Cytogenomics of Frieseomelitta varia (Hymenoptera: Apidae) and the Sharing of a Satellite DNA Family in Several Neotropical Meliponini Genera
Abstract
1. Introduction
2. Materials and Methods
2.1. Analysis of satDNA in the Genome of the Stingless Bee F. varia
2.2. Obtaining Metaphase Chromosomes and Heterochromatin Distribution Patterns in Meliponini Species
2.3. Chromosomal Mapping of the Most Abundant satDNA Family from the F. varia Genome Using Fluorescent In Situ Hybridization (FISH)
3. Results
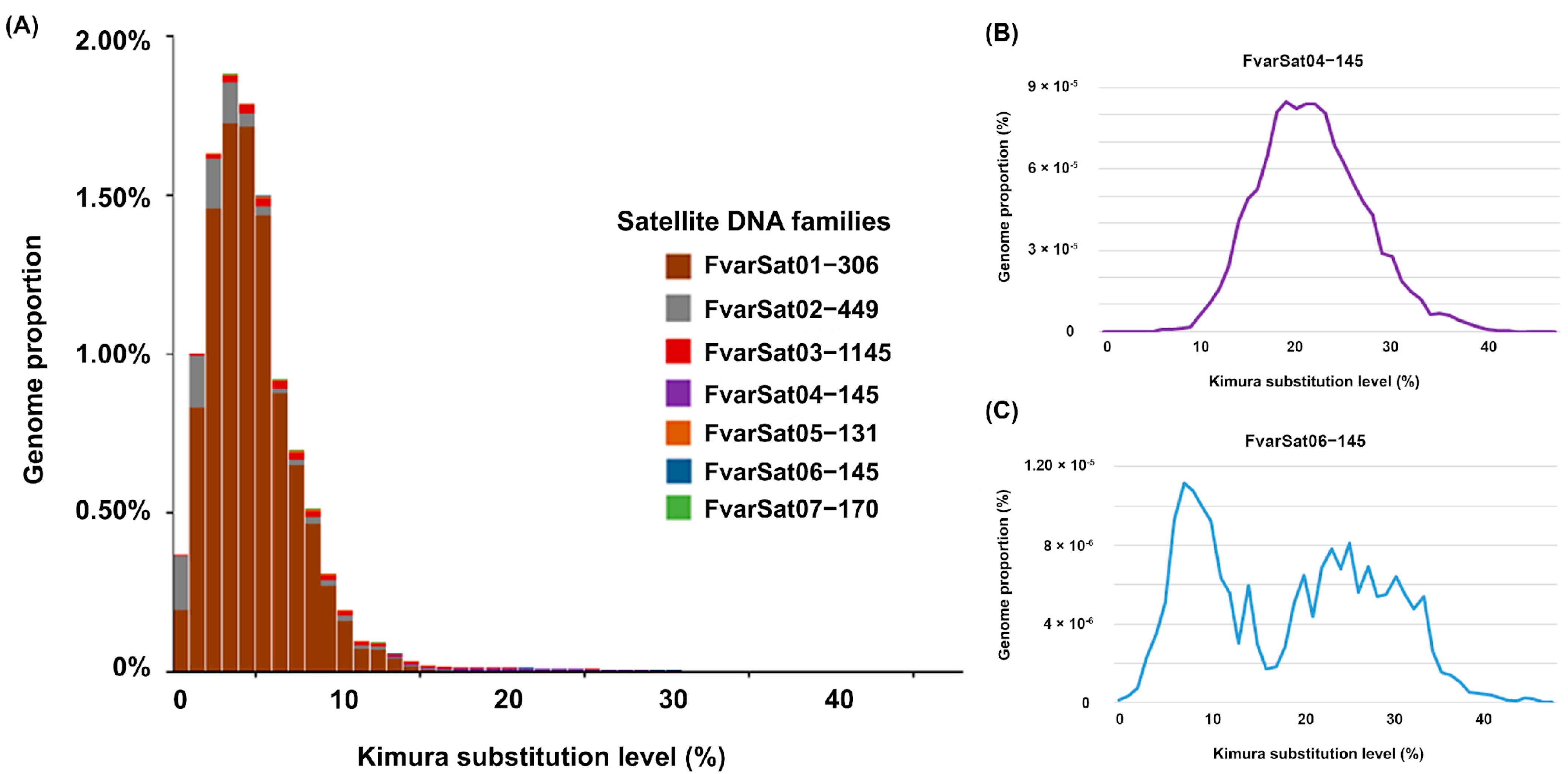
3.1. Genomic Analysis of satDNAs in F. varia
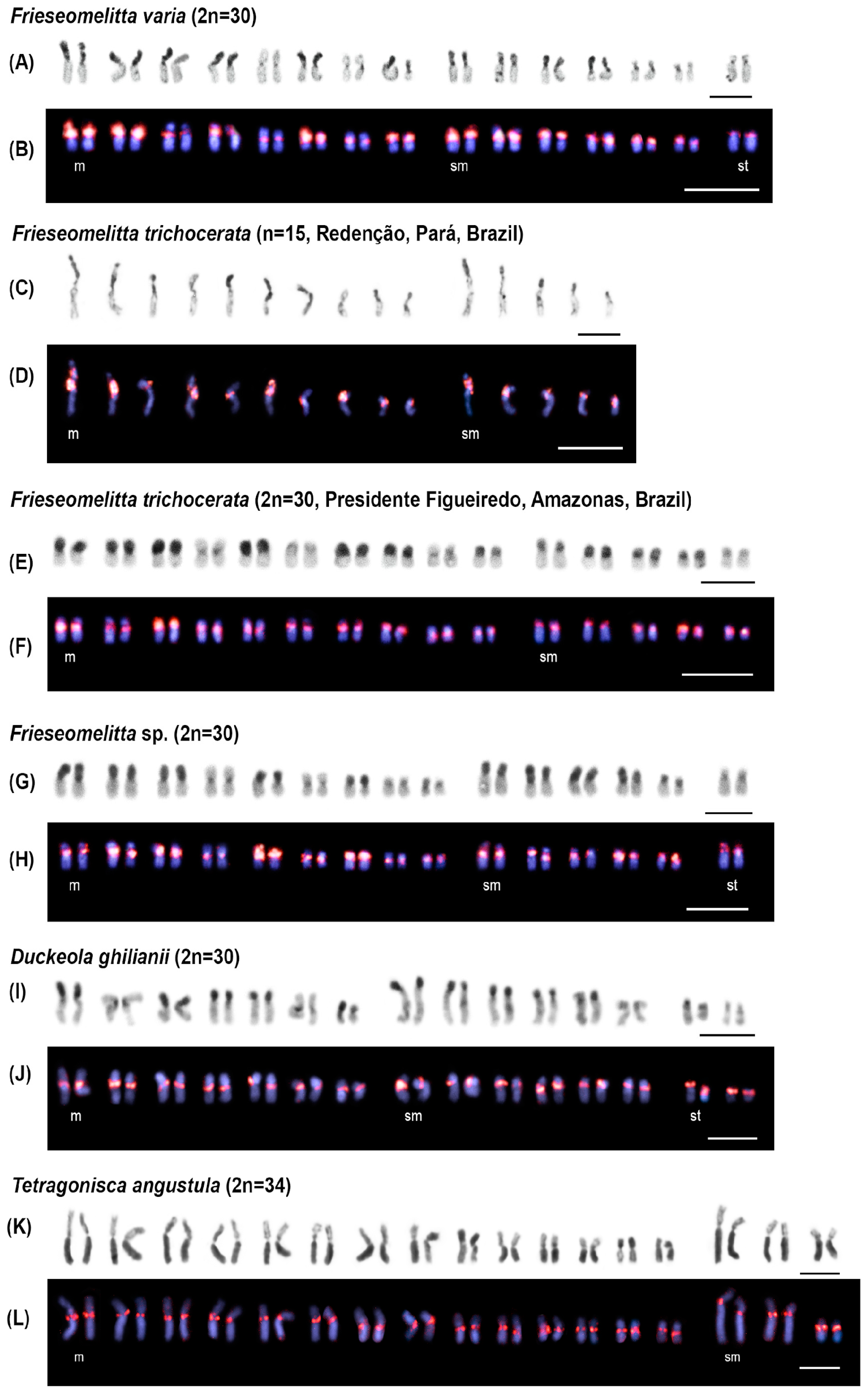
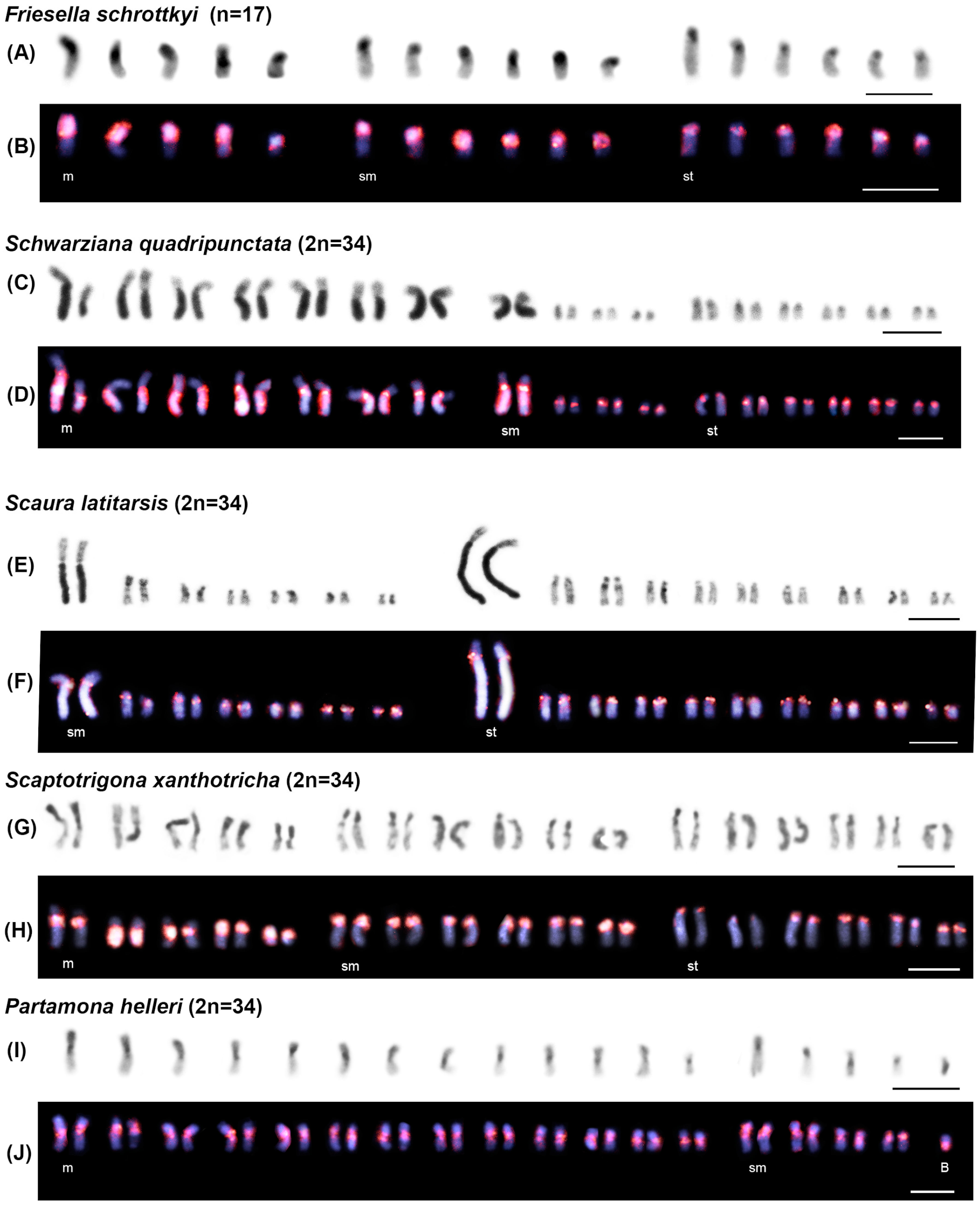
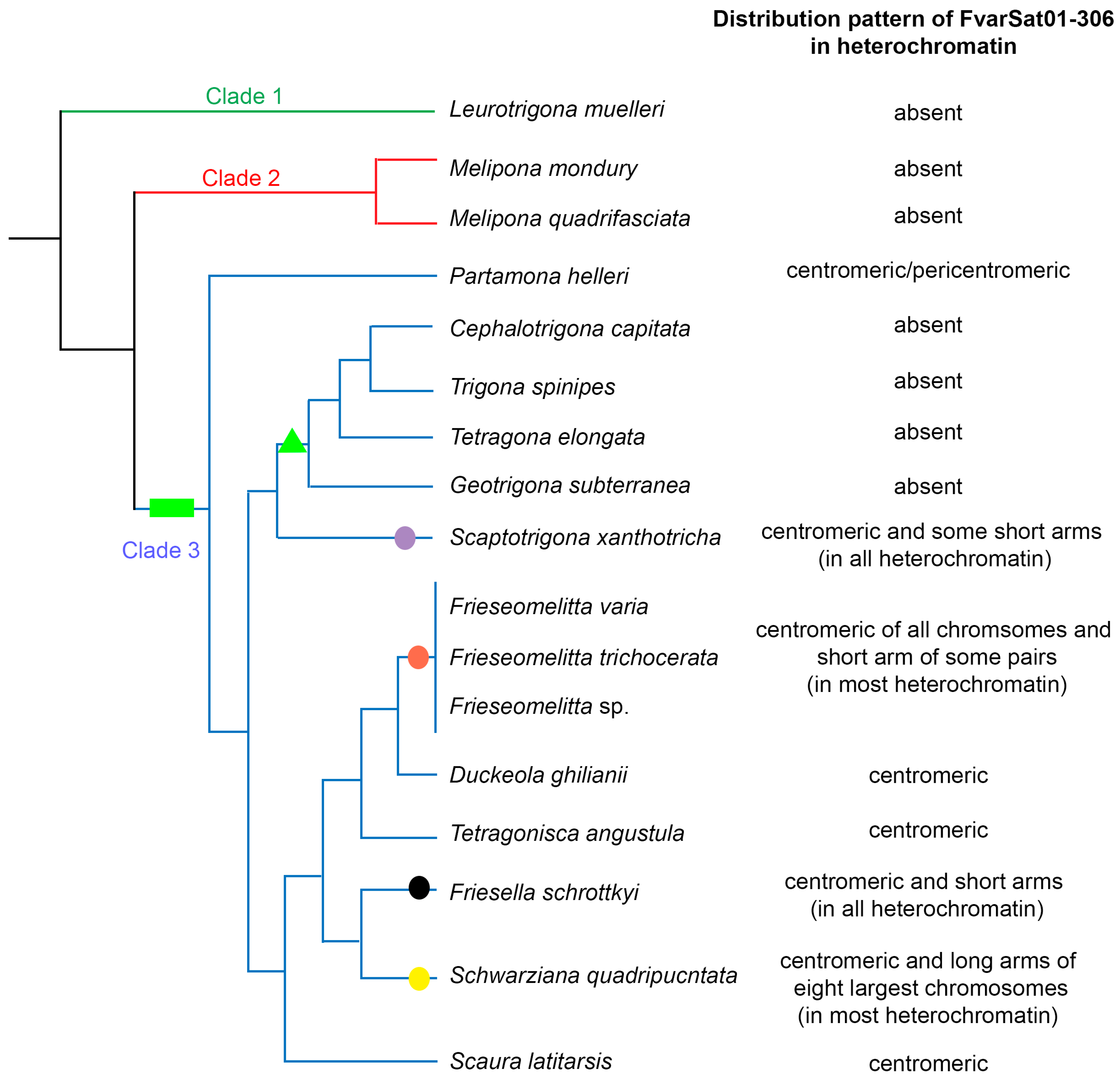
3.2. Chromosomal Analysis: C-Banding and Physical Mapping of the Most Abundant satDNA Family (FvarSat01-306) in Different Meliponini Species
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grewal, S.I.; Jia, S. Heterochromatin revisited. Nat. Rev. Genet. 2007, 8, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Allshire, R.C.; Madhani, H.D. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 2018, 19, 229–244. [Google Scholar] [CrossRef]
- Saksouk, N.; Simboeck, E.; Déjardin, J. Constitutive heterochromatin formation and transcription in mammals. Epigenet. Chromatin 2015, 8, 3. [Google Scholar] [CrossRef]
- Penagos-Puig, A.; Furlan-Magaril, M. Heterochromatin as an Important Driver of Genome Organization. Front. Cell Dev. Biol. 2020, 18, 579137. [Google Scholar] [CrossRef]
- Cunha, M.S.; Soares, F.A.F.; Clarindo, W.R.; Campos, L.A.O.; Lopes, D.M. Robertsonian rearrangements in Neotropical Meliponini karyotype evolution (Hymenoptera: Apidae: Meliponini). Insect Mol. Biol. 2021, 30, 379–389. [Google Scholar] [CrossRef]
- Tavares, M.G.; Carvalho, C.R.; Soares, F.A.F.; Campos, L.A.O. Genome size diversity in stingless bees (Hymenoptera: Apidae, Meliponini). Apidologie 2012, 43, 731–736. [Google Scholar] [CrossRef]
- Lopes, D.M.; Fernandes, A.; Diniz, D.; Scudeler, P.E.S.; Foresti, F.; Campos, L.A.O. Similarity of heterochromatic regions in the stingless bees (Hymenoptera: Meliponini) revealed by chromosome painting. Caryologia 2014, 67, 222–226. [Google Scholar] [CrossRef]
- Piccoli, M.C.A.; Bardella, V.B.; Cabral-de-Mello, D.C. Repetitive DNAs in Melipona scutellaris (Hymenoptera: Apidae: Meliponidae): Chromosomal distribution and test of multiple heterochromatin amplification in the genus. Apidologie 2018, 49, 497–504. [Google Scholar] [CrossRef]
- Cunha, M.S.; Campos, L.A.O.; Lopes, D.M. Insights into the heterochromatin evolution in the genus Melipona (Apidae: Meliponini). Insectes Soc. 2020, 67, 391–398. [Google Scholar] [CrossRef]
- Pereira, J.A.; Salomão, T.M.F.; Lopes, D.M. Different repetitive DNA sequences make up heterochromatin in Meliponini. Apidologie 2020, 51, 855–860. [Google Scholar] [CrossRef]
- Campos, C.L.; Teixeira, G.A.; Lopes, D.M.; Waldschmidt, A.M. Molecular cytogenetics reveals insights into heterochromatin composition and karyotype evolution in species of the Plebeia group (Apidae: Meliponini). Insectes Soc. 2024. submitted. [Google Scholar]
- Pereira, J.A.; Milani, D.; Ferretti, A.B.S.M.; Bardella, V.B.; Cabral-de-Mello, D.C.; Lopes, D.M. The extensive amplification of heterochromatin in Melipona bees revealed by high throughput genomic and chromosomal analysis. Chromosoma 2021, 130, 251–262. [Google Scholar] [CrossRef]
- Pereira, J.A.; Cabral-de-Mello, D.C.; Lopes, D.M. The Satellite DNAs Populating the Genome of Trigona hyalinata and the Sharing of a Highly Abundant satDNA in Trigona Genus. Genes 2023, 14, 418. [Google Scholar] [CrossRef]
- Salser, W.; Bowen, S.; Browne, D.; El-Adli, F.; Fedoroff, N.; Fry, K.; Heindell, H.; Paddock, G.; Poon, R.; Wallace, B.; et al. Investigation of the organization of mammalian chromosomes at the DNA sequence level. Fed. Proc. 1976, 35, 23–35. [Google Scholar] [PubMed]
- Rasmussen, C.; Camargo, J.M.F. A molecular phylogeny and the evolution of nest architecture and behavior in Trigona s.s. (Hymenoptera: Apidae: Meliponini). Apidologie 2008, 39, 102–118. [Google Scholar] [CrossRef]
- De Paula-Freitas, F.C.; Lourenço, A.P.; Nunes, F.M.; Paschoal, A.R.; Abreu, F.C.; Barbin, F.O.; Bataglia, L.; Cardoso-Júnior, C.A.M.; Cervoni, M.S.; Silva, S.R.; et al. The nuclear and mitochondrial genomes of Frieseomelitta varia—A highly eusocial stingless bee (Meliponini) with a permanently sterile worker caste. BMC Genom. 2020, 21, 386. [Google Scholar] [CrossRef]
- Rocha, M.; Pompolo, S.; Campos, L.A.O. Citogenética da Tribo Meliponini (Hymenoptera, Apidae). In Apoidea Neotropica: Homenagem aos 90 anos de Jesus Santiago Moure; UNESC: Criciúma, Brazil, 2003; pp. 311–320. [Google Scholar]
- Santos, J.M.; Diniz, D.; Rodrigues, T.A.S.; Cioffi, M.D.B.; Waldschmidt, A.M. Heterochromatin Distribution and Chromosomal Mapping of Microsatellite Repeats in the Genome of Frieseomelitta Stingless Bees (Hymenoptera: Apidae: Meliponini). Fla. Entomol. 2018, 101, 33–39. [Google Scholar] [CrossRef]
- Carvalho, A.F.; Costa, M.A. Cytogenetic characterization of two species of Frieseomelitta Ihering, 1912 (Hymenoptera, Apidae, Meliponini). Genet. Mol. Biol. 2011, 34, 237–239. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Do Nascimento, R.M.; Carvalho, A.F.; Santana, W.C.; Barth, A.; Costa, M.A. Karyotype diversity of stingless bees of the genus Frieseomelitta (Hymenoptera, Apidae, Meliponini). Caryologia 2020, 73, 121–126. [Google Scholar]
- Rasmussen, C.; Cameron, S.A. Global stingless bee phylogeny supports ancient divergence, vicariance, and long distance dispersal. Biol. J. Linn. Soc. 2010, 99, 206–232. [Google Scholar] [CrossRef]
- Novák, P.; Neumann, P.; Macas, J. Global analysis of repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat. Protoc. 2020, 15, 3745–3776. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Ávila Robledillo, L.; Koblížková, A.; Vrbová, I.; Neumann, P.; Macas, J. TAREAN: A computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res. 2017, 45, e111. [Google Scholar] [CrossRef]
- Drummond, A.; Ashton, B.; Cheung, M.; Helid, J.; Kearse, M.; Moir, R.; Stones-Havas, S.; Thierer, T.; Wilson, S. Geneious v4.8. 2009. Available online: http://www.geneious.com (accessed on 22 February 2022.).
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 2016, 6, 28333. [Google Scholar] [CrossRef]
- Smit, A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. 2013–2015. 2017. Available online: http://www.repeatmasker.org/ (accessed on 22 February 2022).
- Imai, H.T.; Taylor, R.W.; Crozier, R.H. Modes of spontaneous chromosomal mutation and karyotype evolution in ants with reference to the minimum interaction hypothesis. Jpn. J. Genet. 1988, 63, 159–185. [Google Scholar] [CrossRef] [PubMed]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 83, 304–306. [Google Scholar] [CrossRef]
- Imai, H.T. Mutability of constitutive heterochromatin (C-bands) during eukaryotic chromosomal evolution and their cytological meaning. Jpn. J. Genet. 1991, 66, 635–661. [Google Scholar] [CrossRef]
- Pinkel, D.; Straume, T.; Gray, J.W. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc. Natl. Acad. Sci. USA 1986, 83, 2934–2938. [Google Scholar] [CrossRef]
- Camacho, J.P.M.; Cabrero, J.; López-León, M.D.; Martín-Peciña, M.; Perfectti, F.; Garrido-Ramos, M.A.; Ruiz-Ruano, F.J. Satellitome comparison of two oedipodine grasshoppers highlights the contingent nature of satellite DNA evolution. BMC Biol. 2022, 20, 36. [Google Scholar]
- Mestrović, N.; Plohl, M.; Mravinac, B.; Ugarković, D. Evolution of satellite DNAs from the genus Palorus—Experimental evidence for the “library” hypothesis. Mol. Biol. Evol. 1998, 15, 1062–1068. [Google Scholar] [CrossRef]
- Bolsheva, N.L.; Melnikova, N.V.; Kirov, I.V.; Dmitriev, A.A.; Krasnov, G.S.; Amosova, A.V.; Samatadze, T.E.; Yurkevich, O.Y.; Zoshchuk, A.S.; Kudryavtseva, A.V.; et al. Characterization of repeated DNA sequences in genomes of blue-flowered flax. BMC Evol. Biol. 2019, 19, 79–88. [Google Scholar] [CrossRef]
- Palacios-Gimenez, O.M.; Milani, D.; Song, H.; Marti, D.A.; López-León, M.D.; Ruiz-Ruano, F.J.; Camacho, J.P.M.; Cabral-de-Mello, D.C. Eight million years of satellite DNA evolution in grasshoppers of the genus Schistocerca illuminate the ins and outs of the library hypothesis. Genome Biol. Evol. 2020, 12, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.G.; Ruiz-Ruano, F.J. In-depth satellitome analyses of 37 Drosophila species illuminate repetitive DNA evolution in the Drosophila genus. Genome Biol. Evol. 2022, 14, evac064. [Google Scholar]
- Anjos, A.; Milani, D.; Bardella, V.B.; Paladini, A.; Cabral-de-Mello, D.C. Evolution of satDNAs on holocentric chromosomes: Insights from hemipteran insects of the genus Mahanarva. Chromosome Res. 2023, 31, 5. [Google Scholar] [CrossRef]
- Mora, P.; Rico-Porras, J.M.; Palomeque, T.; Montiel, E.E.; Pita, S.; Cabral-de-Mello, D.C.; Lorite, P. Satellitome Analysis of Adalia bipunctata (Coleoptera): Revealing Centromeric Turnover and Potential Chromosome Rearrangements in a Comparative Interspecific Study. Int. J. Mol. Sci. 2024, 25, 9214. [Google Scholar] [CrossRef] [PubMed]
- Félix, A.P.; de Amorim, I.C.; Milani, D.; Cabral-de-Mello, D.C.; Moura, R.C. Differential amplification and contraction of satellite DNAs in the distinct lineages of the beetle Euchroma gigantea. Gene 2024, 927, 148723. [Google Scholar] [CrossRef]
- Cabral-de-Mello, D.C.; Palacios-Gimenez, O.M. Repetitive DNAs: The “invisible” regulators of insect adaptation and speciation. Curr. Opin. Insect Sci. 2024, 67, 101295. [Google Scholar] [CrossRef] [PubMed]
- Fry, K.; Salser, W. Nucleotide sequences of HS-α satellite DNA from kangaroo rat Dipodomys ordii and characterization of similar sequences in other rodents. Cell 1977, 12, 1069–1084. [Google Scholar] [CrossRef]
- Plohl, M.; Meštrović, N.; Mravinac, B. Centromere identity from the DNA point of view. Chromosoma 2014, 123, 313–325. [Google Scholar] [CrossRef] [PubMed]
| Species | Phylogenetic Clades | Locality |
|---|---|---|
| Frieseomelitta varia Lepeletier, 1836 | Clade 3 | Uberlândia, MG, Brazil |
| Frieseomelitta trichocerata Moure, 1990 | Clade 3 | Redenção, PA, Brazil; Presidente Figueiredo, AM, Brazil |
| Frieseomelitta sp. | Clade 3 | Brasília, DF, Brazil |
| Duckeola ghilianii Spinola, 1853 | Clade 3 | Presidente Figueiredo, AM, Brazil |
| Tetragonisca angustula Latreille, 1811 | Clade 3 | Mutum, MG, Brazil |
| Friesella schrottkyi Friese, 1900 | Clade 3 | Rio Paranaíba, MG, Brazil |
| Schwarziana quadripunctata Latreille, 1836 | Clade 3 | Viçosa, MG, Brazil |
| Scaura latitarsis Friese, 1900 | Clade 3 | Presidente Figueiredo, AM, Brazil |
| Scaptotrigona xanthotricha Moure, 1950 | Clade 3 | Viçosa, MG, Brazil |
| Geotrigona subterranea Friese, 1901 | Clade 3 | Passos, MG, Brazil |
| Tetragona elongata Lepeletier & Serville, 1828 | Clade 3 | Viçosa, MG, Brazil |
| Trigona spinipes Fabricius, 1793 | Clade 3 | Januária, MG, Brazil |
| Cephalotrigona capitata Smith, 1854 | Clade 3 | Viçosa, MG, Brazil |
| Partamona helleri Friese, 1900 | Clade 3 | Viçosa, MG, Brazil |
| Melipona quadrifasciata Lepeletier, 1836 | Clade 2 | Viçosa, MG, Brazil |
| Melipona mondury Smith, 1863 | Clade 2 | Viçosa, MG, Brazil |
| Leurotrigona muelleri Friese, 1900 | Clade 1 | Passos, MG, Brazil |
| SatDNA Family | SF | ML | A + T% | Divergence % | Abundance % |
|---|---|---|---|---|---|
| FvarSat01-306 | 306 | 66 | 4.62 | 9.971% | |
| FvarSat02-449 | 449 | 53.2 | 2.92 | 0.803% | |
| FvarSat03-1145 | 1145 | 58.4 | 7.73 | 0.245% | |
| FvarSat04-145 | 1 | 145 | 54.5 | 22.09 | 0.117% |
| FvarSat05-131 | 131 | 72.5 | 8.67 | 0.054% | |
| FvarSat06-145 | 1 | 145 | 59.3 | 18.36 | 0.019% |
| FvarSat07-170 | 170 | 71.2 | 6.13 | 0.014% | |
| Total | 11.223% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vignati, Z.B.M.; Teixeira, G.A.; Cunha, M.S.; Pereira, J.A.; Lopes, D.M. Cytogenomics of Frieseomelitta varia (Hymenoptera: Apidae) and the Sharing of a Satellite DNA Family in Several Neotropical Meliponini Genera. Genes 2025, 16, 86. https://doi.org/10.3390/genes16010086
Vignati ZBM, Teixeira GA, Cunha MS, Pereira JA, Lopes DM. Cytogenomics of Frieseomelitta varia (Hymenoptera: Apidae) and the Sharing of a Satellite DNA Family in Several Neotropical Meliponini Genera. Genes. 2025; 16(1):86. https://doi.org/10.3390/genes16010086
Chicago/Turabian StyleVignati, Zulemara B. M., Gisele A. Teixeira, Marina S. Cunha, Jaqueline A. Pereira, and Denilce M. Lopes. 2025. "Cytogenomics of Frieseomelitta varia (Hymenoptera: Apidae) and the Sharing of a Satellite DNA Family in Several Neotropical Meliponini Genera" Genes 16, no. 1: 86. https://doi.org/10.3390/genes16010086
APA StyleVignati, Z. B. M., Teixeira, G. A., Cunha, M. S., Pereira, J. A., & Lopes, D. M. (2025). Cytogenomics of Frieseomelitta varia (Hymenoptera: Apidae) and the Sharing of a Satellite DNA Family in Several Neotropical Meliponini Genera. Genes, 16(1), 86. https://doi.org/10.3390/genes16010086





