Transcriptome Analysis of Muscle Growth-Related circRNA in the Pacific Abalone Haliotis discus hanna
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Materials
2.2. RNA Extraction and Sequencing
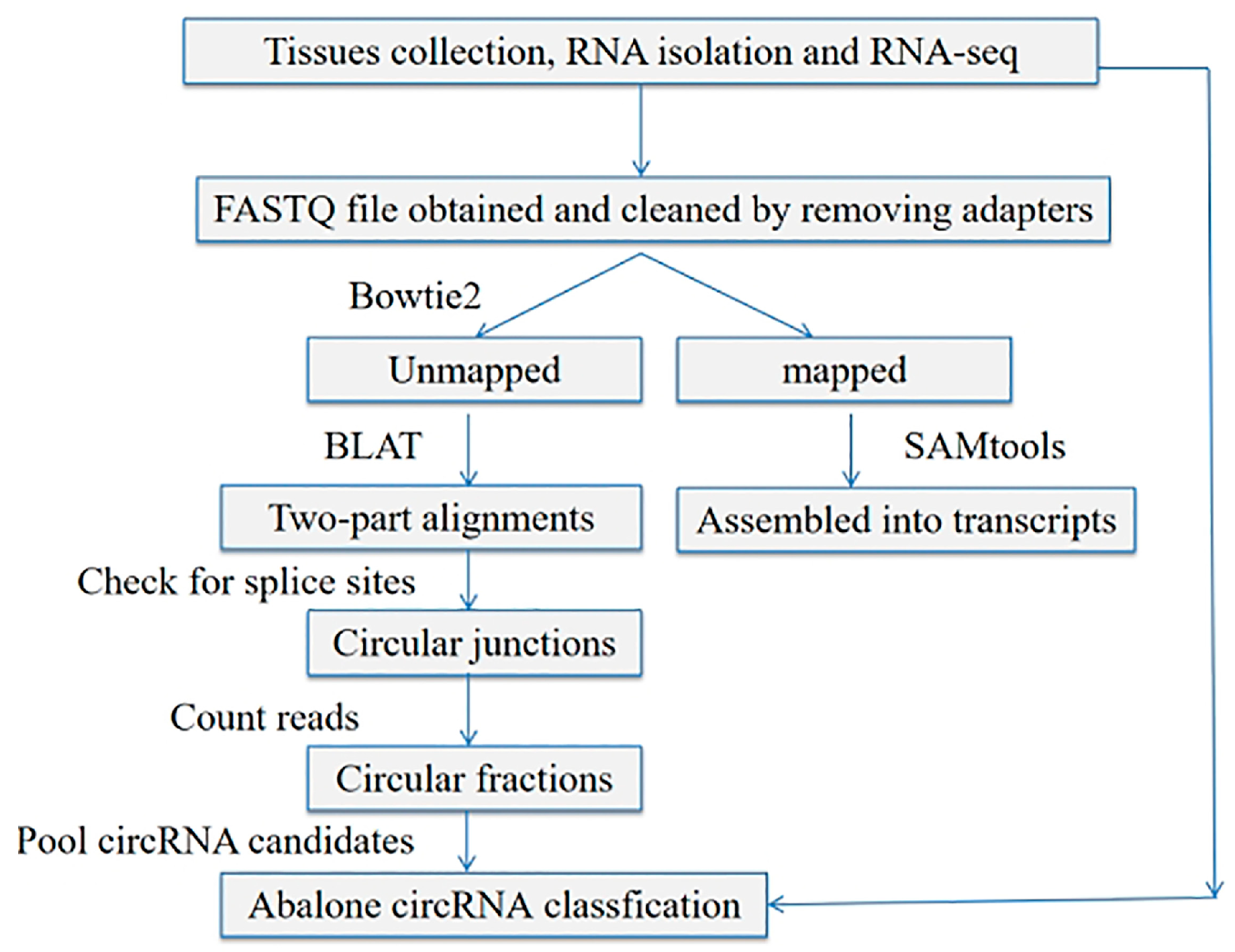
2.3. Identification and Analysis of circRNA
2.4. Construction and Analysis of ceRNA Networks
2.5. Real-Time Quantitative Reverse Transcription PCR (qRT-PCR) Verification
3. Results
3.1. Characteristics of circRNAs
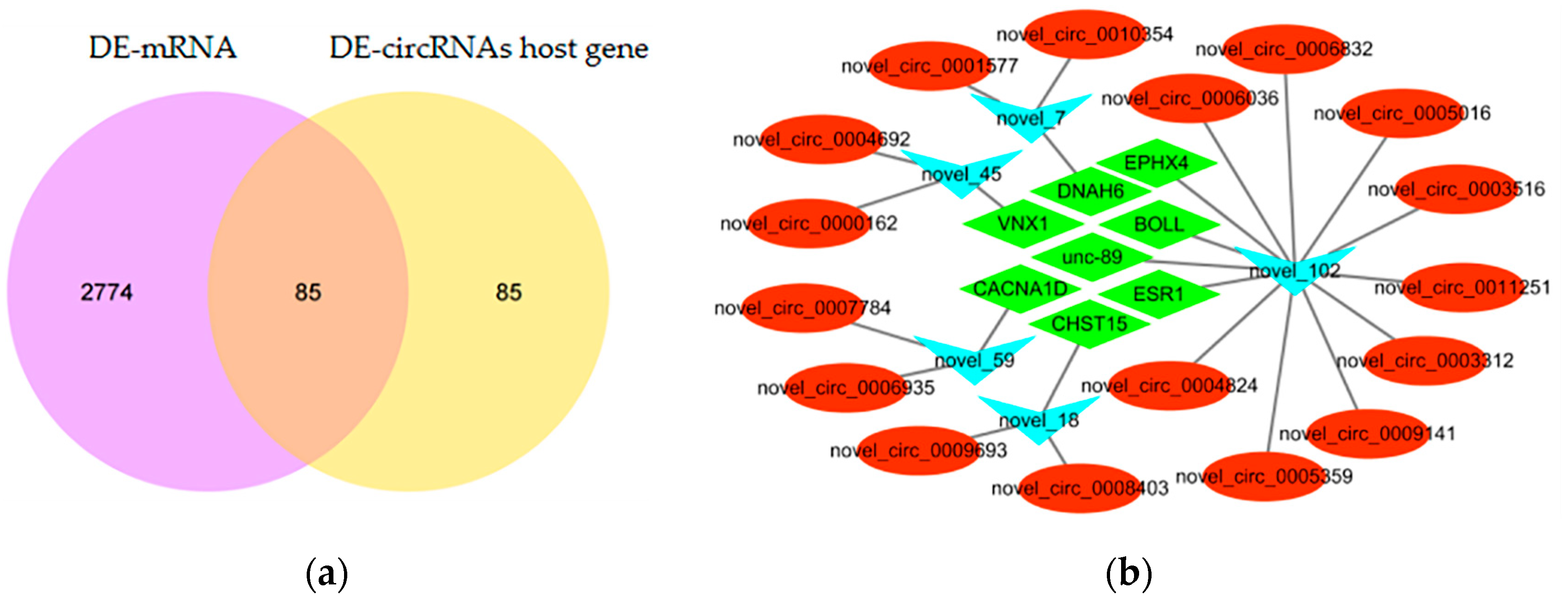
3.2. Differentially Expressed circRNAs and Functional Analysis
3.3. CircRNA-miRNA-mRNA Interaction Network Analysis
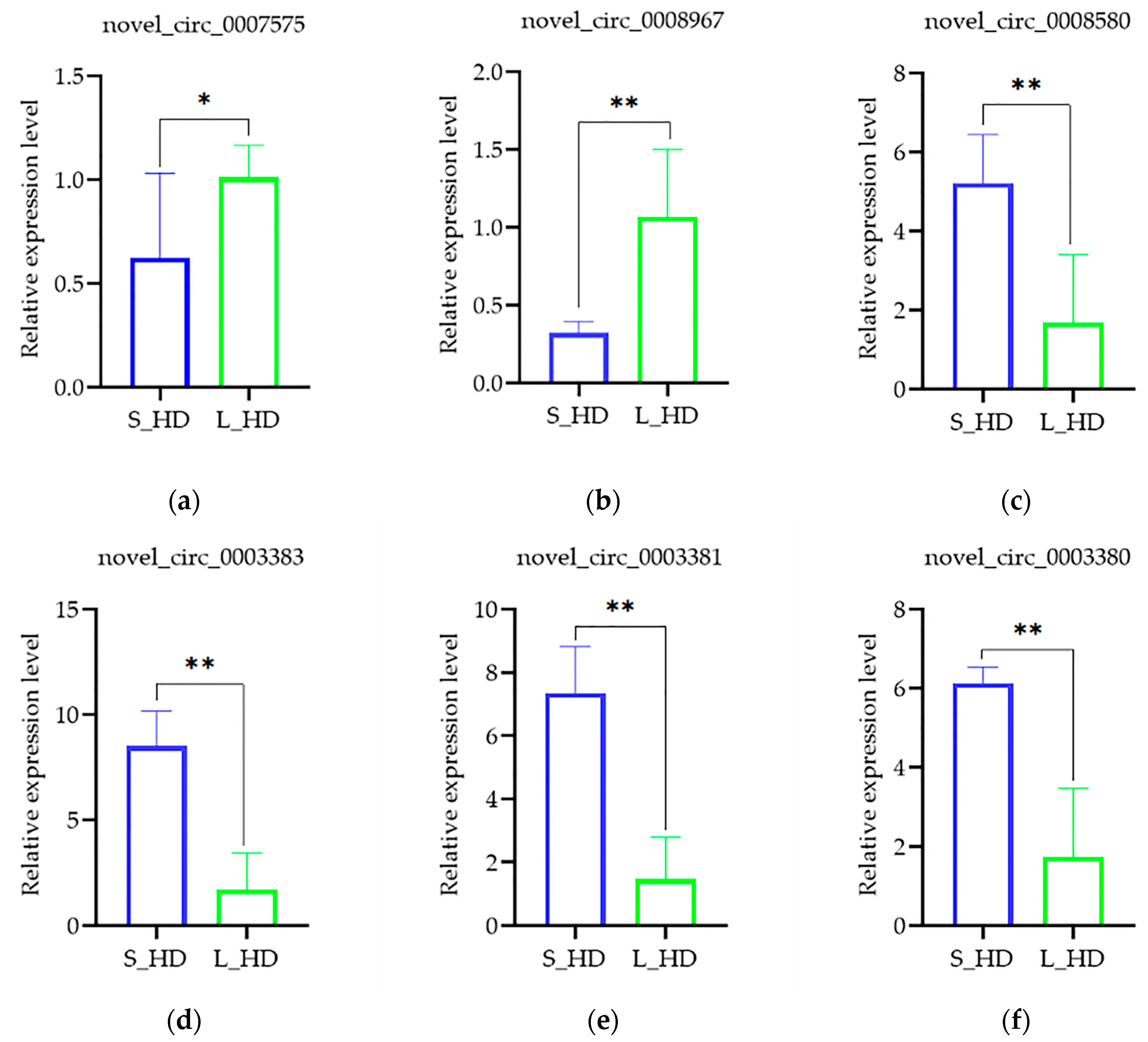
3.4. Verification by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kos, A.; Dijkema, R.; Arnberg, A.C.; van der Meide, P.H.; Schellekens, H. The hepatitis delta (δ) virus possesses a circular RNA. Nature 1986, 323, 558–560. [Google Scholar] [CrossRef] [PubMed]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef] [PubMed]
- Danan, M.; Schwartz, S.; Edelheit, S.; Sorek, R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012, 40, 3131–3142. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Venø, M.T.; Damgaard, C.K.; Kjems, J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016, 44, e58. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef]
- Westholm, J.O.; Miura, P.; Olson, S.; Shenker, S.; Joseph, B.; Sanfilippo, P.; Celniker, S.E.; Graveley, B.R.; Lai, E.C. Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014, 9, 1966–1980. [Google Scholar] [CrossRef]
- Li, Z.Y.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.L.; Zhong, G.L.; Yu, B.; Hu, W.C.; Dai, L.M.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Zhang, Y.; Xue, W.; Li, X.; Zhang, J.; Chen, S.Y.; Zhang, J.L.; Yang, L.; Chen, L.L. The biogenesis of nascent circular RNAs. Cell Rep. 2016, 15, 611–624. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef]
- Rbbani, G.; Nedoluzhko, A.; Galindo-Villegas, J.; Fernandes, J.M.O. Function of circular RNAs in fish and their potential application as biomarkers. Int. J. Mol. Sci. 2021, 22, 7119. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.X.; Dong, L.; Yang, L.S.; Ma, Q.; Liu, F.; Li, Y.J.; Xiong, S. Identification and analysis of circRNA–miRNA–mRNA regulatory network in hepatocellular carcinoma. IET Syst. Biol. 2020, 14, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Reuter, J.A.; Spacek, D.V.; Snyder, M.P. High-throughput sequencing technologies. Mol. Cell 2015, 58, 586–597. [Google Scholar] [CrossRef]
- Wu, P.; Zhou, K.; Zhang, L.; Li, P.; He, M.; Zhang, X.; Ye, H.; Zhang, Q.; Wei, Q.; Zhang, G. High-throughput sequencing reveals crucial miRNAs in skeletal muscle development of Bian chicken. Br. Poult. Sci. 2021, 62, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.C.; Jia, X.W.; Wan, H.F.; Wang, S.H.; Zhang, X.; Zhang, Z.P.; Wang, Y.L. miR-9 and miR-263 regulate the key genes of the ERK pathway in the ovary of mud crab Scylla paramamosain. Mar. Biotechnol. 2020, 22, 594–606. [Google Scholar] [CrossRef]
- Benson, T.W.; Conrad, K.A.; Li, X.S.; Wang, Z.N.; Helsley, R.N.; Schugar, R.C.; Coughlin, T.M.; Wadding-Lee, C.; Fleifil, S.; Russell, H.M.; et al. Gut microbiota-derived trimethylamine N-oxide contributes to abdominal aortic aneurysm through inflammatory and apoptotic mechanisms. Circulation 2023, 147, 1079–1096. [Google Scholar] [CrossRef]
- Liu, S.S.; Chen, G.X.; Xu, H.D.; Zou, W.B.; Yan, W.R.; Wang, Q.Q.; Deng, H.W.; Zhang, H.Q.; Yu, G.J.; He, J.G.; et al. Transcriptome analysis of mud crab (Scylla paramamosain) gills in response to mud crab reovirus (MCRV). Fish Shellfish Immunol. 2017, 60, 545–553. [Google Scholar] [CrossRef]
- Yue, B.L.; Yang, H.Y.; Wang, J.; Ru, W.X.; Wu, J.Y.; Huang, Y.Z.; Lan, X.Y.; Lei, C.Z.; Chen, H. Exosome biogenesis, secretion and function of exosomal miRNAs in skeletal muscle myogenesis. Cell Prolif. 2020, 53, e12857. [Google Scholar] [CrossRef]
- Zhao, X.H.; Ye, J.N.; Lin, X.K.; Xue, H.W.; Zou, X.; Liu, G.B.; Deng, M.; Sun, B.L.; Guo, Y.Q.; Liu, D.W.; et al. Identification of key functional genes and LncRNAs influencing muscle growth and development in Leizhou black goats. Genes 2023, 14, 881. [Google Scholar] [CrossRef]
- Ma, C.; Zhang, L.X.; Wang, X.Y.; He, S.Y.; Bai, J.N.; Li, Q.; Zhang, M.; Zhang, C.; Yu, X.F.; Zhang, J.T.; et al. piRNA-63076 contributes to pulmonary arterial smooth muscle cell proliferation through acyl-CoA dehydrogenase. J. Cell. Mol. Med. 2020, 24, 5260–5273. [Google Scholar] [CrossRef]
- Guo, X.M.; Ford, S.E.; Zhang, F.S. Molluscan aquaculture in China. J. Shellfish Res. 1999, 18, 19–31. [Google Scholar]
- Luo, X.; Ke, C.H.; You, W.W.; Wang, D.X.; Chen, F. Molecular identification of interspecific hybrids between Haliotis discus hannai Ino and Haliotis gigantea Gmelin usingamplified fragment-length polymorphism and microsatellite markers. Aquacult. Res. 2010, 41, 1827–1834. [Google Scholar] [CrossRef]
- Chen, N.; Luo, X.; Gu, Y.T.; Han, G.D.; Dong, Y.W.; You, W.W.; Ke, C.H. Assessment of the thermal tolerance of abalone based on cardiac performance in Haliotis discus hannai, H. gigantea and their interspecific hybrid. Aquaculture 2016, 465, 258–264. [Google Scholar] [CrossRef]
- Guo, Y.F.; Zhao, W.W.; Gao, H.Q.; Wang, S.; Yu, P.M.; Yu, H.S.; Wang, D.; Wang, Q.; Wang, J.X.; Wang, Z.F.; et al. China Fishery Statistical Yearbook; China Agriculture Press: Beijing, China, 2017. [Google Scholar]
- Naipil, C.C.; Muñoz, V.V.; Valdés, J.A.; Molina, A.; Escárate, C.G. RNA interference in Haliotis rufescens myostatin evidences upregulation of insulin signaling pathway. Agri Gene 2016, 1, 93–99. [Google Scholar] [CrossRef]
- Elliott, N.G. Genetic improvement programmes in abalone: What is the future? Aquac. Res. 2000, 31, 51–59. [Google Scholar] [CrossRef]
- Huang, J.F.; Luo, X.; Zeng, L.T.; Huang, Z.K.; Huang, M.Q.; You, W.W.; Ke, C.H. Expression profiling of lncRNAs and mRNAs reveals regulation of muscle growth in the Pacific abalone, Haliotis discus hannai. Sci. Rep. 2018, 8, 16839. [Google Scholar] [CrossRef]
- Liang, G.M.; Yang, Y.L.; Niu, G.L.; Tang, Z.L.; Li, K. Genome-wide profiling of Sus scrofa circular RNAs across nine organs and three developmental stages. DNA Res. 2017, 24, 523–535. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Quan, J.; Kang, Y.; Luo, Z.; Zhao, G.; Li, L.; Liu, Z. Integrated analysis of the responses of a circRNA-miRNA-mRNA ceRNA network to heat stress in rainbow trout (Oncorhynchus mykiss) liver. BMC Genom. 2021, 22, 48. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.F.; Luo, X.; Huang, M.Q.; Liu, G.M.; You, W.W.; Ke, C.H. Identification and characteristics of muscle growth-related microRNA in the Pacific abalone, Haliotis discus hannai. BMC Genom. 2018, 19, 915. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. genome research. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Liu, R.; Li, J.L.; Du, H.R.; Yang, C.W.; Chen, J.L.; Zhao, S.J.; Wang, Q.S. Identification of immune-related circRNA of thymusin chickens. China Anim. Husb. Vet. Med. 2022, 49, 2022–2032. [Google Scholar]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 2017, 66, 22–37.e9. [Google Scholar] [CrossRef]
- Nedoluzhko, A.; Sharko, F.; Rbbani, M.G.; Teslyuk, A.; Konstantinidis, I.; Fernandes, J.M.O. CircParser: A novel streamlined pipeline for circular RNA structure and host gene prediction in non-model organisms. PeerJ 2020, 8, e8757. [Google Scholar] [CrossRef]
- Shen, M.M.; Wu, P.; Li, T.T.; Wu, P.F.; Chen, F.X.; Chen, L.; Xie, K.Z.; Wang, J.Y.; Zhang, G.X. Transcriptome analysis of circRNA and mRNA in theca cells during follicular development in chickens. Genes 2020, 11, 489. [Google Scholar] [CrossRef]
- Goody, M.F.; Henry, C.A. A need for NAD+ in muscle development, homeostasis, and aging. Skelet. Muscle 2018, 8, 9. [Google Scholar] [CrossRef]
- Bassel-Duby, R.; Olson, E.N. Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 2006, 75, 19–37. [Google Scholar] [CrossRef]
- Miska, E.A.; Karlsson, C.; Langley, E.; Nielsen, S.J.; Pines, J.; Kouzarides, T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999, 18, 5099–5107. [Google Scholar] [CrossRef]
- Nie, M.; Deng, Z.L.; Liu, J.M.; Wang, D.Z. Noncoding RNAs, emerging regulators of skeletal muscle development and diseases. Biomed Res. Int. 2015, 2015, 676575. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.F.; Li, H.; Yang, J.M.; Hao, D.; Dong, D.; Huang, Y.Z.; Lan, X.Y.; Plath, M.; Lei, C.Z.; Lin, F.P.; et al. Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis. 2017, 8, e3153. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.H.; Zheng, Q.; Zhu, L.; Xu, L.N.; Sui, M.H.; Zhang, Y.H.; Liu, Y.; Fang, F.G.; Chu, M.X.; Ma, Y.H.; et al. Trend analysis of the role of circular RNA in goat skeletal muscle development. BMC Genom. 2020, 21, 220. [Google Scholar] [CrossRef]
- Shen, X.X.; Liu, Z.H.; Cao, X.N.; He, H.R.; Han, S.S.; Chen, Y.Q.; Cui, C.; Zhao, J.; Li, D.Y.; Wang, Y.; et al. Circular RNA profiling identified an abundant circular RNA circTMTC1 that inhibits chicken skeletal muscle satellite cell differentiation by sponging miR-128-3p. Int. J. Biol. Sci. 2019, 15, 2265–2281. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.J.; Gu, T.; He, Y.J.; Zhou, C.; Hu, Q.; Wang, X.W.; Zheng, E.Q.; Huang, S.X.; Xu, Z.; Yang, J.; et al. Genome-wide analysis of circular RNAs mediated ceRNA regulation in porcine embryonic muscle development. Front. Cell Dev. Biol. 2019, 7, 289. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Mariscal, L.; Dominguez-Calderon, A.; Raya-Sandino, A.; Ortega-Olvera, J.M.; Vargas-Sierra, O.; Martinez-Revollar, G. Tight junctions and the regulation of gene expression. Semin. Cell Dev. Biol. 2014, 36, 213–223. [Google Scholar] [CrossRef]
- Gonzalez-Mariscal, L.; Lechuga, S.; Garay, E. Role of tight junctions in cell proliferation and cancer. Prog. Histochem. Cytochem. 2007, 42, 1–57. [Google Scholar] [CrossRef]
- Nilsson, M.; Fagman, H. Development of the thyroid gland. Development 2017, 144, 2123–2140. [Google Scholar] [CrossRef]
- Andersson, Y.; Sävman, K.; Bläckberg, L.; Hernell, O. Pasteurization of mother’s own milk reduces fat absorption and growth in preterm infants. Acta Paediatr. 2007, 96, 1445–1449. [Google Scholar] [CrossRef]
- Li, G.Z.; Zhou, X.Q.; Jiang, W.D.; Wu, P.; Liu, Y.; Jiang, J.; Kuang, S.Y.; Tang, L.; Shi, H.Q.; Feng, L. Dietary curcumin supplementation enhanced growth performance, intestinal digestion, and absorption and amino acid transportation abilities in on growing grass carp (Ctenopharyngodon idella). Aquac. Res. 2020, 51, 4863–4873. [Google Scholar] [CrossRef]
- Zou, W.G.; Lin, Z.D.; Huang, Y.H.; Limbu, S.M.; Rong, H.; Yu, C.Q.; Lin, F.; Wen, X.B. Effect of dietary vitamin C on growth performance, body composition and biochemical parameters of juvenile Chu’s croaker (Nibea coibor). Aquac. Nutr. 2020, 26, 60–73. [Google Scholar] [CrossRef]
- Ma, S.H.; Meng, Z.P.; Chen, R.; Guan, K.L. The hippo pathway: Biology and pathophysiology. Annu. Rev. Biochem. 2019, 88, 577–604. [Google Scholar] [CrossRef]
- Benian, G.M.; Tinley, T.L.; Tang, X.; Borodovsky, M. The Caenorhabditis elegans gene unc-89, required fpr muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J. Cell Biol. 1996, 132, 835–848. [Google Scholar] [CrossRef] [PubMed]
- Fard, S.S.; Holz, M.K. Regulation of mRNA translation by estrogen receptor in breast cancer. Steroids 2023, 200, 109316. [Google Scholar] [CrossRef]
- Guo, J.T.; Wang, J.F.; Zhang, K.; Yang, Z.M.; Li, B.Z.; Pan, Y.T.; Yu, H.W.; Yu, S.C.; Abbas Raza, S.H.; Kuraz Abebea, B.; et al. Molecular cloning of TPM3 gene in qinchuan cattle and its effect on myoblast proliferation and differentiation. Anim. Biotechnol. 2024, 35, 2345238. [Google Scholar] [CrossRef]
- Li, C.C.; Vargas-Franco, D.; Saha, M.; Davis, R.M.; Manko, K.A.; Draper, I.; Pacak, C.A.; Kang, P.B. Megf10 deficiency impairs skeletal muscle stem cell migration and muscle regeneration. FEBS Open Bio 2021, 11, 114–123. [Google Scholar] [CrossRef]
- Zhao, J.; Shen, X.X.; Cao, X.N.; He, H.R.; Han, S.S.; Chen, Y.Q.; Cui, C.; Wei, Y.H.; Wang, Y.; Li, D.Y.; et al. HDAC4 regulates the proliferation, differentiation and apoptosis of chicken skeletal muscle satellite cells. Animals 2020, 10, 84. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Peng, S.J.; Song, C.C.; Li, H.; Cao, X.K.; Ma, Y.L.; Wang, X.G.; Huang, Y.Z.; Lan, X.Y.; Lei, C.Z.; Chaogetu, B.; et al. Circular RNA SNX29 sponges miR-744 to regulate proliferation and differentiation of myoblasts by activating the Wnt5a/Ca2+ signaling pathway. Mol. Ther. Nucleic Acids 2019, 16, 481–493. [Google Scholar] [CrossRef]
- Kuales, G.; De Mulder, K.; Glashauser, J.; Salvenmoser, W.; Takashima, S.; Hartenstein, V.; Berezikov, E.; Salzburger, W.; Ladurner, P. Boule-like genes regulate male and female gametogenesis in the flatworm Macrostomum lignano. Dev. Biol. 2011, 357, 117–132. [Google Scholar] [CrossRef] [PubMed]


| Primer | Sequence (5′-3′) |
|---|---|
| novel_circ_0007575-qF | GGAGACAAAGTTGACGGGAT |
| novel_circ_0007575-qR | AGCACGACATTTGTACGCAG |
| novel_circ_0008967-qF | AGAGGTGGCATCAGGATCAG |
| novel_circ_0008967-qR | GGGTAGCCATCGATGAGGAA |
| novel_circ_0003381-qF | AGAACAAGGTCTCCAGCACA |
| novel_circ_0003381-qR | TCACTTGTCCCACTAAGCCA |
| novel_circ_0003383-qF | TTGCTGGAAATTGGTGGTCG |
| novel_circ_0003383-qR | CTTGGAAGCAGACGTCAAGG |
| novel_circ_0008580-qF | CTGAAACACTGAAACGTATGGAA |
| novel_circ_0008580-qR | TCAGTGGATGTAATTATCGCGT |
| novel_circ_0003380-qF | TTGAGATCAGACAGCCAGGG |
| novel_circ_0003380-qR | TGATGGGGTTACTCTTGCCA |
| β-actin-qF | GGTATCCTCACCCTCAAGT |
| β-actin-qR | GGGTCATCTTTTCACGGTTG |
| Description | Term_Type | p Value |
|---|---|---|
| Lipid transporter activity | molecular_function | 0.0021589 |
| Lipid transport | biological_process | 0.0021775 |
| Lipid localization | biological_process | 0.0021775 |
| Catalytic activity | molecular_function | 0.0037282 |
| Lipid biosynthetic process | biological_process | 0.0041697 |
| ATP binding | molecular_function | 0.004655 |
| Adenyl ribonucleotide binding | molecular_function | 0.004655 |
| Hydrolase activity | molecular_function | 0.0050891 |
| Adenyl nucleotide binding | molecular_function | 0.00539 |
| Single-organism metabolic process | biological_process | 0.005411 |
| MapTitle | p Value | Gene |
|---|---|---|
| Hypertrophic cardiomyopathy (HCM) | 0.0000976 | Actin, cytoplasmic 1 (Actb), actin, cytoplasmic(Actc), Twitchin (unc-22), muscle M-line assembly protein unc-89 (unc-89), tropomyosin, Fc receptor-like protein 3 (FCRL3) |
| Dilated cardiomyopathy | 0.000131112 | Actb, Actc, unc-22, unc-89, Tropomyosin, FCRL3 |
| Tight junction | 0.000852074 | Janus kinase and microtubule-interacting protein 3 (Jakmip3), Actb, Actc, Janus kinase and microtubule-interacting protein 3 (Jakmip3), circularly permutated Ras protein 1 (Cpras1), band 4.1-like protein 5 (Epb41l5) |
| Vibrio cholerae infection | 0.001678884 | Actb, Actc, V-type proton ATPase catalytic subunit A (Vha68-2), E3 ubiquitin-protein ligase (KCMF1) |
| Viral myocarditis | 0.013475154 | Actb, Actc, inter-α-trypsin inhibitor heavy chain H4 (ITIH4) |
| Thyroid hormone signaling pathway | 0.013793361 | Actb, Actc, estrogen receptor(ESR1), Cpras1, Golgi integral membrane protein 4 (Golim4) |
| Adherens junction | 0.014536358 | Actb, Actc, receptor-type tyrosine-protein phosphatase kappa (PTPRK), multiple epidermal growth factor-like domains protein 10 (MEGF10) |
| Fat digestion and absorption | 0.015652512 | Apolipophorins |
| Vitamin digestion and absorption | 0.017022673 | Deleted in malignant brain tumors 1 (Dmbt1), apolipophorins |
| Collecting duct acid secretion | 0.020650989 | KCMF1 |
| Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 0.025900718 | Actb, Actc |
| Synaptic vesicle cycle | 0.027482768 | KCMF1 |
| Hippo signaling pathway | 0.03606938 | Actb, Actc, Leucine-rich repeat and calponin homology domain-containing protein 3 (Lrch3) |
| Pathogenic Escherichia coli infection | 0.038107063 | Actb, Actc, tubulin β chain |
| Alcoholism | 0.046909914 | Glutamate receptor ionotropic, NMDA 2B (Grin2b), Guanine nucleotide-binding protein subunit β, histone deacetylase 4(HDAC4), Cpras1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; He, J.; She, Z.; Zhou, M.; Li, D.; Chen, J.; Ke, C. Transcriptome Analysis of Muscle Growth-Related circRNA in the Pacific Abalone Haliotis discus hanna. Genes 2025, 16, 65. https://doi.org/10.3390/genes16010065
Huang J, He J, She Z, Zhou M, Li D, Chen J, Ke C. Transcriptome Analysis of Muscle Growth-Related circRNA in the Pacific Abalone Haliotis discus hanna. Genes. 2025; 16(1):65. https://doi.org/10.3390/genes16010065
Chicago/Turabian StyleHuang, Jianfang, Jian He, Zhenghan She, Mingcan Zhou, Dongchang Li, Jianming Chen, and Caihuan Ke. 2025. "Transcriptome Analysis of Muscle Growth-Related circRNA in the Pacific Abalone Haliotis discus hanna" Genes 16, no. 1: 65. https://doi.org/10.3390/genes16010065
APA StyleHuang, J., He, J., She, Z., Zhou, M., Li, D., Chen, J., & Ke, C. (2025). Transcriptome Analysis of Muscle Growth-Related circRNA in the Pacific Abalone Haliotis discus hanna. Genes, 16(1), 65. https://doi.org/10.3390/genes16010065





