Conjunctive BSA-Seq and BSR-Seq to Map the Genes of Yellow Leaf Mutations in Hot Peppers (Capsicum annuum L.)
Abstract
1. Introduction
2. Materials and Methods
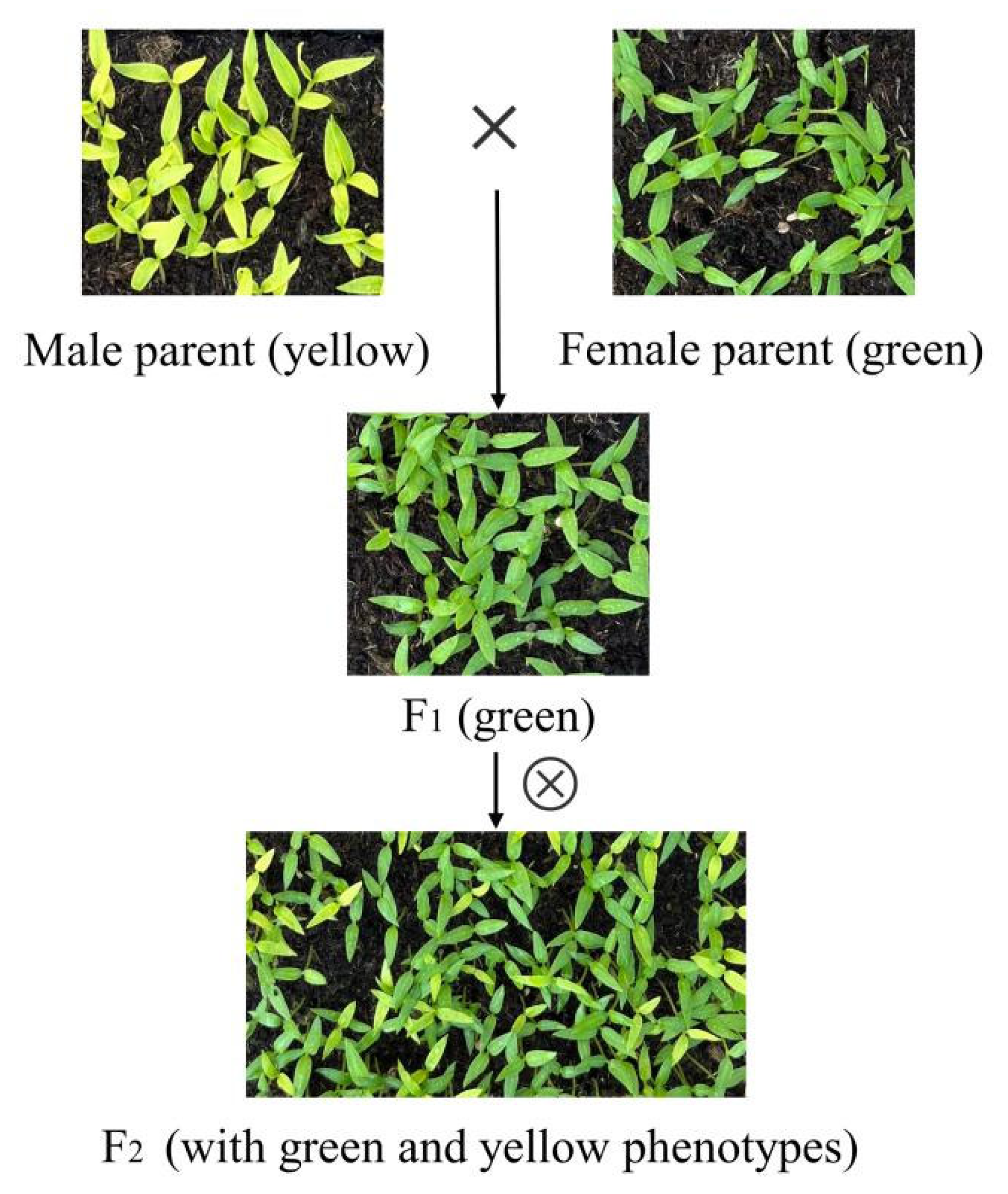
2.1. Plant Materials and Development Conditions
2.2. Establishment of the Illumina Library for Bulked Segregant Analysis
2.3. Investigation of BSA-Seq Data
2.4. Construction of cDNA Library in BSR-Seq
2.5. Investigation of BSR-Seq Data
2.6. qRT-PCR Validation
3. Results
3.1. Bulked Segregant Analysis
3.1.1. Investigation of Sequencing Data of Four DNA Bulks
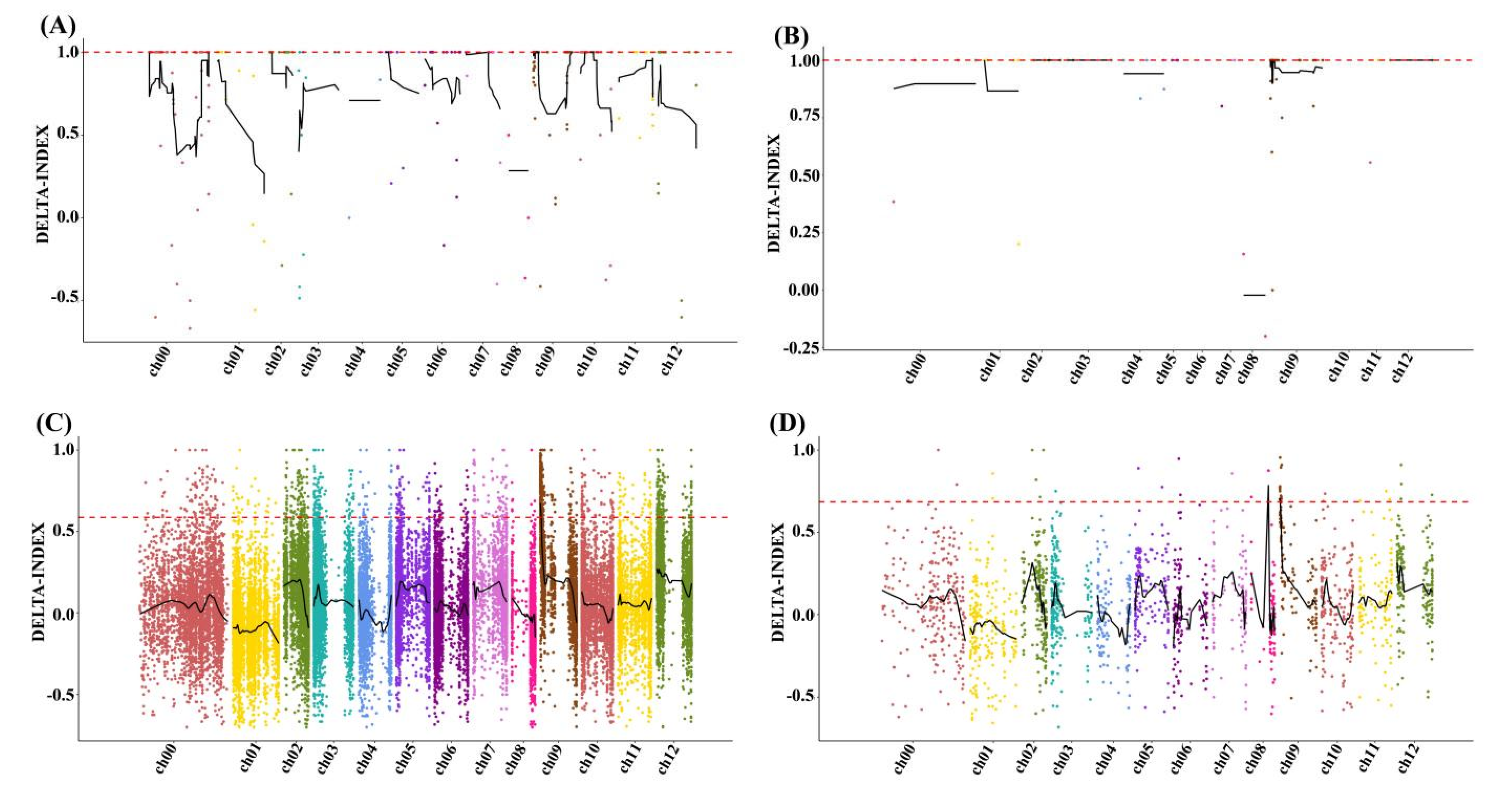
3.1.2. Association Analysis Regarding BSA-Seq
3.2. Bulked Segregant RNA-Seq Analysis
3.2.1. Analysis of Sequencing Data for Four cDNA Bulks
3.2.2. The Comparison of the Gene Expression Files of Two Set Pairs of cDNA Sequencing Bulks
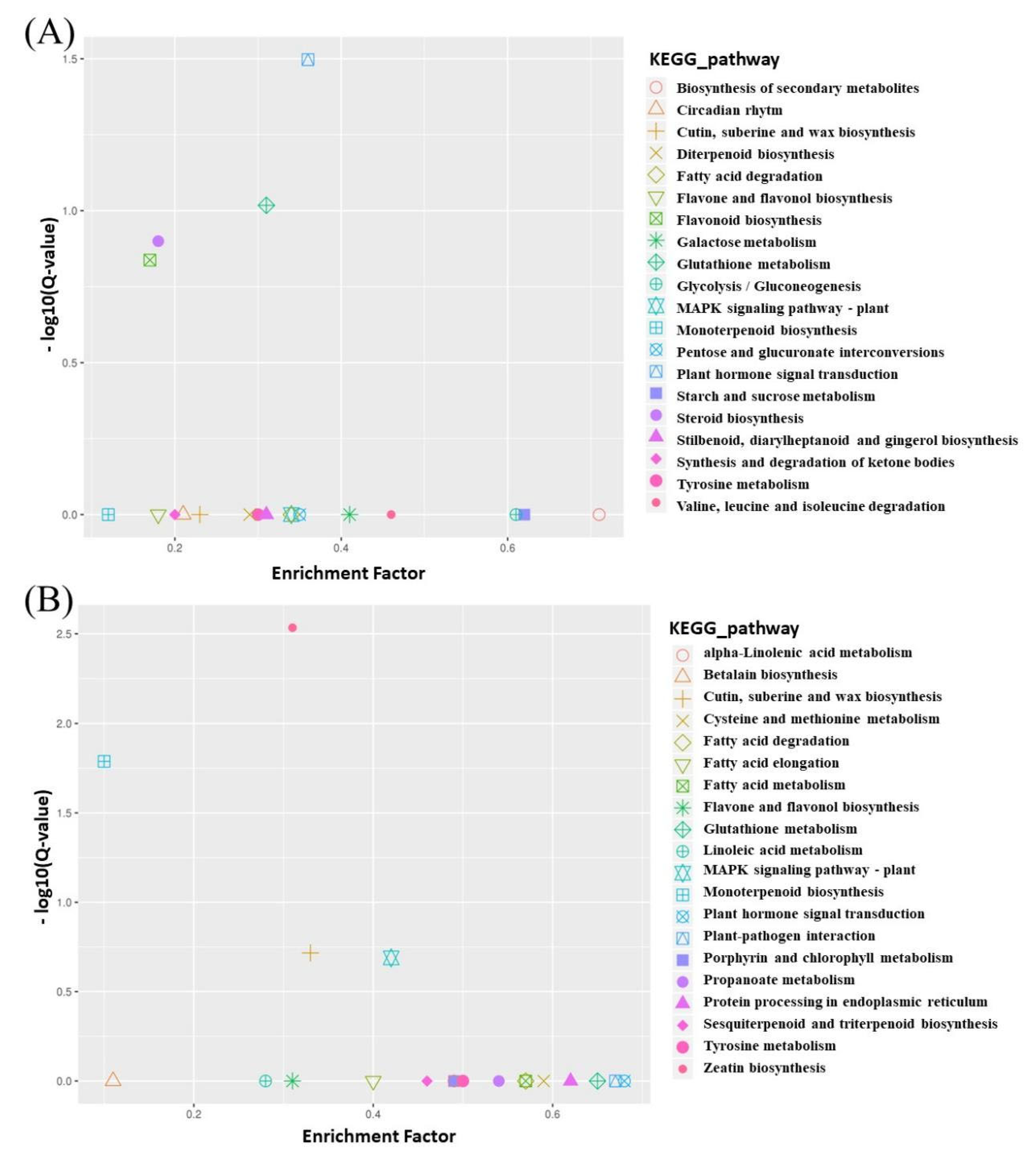
3.2.3. Association Analysis of BSR-Seq Data
3.3. Morphological Genetic Analysis and Candidate Genes of Yellow Leaves
3.4. qRT-PCR Validation of the Expression Pattern of Candidates in Hot Peppers
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zou, X.; Zhu, F. Origin, Evolution and Cultivation History of the Pepper. Acta Hortic. Sin. 2022, 49, 371–1381. [Google Scholar]
- Wu, Y.H. Important Research Advances in the Selected Areas of Pepper Sciences in 2021. J. China Capsicun 2022, 20, 1–11. [Google Scholar]
- Li, W.; Xin, Y.I.; Wang, D.; Jing, X.Y.; Zhou, Q.; Su, X.Q.; Ning, K.; Chen, F.; Hu, Q.; Xu, J. Nannochloropsis plastid and mito chondrial phylogenomes reveal organelle diversification mech anism and intragenus phylotyping strategy in microalgae. BMC Genom. 2013, 14, 534. [Google Scholar]
- Zeng, S. Cytological, biochemical, and transcriptomic analyses of a novel yellow leaf variation in a Paphiopedilum (Orchidaceae) SCBG COP15. Genes 2021, 13, 71. [Google Scholar] [CrossRef]
- Ma, Y.Q. Analysis on the situation of pepper industry in China. J. China Capsicum 2011, 9, 1–5. [Google Scholar]
- Deng, X.J.; Zhang, H.Q.; Wang, Y.; He, F.; Liu, J.L.; Xiao, X.; Shu, Z.F.; Li, W.; Wang, G.H.; Wang, G.L. Mapped clone and functional analysis of leaf-color gene Ygl7 in a rice hybrid (Oryza sativa L. ssp. indica). PLoS ONE 2014, 9, e99564. [Google Scholar] [CrossRef] [PubMed]
- Beale, S.I.; Appleman, D. Chlorophyll synthesis in chlorella regulation by degree of light limitation of growth. Plant Physiol. 1971, 47, 230–235. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brestic, M.; Zivcak, M.; Kunderlikova, K.; Allakhverdiev, S. High temperature specifically affects the photoprotective responses of chlorophyll b-deficient wheat mutant lines. Photosynth. Res. 2016, 130, 251–266. [Google Scholar] [CrossRef]
- Wang, L.; Yue, C.; Cao, H.L.; Zhou, Y.H.; Zeng, J.M.; Yang, Y.J.; Wang, X.C. Biochemical and transcriptome analysis of a novel chlorophyll-deficient chlorina tea plant cultivar. BMC Plant Biol. 2014, 14, 352. [Google Scholar] [CrossRef]
- Wu, Z.M.; Zhang, X.; Wang, J.L.; Wan, J.M. Leaf chloroplast ultrastructure and photosynthetic properties of a chlorophyll-deficient mutant of rice. Photosynthetica 2014, 52, 217–222. [Google Scholar] [CrossRef]
- Zhu, L.X.; Zeng, X.H.; Chen, Y.L.; Yang, Z.H.; Qi, L.P.; Pu, Y.Y.; Yi, B.; Wen, J.; Ma, C.Z.; Shen, J.X. Genetic characterisation and fine mapping of a chlorophyll-deficient mutant (BnaC. ygl) in Brassica napus. Mol. Breed. 2014, 34, 603–614. [Google Scholar] [CrossRef]
- Zhao, M.H.; Li, X.; Zhang, X.X.; Zhang, H.; Zhao, X.Y. Mutation Mechanism of Leaf Color in Plants: A Review. Forests 2020, 11, 851. [Google Scholar] [CrossRef]
- Eckhardt, U.; Grimm, B.; Hörtensteiner, S. Recent advances in chlorophyll biosynthesis and breakdown in higher plants. Plant Mol. Biol. 2004, 56, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.l.; Li, I.X. Areview: Molecular mechanism of plant yellow leaf mutation. J. South. Agric. 2017, 48, 1358–1366. [Google Scholar]
- Christ, B.; Hörtensteiner, S. Mechanism and Significance of Chlorophyll Breakdown. J. Plant Growth Regul. 2014, 33, 4–20. [Google Scholar] [CrossRef]
- Stefan, H. NCC malonyltransferase catalyses the finalstep of chlorophyll breakdown in rape (Brassica napus). Phytochemistry 1998, 49, 953–956. [Google Scholar]
- Stefan, H. Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci. 2009, 14, 155–162. [Google Scholar]
- Stefan, H. Update on the biochemistry of chlorophyll breakdown. Plant Mol. Biol. 2013, 82, 505–517. [Google Scholar]
- Stefan, H.; Kra¨utler, B. Chlorophyll breakdown in higher plants. Biochim. Biophys. Acta 2011, 1807, 977–988. [Google Scholar]
- Park, S.Y.; Yu, J.W.; Park, J.S.; Li, J.; Yoo, S.C.; Lee, N.Y.; Lee, S.K.; Jeong, S.W.; Seo, H.S.; Koh, H.J.; et al. The senescence-induced STAYGREEN protein regulates chlorophyll degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef]
- Thomas, H.; Howarth, C.J. Five ways to stay green. J. Exp. Bot. 2000, 51, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Schelbert, S.; Park, S.Y.; Han, S.H.; Lee, B.D.; Céline, B.A.; Kessler, F.; Stefan, H.; Paeka, N.C. STAY-GREEN andchlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell 2012, 24, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Kim, Y.S.; Yoo, S.C.; Stefan, H.; Paek, N.C. 7-Hydroxymethylchlorophyll a reductase functions in metabolic channeling of chlorophyll breakdown intermediates during leaf senescence. Biochem. Biophys. Res. Commun. 2013, 430, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.Q.; Li, F.M.; Xuan, X.X.; Ahiakpa, J.K.; Tao, J.B.; Zhang, X.Y.; Ge, P.F.; Wang, Y.R.; Gai, W.X.; Zhang, Y.Y. The genetic basis and improvement of photosynthesis in tomato. Hortic. Plant J. 2024, in press. [Google Scholar] [CrossRef]
- Mei, X.; Zhang, K.; Lin, Y.; Su, H.; Lin, C.; Chen, B.; Yang, H.; Zhang, L. Metabolic and Transcriptomic Profiling Reveals Etiolated Mechanism in Huangyu Tea (Camellia sinensis) Leaves. Int. J. Mol. Sci. 2022, 23, 15044. [Google Scholar] [CrossRef]
- Xue, T.Y.; Wan, H.P.; Chen, J.D.; He, S.X.; Lujin, C.Z.; Xia, M.; Wang, S.S.; Dai, X.G.; Zeng, C.L. Genome-wide identification and expression analysis of the chlorophyll a/b binding protein gene family in oilseed (Brassica napus L.) under salt stress conditions. Plant Stress. 2024, 11, 100339. [Google Scholar] [CrossRef]
- Klindworth, D.L. Williams and M.E. Duysen. Genetic Analysis of chlorina mutants of durum-wheat. Crop Sci. 1995, 35, 431–436. [Google Scholar] [CrossRef]
- Watanabe, N.; Koval, S.E. Mapping of chlorina mutant geneson the long arm of homoeologous group 7 chromosomes in common wheat with partial deletion lines. Euphytica 2003, 129, 259–265. [Google Scholar] [CrossRef]
- Li, N.; Jia, J.; Xia, C.; Liu, X.; Kong, X. Characterization and mapping of novel chlorophyll deficient mutant genes in durum wheat. Breed. Sci. 2013, 63, 169–175. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, X.; He, B.; Diao, L.; Sheng, S.; Wang, J.; Guo, X.; Su, N.; Wang, L.; Jiang, L.; et al. A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol. 2007, 145, 29–40. [Google Scholar] [CrossRef]
- Zhang, H.; Li, J.; Yoo, J.H.; Yoo, S.C.; Cho, S.H.; Koh, H.J.; Seo, H.S.; Paek, N.C. Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol. Biol. 2006, 62, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.T.; Dong, X.Y.; Yuan, X.; Hong, Y.Y.; Zhang, L.L.; Zhang, X.; Chen, S.X. Identification and characterization of CsSRP43, a major gene controlling leaf yellowing in cucumber. Hortic. Res. 2022, 9, 212. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Ding, Y.; Yu, J.; Zu, M. Study on Leaf Ultrastructure of the Thermo-sensitive Chlorophyll Deficient Mutant in Rice. Plant Sci. J. 2001, 19, 1–5. [Google Scholar]
- Su, X.J.; Wang, P.H. Study on the mechanism of albinism in wheat mutants. J. Northwest AF Univ. 1990, 18, 73–77. [Google Scholar]
- Keck, R.W.; Dilley, R.A.; Allen, C.F.; Biggs, S. Chloroplast composition and structure differences in a soybean mutant. Plant Physiol. 1970, 46, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Jing, L. Chlorophyll contents and chloroplast ultrastructure of chlorophyll deficient mutant in B. napus. Chin. J. Oil Crop Sci. 2000, 22, 27–29. [Google Scholar]
- Guo, Y.M.; Gu, X.F.; Zhang, C.Z.; Fang, X.; Zhang, S.; Xu, C. Genetic mechanism of cucumber leaf mutant. Acta Hortic. Sin. 2003, 30, 409–412. [Google Scholar]
- Shao, Q.; Yu, Z.Y.; Li, X.G. Studies on internal physiological and biochemical changes of xantha mutant in melon leaves. China Veg. 2013, 1, 59–65. [Google Scholar]
- Yao, J.G.; Zhang, H.; Xu, X.Y.; Zhang, L.L.; Li, J.F. Studies on weak light tolerance of tomato leaf color mutant. China Veg. 2010, 1, 31–35. [Google Scholar]
- Yang, S.; Zhang, Z.Q.; Chen, W.C. Genetic analysis and physiological characteristics of yellow leaf mutant in pepper. J. Hunan Agric. Univ. 2020, 46, 48–52. [Google Scholar]
- Ohlrogge, J.B.; Jaworski, J.G. Regulation of fatty acid synthesis. Annual Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 109–136. [Google Scholar] [CrossRef]
- Sun, X.; Chongde, W.; Xiang, N.; Li, X.; Yang, S.; Du, J.; Yang, Y. Activation of secondary cell wall biosynthesis by mir319-targeted tcp4 transcription factor. Plant Biotechnol. J. 2017, 15, 1284–1294. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Xu, R.; Chen, J.; Liu, A. β-Ketoacyl-acyl Carrier Protein Synthase I (KASI) Plays Crucial Roles in the Plant Growth and Fatty Acids Synthesis in Tobacco. Int. J. Mol. Sci. 2016, 17, 1287. [Google Scholar] [CrossRef]
- Carlsson, A.S.; LaBrie, S.T.; Kinney, A.J.; Wettstein-Knowles, P.; Browse, J. A kas2 cdna complements the phenotypes of the Arabidopsis fab1 mutant that differs in a single residue bordering the substrate binding pocket. Plant J. 2002, 29, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.S.; Dai, Z.L.; Bosland, P.W.; Wang, Q.; Sun, C.Q.; Zhang, Z.C.; Ma, Z.H. Characterizing and marker-assisting a novel chili pepper (Capsicum annuum L.) yellow bud mutant with cytoplasmic male sterility. Genet. Mol. Res. 2017, 16, gmr16019459. [Google Scholar] [CrossRef]
- Wu, L.; Cui, Y.; Xu, Z.; Xu, Q. Identification of Multiple Grain Shape-Related Loci in Rice Using Bulked Segregant Analysis with High-Throughput Sequencing. Front. Plant Sci. 2020, 11, 303. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Bi, B.; Xu, X.; Li, B.; Tian, S.; Wang, J.; Zhang, H.; Wang, G.; Han, Y.; McElroy, J.S. Rapid identification of a candidate nicosulfuron sensitivity gene (Nss) in maize (Zea mays L.) via combining bulked segregant analysis and RNA-seq. Theor. Appl. Genet. 2019, 132, 1351–1361. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Yang, H.; Yuan, Y.; Zhu, H.; Zhang, M.; Wei, X.; Sun, D.; Wang, X.; Yang, S.; Yang, L. Comparative Transcriptome Analysis Provides Insights into Yellow Rind Formation and Preliminary Mapping of the Clyr (Yellow Rind) Gene in Watermelon. Front. Plant Sci. 2020, 11, 192. [Google Scholar] [CrossRef]
- Li, R.; Hou, Z.; Gao, L.; Xiao, D.; Hou, X.; Zhang, C.; Yan, J.; Song, L. Conjunctive Analyses of BSA-Seq and BSR-Seq to Reveal the Molecular Pathway of Leafy Head Formation in Chinese Cabbage. Plants 2019, 8, 603. [Google Scholar] [CrossRef]
- Liu, H.; Su, W.W.; Li, X.Y.; Chao, K.X.; Wang, M.N.; Yue, W.Y.; Wang, B.T.; Li, Q. Rapid mapping of a stripe rust resistance gene YrZl31 using bulked segregant analysis combined with high-throughput single-nucleotide polymorphism genotyping arrays. Crop Prot. 2020, 134, 105174. [Google Scholar] [CrossRef]
- Dai, D.; Huang, L.; Zhang, X.; Zhang, S.; Yuan, Y.; Wu, G.; Hou, Y.; Yuan, X.; Chen, X.; Xue, C. Identification of a Branch Number Locus in Soybean Using BSA-Seq and GWAS Approaches. Int. J. Mol. Sci. 2024, 25, 873. [Google Scholar] [CrossRef]
- Wang, G.; Chen, B.; Du, H.; Zhang, F.; Zhang, H.; Wang, Y.; He, H.; Geng, S.; Zhang, X. Genetic mapping of anthocyanin accumulation-related genes in pepper fruits using a combination of SLAF-seq and BSA. PLoS ONE 2018, 13, e0204690. [Google Scholar] [CrossRef] [PubMed]
- Wenger, J.W.; Schwartz, K.; Sherlock, G. Bulk segregant analysis by high throughput sequencing reveals a novel xylose utilization gene from Saccharomyces cerevisiae. PLoS Genet. 2010, 6, e1000942. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yeh, C.T.; Tang, H.M.; Nettleton, D.; Schnable, P.S. Gene Mapping via Bulked Segregant RNA-Seq (BSR-Seq). PLoS ONE 2012, 7, e36406. [Google Scholar] [CrossRef]
- Françoise, M.; Sari, K.; Tapio, E.P.; Marja-Liisa, S. Contribution of omega-3 fatty acid desaturase and 3-ketoacyl-ACP synthase II (KASII) genes in the modulation of glycerolipid fatty acid composition during cold acclimation in birch leaves. J. Exp. Bot. 2006, 57, 897–909. [Google Scholar]
- Hao, Q.T. Identification and Function Analysis of Upland Cotton β-Ketoacyl-ACP Synthetase II (KASII) Family Genes. Master’s Thesis, Shanxi Agricultural University, Jinzhong, China, 2018; pp. 2–10. [Google Scholar]
- Harwood, J.L. Recent advances in the biosynthesis of plant fatty acids. Biochim. Biophys. Acta 1996, 1301, 7–56. [Google Scholar] [CrossRef]
- Wu, J.; Lightner, J.; Warwick, N.; Browse, J. Low-Temperature Damage and Subsequent Recovery of fab1 Mutant Arabidopsis Exposed to 2[deg]C. Plant Physiol. 1997, 113, 347–356. [Google Scholar] [CrossRef]
- Hirokazu, H.; Jong-In, P.; Makoto, E.; Yoshinobu, T.; Tomohiko, K.; Yoshimitsu, T.; Go, S.; Makiko, K.K.; Masao, W. Expression and developmental function of the 3-ketoacyl-ACP synthase2 gene in Arabidopsis thaliana. Genes Genet. Syst. 2008, 83, 143–152. [Google Scholar]
- Peltier, J.B.; Emanuelsson, O.; Kalume, D.E.; Ytterberg, J.; Friso, G.; Rudella, A.; Liberles, D.A.; Söderberg, L.; Roepstorff, P.; Heijne, G.; et al. Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 2002, 14, 211–236. [Google Scholar] [CrossRef]
- Giuli, F.; Lisa, G.; Jimmy, Y.A.; Jean-Benoit, P.; Andrea, R.; Qi, S.; Klaas, J.W. In-Depth Analysis of the Thylakoid Membrane Proteome of Arabidopsis thaliana Chloroplasts: New Proteins, New Functions, and a Plastid Proteome Database. Plant Cell 2004, 16, 478–499. [Google Scholar]
- Liu, J.; Last, R.L. A chloroplast thylakoid lumen protein is required for proper photosynthetic acclimation of plants under fluctuating light environments. Proc. Nat. Acad. Sci. USA 2017, 114, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.X.; Mao, H.T.; Lin, S.; Din, A.M.U.; Yin, X.Y.; Yuan, M.; Zhang, Z.W.; Yuan, S.; Zhang, H.Y.; Chen, Y.E. Different Photosynthetic Response to High Light in Four Triticeae Crops. Int. J. Mol. Sci. 2023, 24, 1569. [Google Scholar] [CrossRef] [PubMed]
- Oindrila, B.; Irma, O.; Nathan, H.; Linda, L.W. The tomato chloroplast stromal proteome compendium elucidated by leveraging a plastid protein-localization prediction Atlas. Front. Plant Sci. 2023, 14, 2013. [Google Scholar]
- Zhou, R.; Yu, X.; Zhao, T.M.; Carl-Otto, O.; Eva, R.; Wu, Z. Physiological analysis and transcriptome sequencing reveal the effects of combined cold and drought on tomato leaf. BMC Plant Biol. 2019, 19, 377. [Google Scholar] [CrossRef] [PubMed]



| Bulk | Clean Reads | Clean Data | Q30 (%) | Genome Coverage 10× (%) | Average Depth (×) | SNP Number | Alignment Efficiency (%) |
|---|---|---|---|---|---|---|---|
| R01 | 125,450,705 | 37,635,211,500 | 91.67 | 68.5 | 21.3478 | 7,131,298 | 99.13 |
| R02 | 129,325,381 | 38,797,614,300 | 92.67 | 68.91 | 21.6186 | 7,464,055 | 99.41 |
| R03 | 235,759,304 | 70,727,791,200 | 93.14 | 96.82 | 33.7568 | 10,429,419 | 99.21 |
| R04 | 241,371,002 | 72,411,300,600 | 92.9 | 96.58 | 46.74 | 10,407,663 | 99.16 |
| Method | Chromosome | Start | End | Size (M) | Genes |
|---|---|---|---|---|---|
| SNP index | 2 | 15,717,687 | 15,817,687 | 0.1 | 4 |
| 2 | 3,098,963 | 3,718,171 | 0.62 | 2 | |
| 2 | 7,488,745 | 7,588,745 | 0.1 | 1 | |
| 5 | 2,844,929 | 2,944,929 | 0.1 | 7 | |
| 7 | 6,169,571 | 6,731,315 | 0.56 | 2 | |
| 9 | 18,288,580 | 20,398,403 | 2.11 | 31 | |
| 9 | 22,093,862 | 23,040,848 | 0.95 | 8 | |
| 9 | 227,478,236 | 227,578,236 | 0.1 | 3 | |
| 9 | 2,6013,861 | 26,524,148 | 0.51 | 4 | |
| 9 | 4,828,484 | 15,906,110 | 11.08 | 324 | |
| 9 | 98,520 | 1,575,124 | 1.48 | 59 | |
| 10 | 50,118,507 | 50,271,184 | 0.15 | 1 | |
| 10 | 96,017,825 | 96,117,825 | 0.1 | 1 | |
| Small indel index | 1 | 15,142,615 | 15,242,615 | 0.1 | 3 |
| 2 | 113,098,665 | 113,198,665 | 0.1 | 1 | |
| 2 | 162,781,799 | 162,881,799 | 0.1 | 5 | |
| 2 | 93,723,980 | 93,823,980 | 0.1 | 1 | |
| 3 | 12,350,622 | 12,450,622 | 0.1 | 8 | |
| 3 | 204,823,413 | 204,923,413 | 0.1 | 2 | |
| 3 | 35,380,979 | 35,480,979 | 0.1 | 6 | |
| 5 | 34,832,217 | 34,932,217 | 0.1 | 1 | |
| 9 | 102,543 | 202,543 | 0.1 | 2 |
| Bulk | Clean Reads | Clean Data | Q30 (%) | SNP Number | Alignment Efficiency (%) |
|---|---|---|---|---|---|
| T01 | 20,530,863 | 6,159,258,900 | 91.79 | 115,475 | 89.74 |
| T02 | 22,736,055 | 6,820,816,500 | 91.83 | 122,203 | 91.53 |
| T03 | 59,034,487 | 17,710,346,100 | 92.13 | 168,598 | 91.69 |
| T04 | 55,912,808 | 16,773,842,400 | 92.21 | 163,342 | 91.71 |
| Method | Chromosome | Start | End | Size (M) | Genes |
|---|---|---|---|---|---|
| SNP index | 9 | 3,520,729 | 6,351,360 | 2.83 | 99 |
| 9 | 82,366 | 2,105,241 | 2.02 | 64 | |
| Indel index | 8 | 112,304,151 | 112,404,151 | 0.10 | 2 |
| 9 | 147,534 | 1,565,785 | 1.42 | 58 |
| Method | Chromosome | Start | End | Size (M) | Genes |
|---|---|---|---|---|---|
| SNP index | 9 | 98,520 | 1,575,124 | 1.48 | 44 |
| 9 | 3,520,729 | 6,351,360 | 2.83 | 68 | |
| Indel index | 9 | 147,534 | 202,543 | 0.05 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, G.; Zhang, C.; Shan, X.; Zhang, Z.; Wang, W.; Lu, W.; Dai, Z.; E, L.; Wang, Y.; Ma, Z.; et al. Conjunctive BSA-Seq and BSR-Seq to Map the Genes of Yellow Leaf Mutations in Hot Peppers (Capsicum annuum L.). Genes 2024, 15, 1115. https://doi.org/10.3390/genes15091115
Sun G, Zhang C, Shan X, Zhang Z, Wang W, Lu W, Dai Z, E L, Wang Y, Ma Z, et al. Conjunctive BSA-Seq and BSR-Seq to Map the Genes of Yellow Leaf Mutations in Hot Peppers (Capsicum annuum L.). Genes. 2024; 15(9):1115. https://doi.org/10.3390/genes15091115
Chicago/Turabian StyleSun, Guosheng, Changwei Zhang, Xi Shan, Zhenchao Zhang, Wenlong Wang, Wenjun Lu, Zhongliang Dai, Liu E, Yaolong Wang, Zhihu Ma, and et al. 2024. "Conjunctive BSA-Seq and BSR-Seq to Map the Genes of Yellow Leaf Mutations in Hot Peppers (Capsicum annuum L.)" Genes 15, no. 9: 1115. https://doi.org/10.3390/genes15091115
APA StyleSun, G., Zhang, C., Shan, X., Zhang, Z., Wang, W., Lu, W., Dai, Z., E, L., Wang, Y., Ma, Z., & Hou, X. (2024). Conjunctive BSA-Seq and BSR-Seq to Map the Genes of Yellow Leaf Mutations in Hot Peppers (Capsicum annuum L.). Genes, 15(9), 1115. https://doi.org/10.3390/genes15091115








