (MIM: # 105650) AD | NM_001022.4:c.185G>A p.(Arg62Gln) | NA | HET | De novo | Pathogenic (PS2; PS3_supporting; PS4; PM1; PM2_supporting; PM5_strong; PP3_moderate) | [52] |
| 20 | Multiple malformations | Multisystemic condition | TBX1 | Velocardiofacial syndrome (MIM: # 192430) AD | NM_080647.1:c.698C>T p.(Ser233Phe) | NA | HET | Maternal | Likely Pathogenic (PM1; PM2_supporting; PM5_supporting; PP3_strong) | [42] |
| 21 | Multiple malformations | Multisystemic condition | HRAS | Costello syndrome (MIM: # 218040) AD | NM_005343.4:c.37G>T p.(Gly13Cys) | NA | HET | De novo | Pathogenic (PS2_very strong; PS4; PM1; PM2_supporting; PM5_strong; PP3_moderate) | NA |
| 22 | Multiple malformations | Multisystemic condition | KMT2D | Kabuki syndrome 1 (MIM: # 147920) AD | NM_003482.4:c.643_644delCCinsTG p.(Pro215*) | NA | HET | De novo | Pathogenic (PVS1; PS2; PM2_supporting) | NA |
| 23 | Multiple malformations | Multisystemic condition | PEX1 | Peroxisome biogenesis disorder 1A (Zellweger) (MIM: # 214100) AR | NM_000466.3:c.2760delA p.(Ala921Leu fs*40) | 6.214 × 10−7 | HOM | Maternal and paternal | Pathogenic (PVS1; PM2_supporting; PM3) | [53] |
| Section B: Identification of Variants of Uncertain Significance | ||||||||||
| Foetal ID | Diagnostic Group | Phenotypic Group | Gene | Associated Disease and Inheritance Pattern | DNA Change | Frequency | Zygosity | Inheritance | ACMG/AMP Classification | Ref. |
| 24 | Single major malformation | CNS anomalies | ZNF292 | Intellectual developmental disorder, autosomal dominant 64 (MIM: # 619188) AD | NM_015021.3:c.6325A>C p.(Ser2109Arg) | NA | HET | De novo | Variant of Uncertain Significance (PS2; PM2_supporting) | NA |
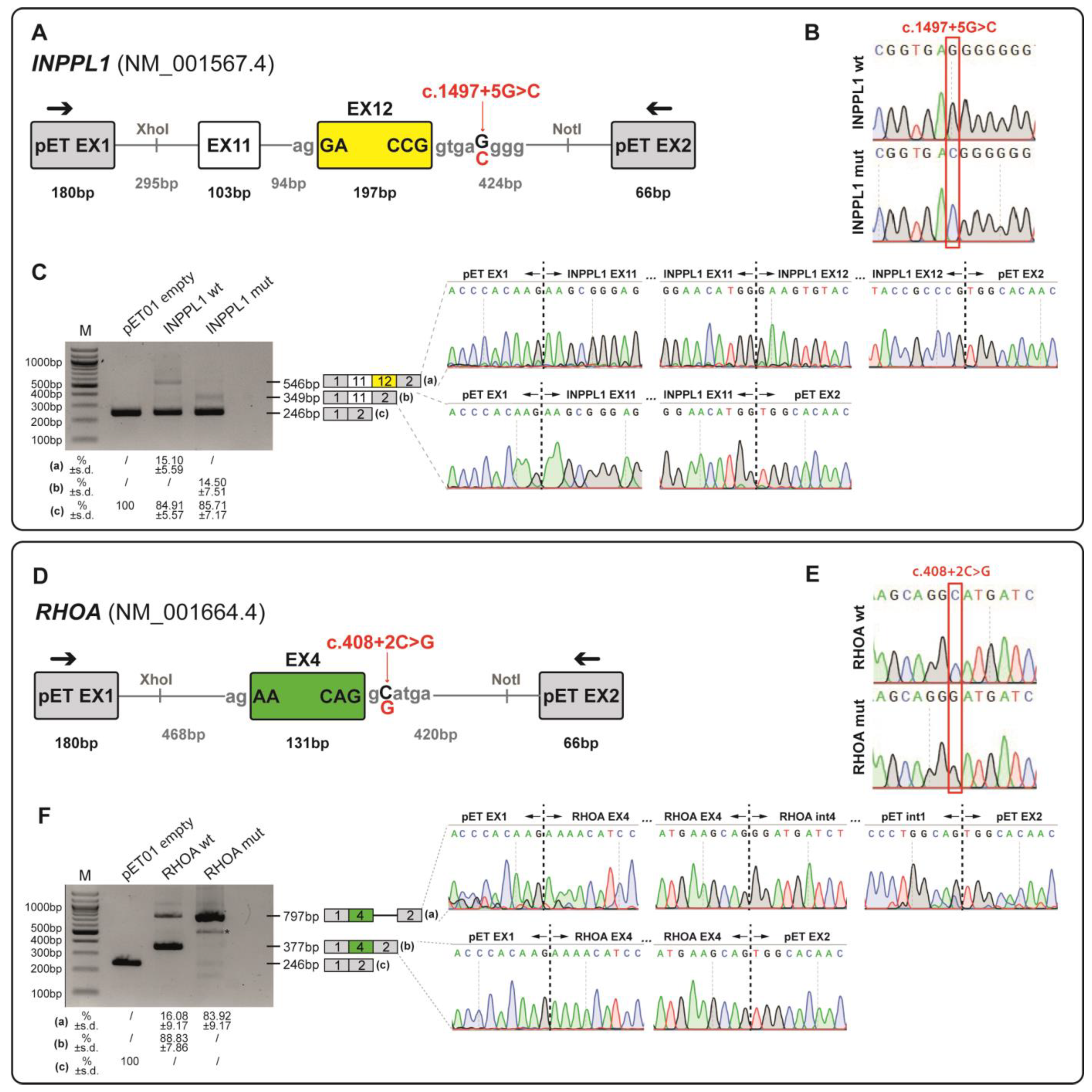
| 25 | Single major malformation | Skeletal anomalies | INPPL1 | Opsismodysplasia (MIM: # 258480) AR | NM_001567.4:c.1497+5G>C | 0.00001371 | HOM | Maternal and paternal | Variant of Uncertain Significance (PM2_supporting; PM3_supporting; PP3) | NA |
| 26 | Single major malformation | Cardiac anomalies | NOTCH2 | Alagille syndrome 2 (MIM: # 610205) AD | NM_024408.4:c.5177G>A p.(Arg1726His) | NA | HET | Paternal | Variant of Uncertain Significance (PM2_supporting) | [37] |
| 27 | Single major malformation | Fluid accumulation | NOTCH2 | Alagille syndrome 2 (MIM: # 610205) AD | NM_024408.4:c.5177G>A p.(Arg1726His) | NA | HET | Paternal | Variant of Uncertain Significance (PM2_supporting) | [37] |
| 28 | Multiple malformations | Multisystemic condition | RHOA | Ectodermal dysplasia with facial dysmorphism and acral, ocular, and brain anomalies (MIM: # 618727) Somatic mosaicism | NM_001664.4:c.408+2C>G | NA | HET | De novo | Variant of Uncertain Significance (PS2; PM2_supporting; PP3) | NA |
| 29 | Multiple malformations | Multisystemic condition | COL6A1 | Ullrich congenital muscular dystrophy 1A (MIM: # 254090) AR | NM_001848.3:c.1712A>C p.(Lys571Thr) | 0.0002714 | COMP HET | Maternal | Variant of Uncertain Significance (PM2_supporting; PP3_moderate) | [38] |
| NM_001848.3:c.751G>A p.(Glu251Lys) | 0.0002554 | Paternal | Variant of Uncertain Significance (PM2_supporting; PP3) | [39] | ||||||
3.4. Functional Validation of INPPL1 and RHOA Splicing Variants Through Minigene Assay
3.4.1. Minigene Splicing Assay of NM_001567.4(INPPL1):c.1497+5G>C Variant
3.4.2. Minigene Splicing Assay of NM_001664.4(RHOA):c.408+2C>G Variant
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACMG/AMP | American College of Medical Genetics/Association for Molecular Pathology |
| AD | Autosomal Dominant |
| AR | Autosomal Recessive |
| CMA | Chromosomal Microarray |
| CNS | Central nervous system |
| CNV | Copy Number Variant |
| COMP HET | Compound Heterozygous |
| CT | Computed Tomography |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| HEK | Human Embryonic Kidney cells |
| HET | Heterozygous |
| HGMD | Human Gene Mutation Database |
| HGVS | Human Genome Variation Society |
| HOM | Homozygous |
| HPO | Human Phenotype Ontology |
| MIM | Mendelian Inheritance in Man |
| MRI | Magnetic Resonance Imaging |
| MUT | Mutated |
| NGS | Next Generation Sequencing |
| NT | Nuchal Translucency |
| QF-PCR | Quantitative Fluorescence Polymerase Chain Reaction |
| RT-PCR | Reverse Transcription Polymerase Chain Reaction |
| SNP | Single Nucleotide Polymorphism |
| VUS | Variants of Uncertain Significance |
| WES | Whole-Exome Sequencing |
| WT | Wild-Type |
| XL | X-linked |
| XLD | X-linked Dominant |
References
- Al-Dewik, N.; Samara, M.; Younes, S.; Al-jurf, R.; Nasrallah, G.; Al-Obaidly, S.; Salama, H.; Olukade, T.; Hammuda, S.; Marlow, N.; et al. Prevalence, predictors, and outcomes of major congenital anomalies: A population-based register study. Sci. Rep. 2023, 13, 2198. [Google Scholar] [CrossRef]
- Nguengang Wakap, S.; Lambert, D.M.; Olry, A.; Rodwell, C.; Gueydan, C.; Lanneau, V.; Murphy, D.; Le Cam, Y.; Rath, A. Estimating cumulative point prevalence of rare diseases: Analysis of the Orphanet database. Eur. J. Hum. Genet. 2020, 28, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Yates, C.L.; Monaghan, K.G.; Copenheaver, D.; Retterer, K.; Scuffins, J.; Kucera, C.R.; Friedman, B.; Richard, G.; Juusola, J. Whole-exome sequencing on deceased fetuses with ultrasound anomalies: Expanding our knowledge of genetic disease during fetal development. Genet. Med. 2017, 19, 1171–1178. [Google Scholar] [CrossRef] [PubMed]
- Petrovski, S.; Aggarwal, V.; Giordano, J.L.; Stosic, M.; Wou, K.; Bier, L.; Spiegel, E.; Brennan, K.; Stong, N.; Jobanputra, V.; et al. Whole-exome sequencing in the evaluation of fetal structural anomalies: A prospective cohort study. Lancet 2019, 393, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yin, F.; Chai, Y.; Jin, J.; Zhang, P.; Tan, Q.; Chen, Z. Prenatal Diagnosis of Fetuses with Ultrasound Anomalies by Whole-Exome Sequencing in Luoyang City, China. Front Genet. 2024, 14, 1301439. Available online: https://www.frontiersin.org/journals/genetics/articles/10.3389/fgene.2023.1301439 (accessed on 31 December 2024). [CrossRef]
- Sadlecki, P.; Walentowicz-Sadlecka, M. Prenatal Diagnosis of Fetal Defects and Its Implications on the Delivery Mode. Open Med. 2023, 18, 20230704. [Google Scholar] [CrossRef]
- Salomon, L.J.; Alfirevic, Z.; Berghella, V.; Bilardo, C.M.; Chalouhi, G.E.; Da Silva Costa, F.; Hernandez-Andrade, E.; Malinger, G.; Munoz, H.; Paladini, D.; et al. ISUOG Practice Guidelines (updated): Performance of the routine mid-trimester fetal ultrasound scan. Ultrasound Obstet. Gynecol. 2022, 59, 840–856. [Google Scholar] [CrossRef]
- Prayer, D.; Malinger, G.; De Catte, L.; De Keersmaecker, B.; Gonçalves, L.F.; Kasprian, G.; Laifer-Narin, S.; Lee, W.; Millischer, A.; Platt, L.; et al. ISUOG Practice Guidelines (updated): Performance of fetal magnetic resonance imaging. Ultrasound Obstet. Gynecol. 2023, 61, 278–287. [Google Scholar] [CrossRef]
- Ghi, T.; Sotiriadis, A.; Calda, P.; Da Silva Costa, F.; Raine-Fenning, N.; Alfirevic, Z.; McGillivray, G. ISUOG Practice Guidelines: Invasive procedures for prenatal diagnosis. Ultrasound Obstet. Gynecol. 2016, 48, 256–268. [Google Scholar] [CrossRef]
- Shaffer, L.G.; Bejjani, B.A. A cytogeneticist’s perspective on genomic microarrays. Hum. Reprod. Update 2004, 10, 221–226. [Google Scholar] [CrossRef]
- American College of Obstetricians and Gynecologists Committee on Genetics. Committee Opinion No. 581: The Use of Chromosomal Microarray Analysis in Prenatal Diagnosis. Obstet. Gynecol. 2013, 122, 1374–1377. Available online: https://pubmed.ncbi.nlm.nih.gov/24264715/ (accessed on 31 December 2024). [CrossRef]
- Dugoff, L.; Norton, M.E.; Kuller, J.A. The use of chromosomal microarray for prenatal diagnosis. Am. J. Obstet. Gynecol. 2016, 215, B2–B9. [Google Scholar] [CrossRef]
- Almubarak, A.; Zhang, D.; Kosak, M.; Rathwell, S.; Doonanco, J.; Eaton, A.J.; Kannu, P.; Lazier, J.; Lui, M.; Niederhoffer, K.Y.; et al. Prenatal Genetic Testing in the Era of Next Generation Sequencing: A One-Center Canadian Experience. Genes 2022, 13, 2019. [Google Scholar] [CrossRef] [PubMed]
- Olayiwola, J.O.; Marhabaie, M.; Koboldt, D.; Matthews, T.; Siemon, A.; Mouhlas, D.; Porter, T.; Kyle, G.; Myers, C.; Mei, H.; et al. Clinically significant findings in a decade-long retrospective study of prenatal chromosomal microarray testing. Mol. Genet. Genom. Med. 2024, 12, e2349. [Google Scholar] [CrossRef] [PubMed]
- Mellis, R.; Oprych, K.; Scotchman, E.; Hill, M.; Chitty, L.S. Diagnostic yield of exome sequencing for prenatal diagnosis of fetal structural anomalies: A systematic review and meta-analysis. Prenat Diagn. 2022, 42, 662–685. [Google Scholar] [CrossRef] [PubMed]
- Shamseldin, H.E.; Kurdi, W.; Almusafri, F.; Alnemer, M.; Alkaff, A.; Babay, Z.; Alhashem, A.; Tulbah, M.; Alsahan, N.; Khan, R.; et al. Molecular autopsy in maternal-fetal medicine. Genet. Med. 2018, 20, 420–427. [Google Scholar] [CrossRef]
- Quinlan-Jones, E.; Lord, J.; Williams, D.; Hamilton, S.; Marton, T.; Eberhardt, R.Y.; Rinck, G.; Prigmore, E.; Keelagher, R.; McMullan, D.J.; et al. Molecular autopsy by trio exome sequencing (ES) and postmortem examination in fetuses and neonates with prenatally identified structural anomalies. Genet. Med. 2019, 21, 1065–1073. [Google Scholar] [CrossRef]
- Peyronnet, V.; Anselem, O.; Loeuillet, L.; Roux, N.; Tsatsaris, V. Diagnostic value of fetal autopsy after early termination of pregnancy for fetal anomalies. PLoS ONE 2022, 17, e0275674. [Google Scholar] [CrossRef]
- Byrne, A.B.; Arts, P.; Ha, T.T.; Kassahn, K.S.; Pais, L.S.; O’Donnell-Luria, A.; Babic, M.; Frank, M.S.B.; Feng, J.; Wang, P.; et al. Genomic autopsy to identify underlying causes of pregnancy loss and perinatal death. Nat. Med. 2023, 29, 180–189. [Google Scholar] [CrossRef]
- Daum, H.; Meiner, V.; Elpeleg, O.; Harel, T.; Bar-Or, L.; Eilat, A.; Fahham, D.; Gur, M.; Hacohen, N.; Kimchi, A. Fetal exome sequencing: Yield and limitations in a tertiary referral center. Ultrasound Obstet. Gynecol. 2019, 53, 80–86. [Google Scholar] [CrossRef]
- Gold, N.B.; Nadel, A.; Green, R.C. Ready or not, genomic screening of fetuses is already here. Genet. Med. 2024, 26, 01008. [Google Scholar] [CrossRef]
- Bromley, B.; Platt, L.D. First-Trimester Ultrasound Screening in Routine Obstetric Practice. Obstet. Gynecol. 2024, 143, 730–744. Available online: https://journals.lww.com/greenjournal/fulltext/2024/06000/first_trimester_ultrasound_screening_in_routine.4.aspx (accessed on 31 December 2024). [CrossRef]
- Carmen Prodan, N.; Hoopmann, M.; Jonaityte, G.; Oliver Kagan, K. How to do a second trimester anomaly scan. Arch. Gynecol. Obstet. 2023, 307, 1285–1290. [Google Scholar] [CrossRef]
- Guimaraes, C.V.A.; Dahmoush, H.M. Fetal Brain Anatomy. Neuroimaging Clin. N. Am. 2022, 32, 663–681. Available online: https://www.sciencedirect.com/science/article/pii/S1052514922000259 (accessed on 31 December 2024). [CrossRef]
- Aggarwal, S.; Tandon, A.; Das Bhowmik, A.; Safarulla, J.M.N.J.; Dalal, A. A Dysmorphology Based Systematic Approach Toward Perinatal Genetic Diagnosis in a Fetal Autopsy Series. Fetal Pediatr. Pathol. 2018, 37, 49–68. [Google Scholar] [CrossRef]
- Venkataswamy, C.; Gurusamy, U.; Lakshmi, S.V. Second-Trimester Fetal Autopsy: A Morphological Study with Prenatal USG Correlations and Clinical Implications. J. Lab. Physicians 2018, 10, 338–345. Available online: http://www.thieme-connect.com/products/ejournals/abstract/10.4103/JLP.JLP_134_17 (accessed on 31 December 2024). [CrossRef]
- Conlin, L.K.; Thiel, B.D.; Bonnemann, C.G.; Medne, L.; Ernst, L.M.; Zackai, E.H.; Deardorff, M.A.; Krantz, I.D.; Halkonarson, H.; Spinner, N.B. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum. Mol Genet. 2010, 19, 1263–1275. [Google Scholar] [CrossRef] [PubMed]
- Spedicati, B.; Santin, A.; Nardone, G.G.; Rubinato, E.; Lenarduzzi, S.; Graziano, C.; Garavelli, L.; Miccoli, S.; Bigoni, S.; Morgan, A.; et al. The Enigmatic Genetic Landscape of Hereditary Hearing Loss: A Multistep Diagnostic Strategy in the Italian Population. Biomedicines 2023, 11, 703. [Google Scholar] [CrossRef] [PubMed]
- Highnam, G.; Wang, J.J.; Kusler, D.; Zook, J.; Vijayan, V.; Leibovich, N.; Mittelman, D. An analytical framework for optimizing variant discovery from personal genomes. Nat. Commun. 2015, 6, 6275. [Google Scholar] [CrossRef] [PubMed]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef]
- Limongelli, I.; Marini, S.; Bellazzi, R. PaPI: Pseudo amino acid composition to score human protein-coding variants. BMC Bioinform. 2015, 16, 123. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7–20. [Google Scholar]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed]
- Jian, X.; Boerwinkle, E.; Liu, X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014, 42, 13534–13544. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef]
- Jaganathan, K.; Kyriazopoulou Panagiotopoulou, S.; McRae, J.F.; Darbandi, S.F.; Knowles, D.; Li, Y.I.; Kosmicki, J.A.; Arbelaez, J.; Cui, W.; Schwartz, G.B.; et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell 2019, 176, 535–548.e24. [Google Scholar] [CrossRef] [PubMed]
- Sevim Bayrak, C.; Zhang, P.; Tristani-Firouzi, M.; Gelb, B.D.; Itan, Y. De novo variants in exomes of congenital heart disease patients identify risk genes and pathways. Genome Med. 2020, 12, 9. [Google Scholar] [CrossRef]
- Lampe, A.K.; Dunn, D.M.; von Niederhausern, A.C.; Hamil, C.; Aoyagi, A.; Laval, S.H.; Marie, S.K.; Chu, M.-L.; Swoboda, K.; Muntoni, F.; et al. Automated Genomic Sequence Analysis of the Three Collagen VI Genes: Applications to Ullrich Congenital Muscular Dystrophy and Bethlem Myopathy. J. Med. Genet. 2005, 42, 108. Available online: http://jmg.bmj.com/content/42/2/108.abstract (accessed on 31 December 2024).
- Gupta, P.; Anne, R.P.; Deshabhotla, S.K.; Nerakh, G. An Infant with Blended Phenotype of Zellweger Spectrum Disorder and Congenital Muscular Dystrophy. Ann. Indian Acad. Neurol. 2021, 24, 759–760. Available online: https://journals.lww.com/annalsofian/fulltext/2021/24050/an_infant_with_blended_phenotype_of_zellweger.18.aspx (accessed on 31 December 2024). [CrossRef]
- Mirzaa, G.M.; Chong, J.X.; Piton, A.; Popp, B.; Foss, K.; Guo, H.; Harripaul, R.; Xia, K.; Scheck, J.; Aldinger, K.A.; et al. De novo and inherited variants in ZNF292 underlie a neurodevelopmental disorder with features of autism spectrum disorder. Genet. Med. 2020, 22, 538–546. [Google Scholar] [CrossRef]
- Lopes-Marques, M.; Mort, M.; Carneiro, J.; Azevedo, A.; Amaro, A.P.; Cooper, D.N.; Azevedo, L. Meta-analysis of 46,000 germline de novo mutations linked to human inherited disease. Hum. Genom. 2024, 18, 20. [Google Scholar] [CrossRef]
- Zhang, M.; Li, F.X.; Liu, X.Y.; Hou, J.Y.; Ni, S.H.; Wang, J.; Zhao, C.; Zhang, W.; Kong, Y.; Huang, R.; et al. TBX1 loss-of-function mutation contributes to congenital conotruncal defects. Exp. Ther. Med. 2018, 15, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Vabres, P.; Sorlin, A.; Kholmanskikh, S.S.; Demeer, B.; St-Onge, J.; Duffourd, Y.; Kuentz, P.; Courcet, J.-B.; Carmignac, V.; Garret, P.; et al. Postzygotic inactivating mutations of RHOA cause a mosaic neuroectodermal syndrome. Nat. Genet. 2019, 51, 1438–1441. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Krishnappa, J.; Lin, G.; Kavalloor, N.; Lim, J.Y.; Goh, C.Y.J.; Jamuar, S.S.; Thomas, T.; Tan, E.C. Microcephaly with a simplified gyral pattern in a child with a de novo TUBA1A variant. Am. J. Med. Genet. A 2020, 182, 576–578. [Google Scholar] [CrossRef] [PubMed]
- Allou, L.; Lambert, L.; Amsallem, D.; Bieth, E.; Edery, P.; Destrée, A.; Rivier, F.; Amor, D.; Thompson, E.; Nicholl, J.; et al. 14q12 and severe Rett-like phenotypes: New clinical insights and physical mapping of FOXG1-regulatory elements. Eur. J. Hum. Genet. 2012, 20, 1216–1223. [Google Scholar] [CrossRef]
- Leclerc-Mercier, S.; Dufernez, F.; Fraitag, S.; Coulombe, J.; Dompmartin, A.; Barreau, M.; Bozon, D.; Lamazière, A.; Bonnefont, J.-P.; Khalifa, E.; et al. Keratotic follicular plugs with calcifications in Conradi–Hünermann–Happle syndrome: Histological, biochemical and genetic testing correlation. Br. J. Dermatol. 2015, 173, 1316–1318. [Google Scholar] [CrossRef]
- Stals, K.L.; Wakeling, M.; Baptista, J.; Caswell, R.; Parrish, A.; Rankin, J.; Tysoe, C.; Jones, G.; Gunning, A.C.; Lango Allen, H.; et al. Diagnosis of lethal or prenatal-onset autosomal recessive disorders by parental exome sequencing. Prenat. Diagn. 2018, 38, 33–43. [Google Scholar] [CrossRef]
- Groener, J.; Maaswinkel-Mooy, P.; Smit, V.; Hoeven, M.V.D.; Bakker, J.; Campos, Y.; d’Azzo, A. New mutations in Two Dutch Patients with Early Infantile Galactosialidosis. Mol Genet Metab. 2003, 78, 222–228. Available online: https://www.sciencedirect.com/science/article/pii/S1096719203000052 (accessed on 31 December 2024). [CrossRef]
- van der Sluijs, P.J.; Jansen, S.; Vergano, S.A.; Adachi-Fukuda, M.; Alanay, Y.; AlKindy, A.; Baban, A.; Bayat, A.; Beck-Wodl, S.; Berry, K.; et al. The ARID1B Spectrum in 143 Patients: From Nonsyndromic Intellectual Disability to Coffin–Siris Syndrome. Genet. Med. 2019, 21, 1295–1307. Available online: https://www.sciencedirect.com/science/article/pii/S1098360021016464 (accessed on 31 December 2024). [CrossRef]
- Wilkie, A.O.M.; Slaney, S.F.; Oldridge, M.; Poole, M.D.; Ashworth, G.J.; Hockley, A.D.; Hayward, R.D.; David, D.J.; Pulleyn, L.J.; Rutland, P.; et al. Apert syndrome results from localized mutations of FGFR2 and is allelic with Crouzon syndrome. Nat. Genet. 1995, 9, 165–172. [Google Scholar] [CrossRef]
- Sandomirsky, K.; Marchi, E.; Gavin, M.; Amble, K.; Lyon, G.J. Phenotypic variability and gastrointestinal manifestations/interventions for growth in NAA10-related neurodevelopmental syndrome. Am. J. Med. Genet. A 2023, 191, 1293–1300. [Google Scholar] [CrossRef] [PubMed]
- Cmejla, R.; Blafkova, J.; Stopka, T.; Zavadil, J.; Pospisilova, D.; Mihal, V.; Petrtylova, K.; Jelinek, J. Ribosomal Protein S19 Gene Mutations in Patients with Diamond-Blackfan Anemia and Identification of Ribosomal Protein S19 Pseudogenes. Blood Cells Mol. Dis. 2000, 26, 124–132. Available online: https://www.sciencedirect.com/science/article/pii/S1079979600902869 (accessed on 31 December 2024). [CrossRef]
- Ebberink, M.S.; Mooijer, P.A.W.; Gootjes, J.; Koster, J.; Wanders, R.J.A.; Waterham, H.R. Genetic classification and mutational spectrum of more than 600 patients with a Zellweger syndrome spectrum disorder. Hum. Mutat. 2011, 32, 59–69. [Google Scholar] [CrossRef]
- Carss, K.J.; Hillman, S.C.; Parthiban, V.; McMullan, D.J.; Maher, E.R.; Kilby, M.D.; Hurles, M.E. Exome sequencing improves genetic diagnosis of structural fetal abnormalities revealed by ultrasound. Hum. Mol. Genet. 2014, 23, 3269–3277. [Google Scholar] [CrossRef]
- Boissel, S.; Fallet-Bianco, C.; Chitayat, D.; Kremer, V.; Nassif, C.; Rypens, F.; Delrue, M.-A.; Dal Soglio, D.; Oligny, L.L.; Patey, N.; et al. Genomic study of severe fetal anomalies and discovery of GREB1L mutations in renal agenesis. Genet. Med. 2018, 20, 745–753. [Google Scholar] [CrossRef]
- Takayasu, H.; Masumoto, K.; Hagiwara, K.; Sasaki, T.; Ono, K.; Jimbo, T.; Uesugi, T.; Gotoh, C.; Urita, Y.; Shinkai, T.; et al. Increased pulmonary RhoA expression in the nitrofen-induced congenital diaphragmatic hernia rat model. J. Pediatr. Surg. 2015, 50, 1467–1471. [Google Scholar] [CrossRef]
- Morton, S.U.; Shimamura, A.; Newburger, P.E.; Opotowsky, A.R.; Quiat, D.; Pereira, A.C.; Jin, S.C.; Gurvitz, M.; Brueckner, M.; Chung, W.K.; et al. Association of Damaging Variants in Genes With Increased Cancer Risk Among Patients with Congenital Heart Disease. JAMA Cardiol. 2021, 6, 457–462. [Google Scholar] [CrossRef]
| Diagnostic Group | Phenotypic Group | Total Number (%) | Males—n (%) | Females—n (%) | p-Value |
|---|---|---|---|---|---|
| Single major malformation | CNS anomalies | 21 (24.4) | 11 (52.4) | 10 (47.6) | 0.52 |
| Single major malformation | Skeletal anomalies | 7 (8.1) | 3 (42.9) | 4 (57.1) | 0.43 |
| Single major malformation | Cardiac anomalies | 5 (5.8) | 3 (60) | 2 (40) | 0.60 |
| Single major malformation | Urogenital anomalies | 4 (4.7) | 3 (75) | 1 (25) | 0.75 |
| Single major malformation | Fluid accumulation | 5 (5.8) | 3 (60) | 2 (40) | 0.60 |
| Multiple malformations | Multisystemic condition | 44 (51.2) | 28 (63.6) | 16 (36.4) | 0.64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spedicati, B.; Pianigiani, G.; Santin, A.; Gasparini, V.R.; Falcomer, I.; Feresin, A.; Bonati, M.T.; Mazzà, D.; Paccagnella, E.; Pasquetti, D.; et al. Beyond the Curtains: Identification of the Genetic Cause of Foetal Developmental Abnormalities Through the Application of Molecular Autopsy. Genes 2025, 16, 1167. https://doi.org/10.3390/genes16101167
Spedicati B, Pianigiani G, Santin A, Gasparini VR, Falcomer I, Feresin A, Bonati MT, Mazzà D, Paccagnella E, Pasquetti D, et al. Beyond the Curtains: Identification of the Genetic Cause of Foetal Developmental Abnormalities Through the Application of Molecular Autopsy. Genes. 2025; 16(10):1167. https://doi.org/10.3390/genes16101167
Chicago/Turabian StyleSpedicati, Beatrice, Giulia Pianigiani, Aurora Santin, Vanessa Rebecca Gasparini, Ilaria Falcomer, Agnese Feresin, Maria Teresa Bonati, Daniela Mazzà, Elisa Paccagnella, Domizia Pasquetti, and et al. 2025. "Beyond the Curtains: Identification of the Genetic Cause of Foetal Developmental Abnormalities Through the Application of Molecular Autopsy" Genes 16, no. 10: 1167. https://doi.org/10.3390/genes16101167
APA StyleSpedicati, B., Pianigiani, G., Santin, A., Gasparini, V. R., Falcomer, I., Feresin, A., Bonati, M. T., Mazzà, D., Paccagnella, E., Pasquetti, D., Rubinato, E., Granata, C., Murru, F. M., Pinamonti, M., Bussani, R., Fantasia, I., Stampalija, T., Gasparini, P., Zampieri, S., & Girotto, G. (2025). Beyond the Curtains: Identification of the Genetic Cause of Foetal Developmental Abnormalities Through the Application of Molecular Autopsy. Genes, 16(10), 1167. https://doi.org/10.3390/genes16101167








