Full-Length Transcriptome Construction and Systematic Characterization of Virulence Factor-Associated Isoforms in Vairimorpha (Nosema) Ceranae
Abstract
1. Introduction
2. Materials and Methods
2.1. Bee and Fungus
2.2. Preparation of V. ceranae Spores
2.3. cDNA Library Construction and PacBio SMRT Sequencing
2.4. cDNA Library Construction and Illumina Sequencing
2.5. Quality Control and Full-Length Transcript Identification
2.6. Genomic Mapping
2.7. Annotation of Full-Length Transcripts
2.8. Analysis and Validation of AS Events
2.9. Structural Optimization of Annotated Genes in the Reference Genome
2.10. Identification of lncRNAs
3. Results
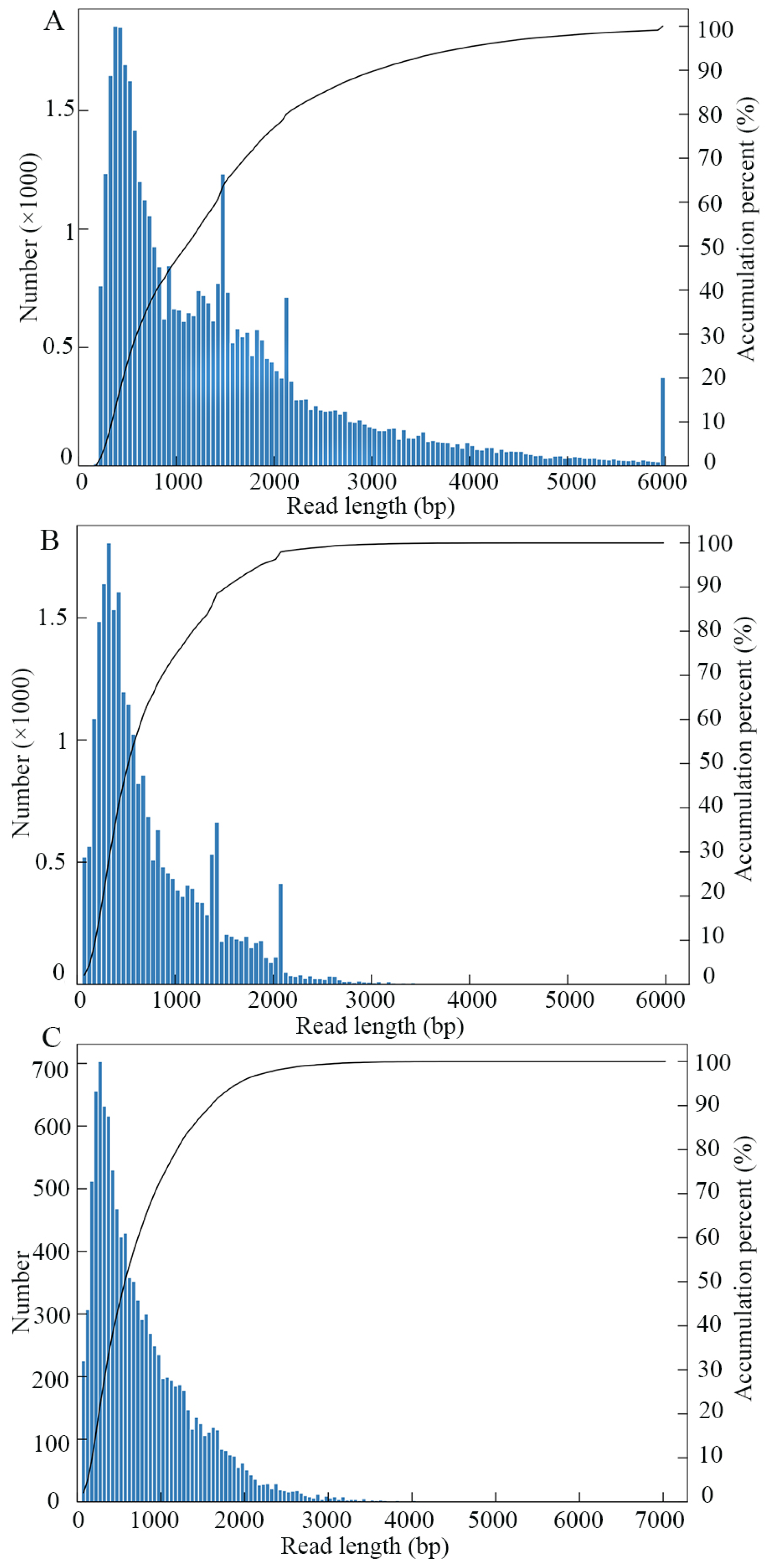
3.1. Quality Control and Identification of Full-Length Transcripts
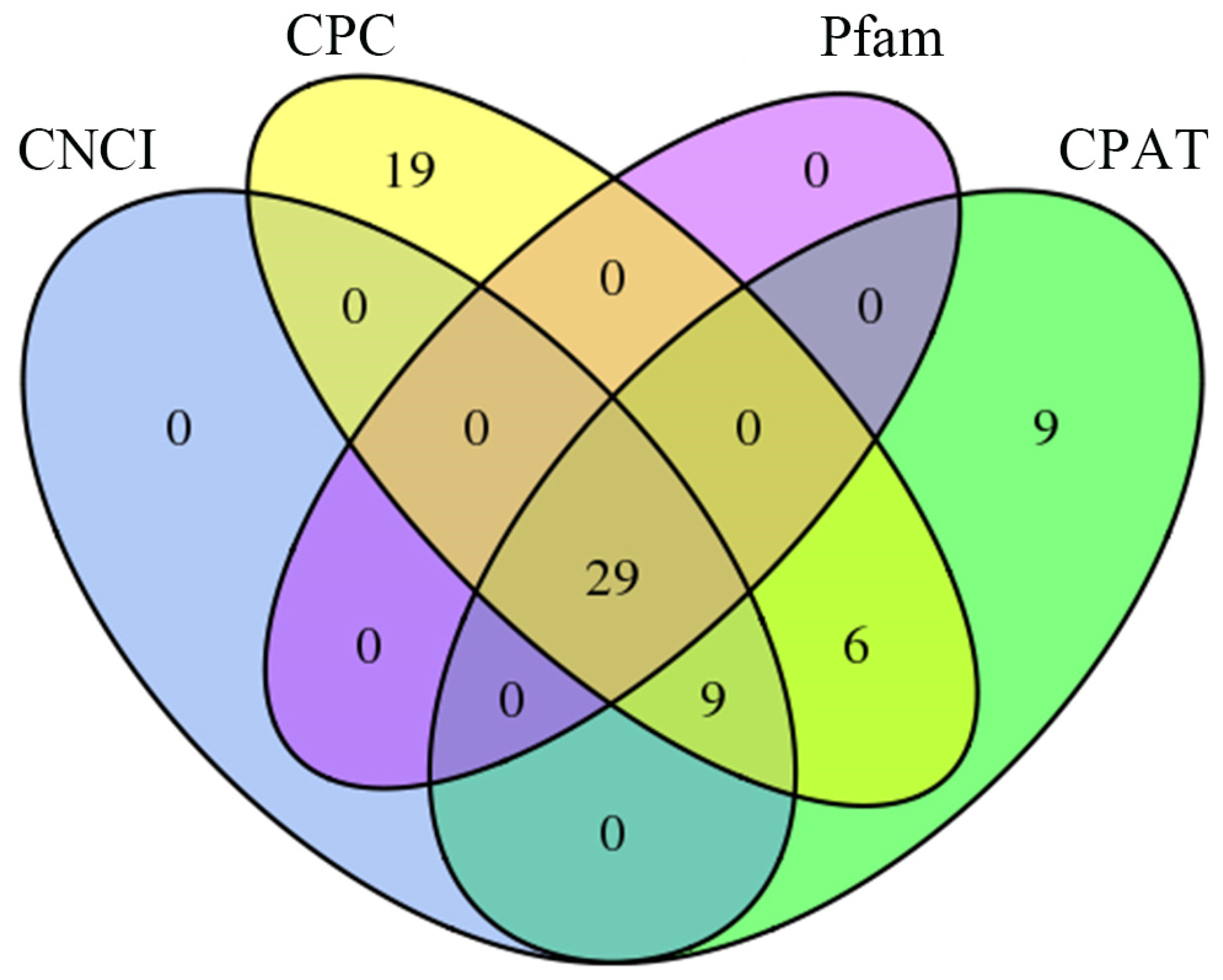
3.2. Functional Annotation of V. ceranae Full-Length Transcripts
3.3. Analysis of Novel Transcripts Related to V. ceranae Virulence Factors and Energy Metabolism
3.4. Identification of lncRNAs
3.5. Structural Optimization of Annotated Genes on the V. ceranae Reference Genome
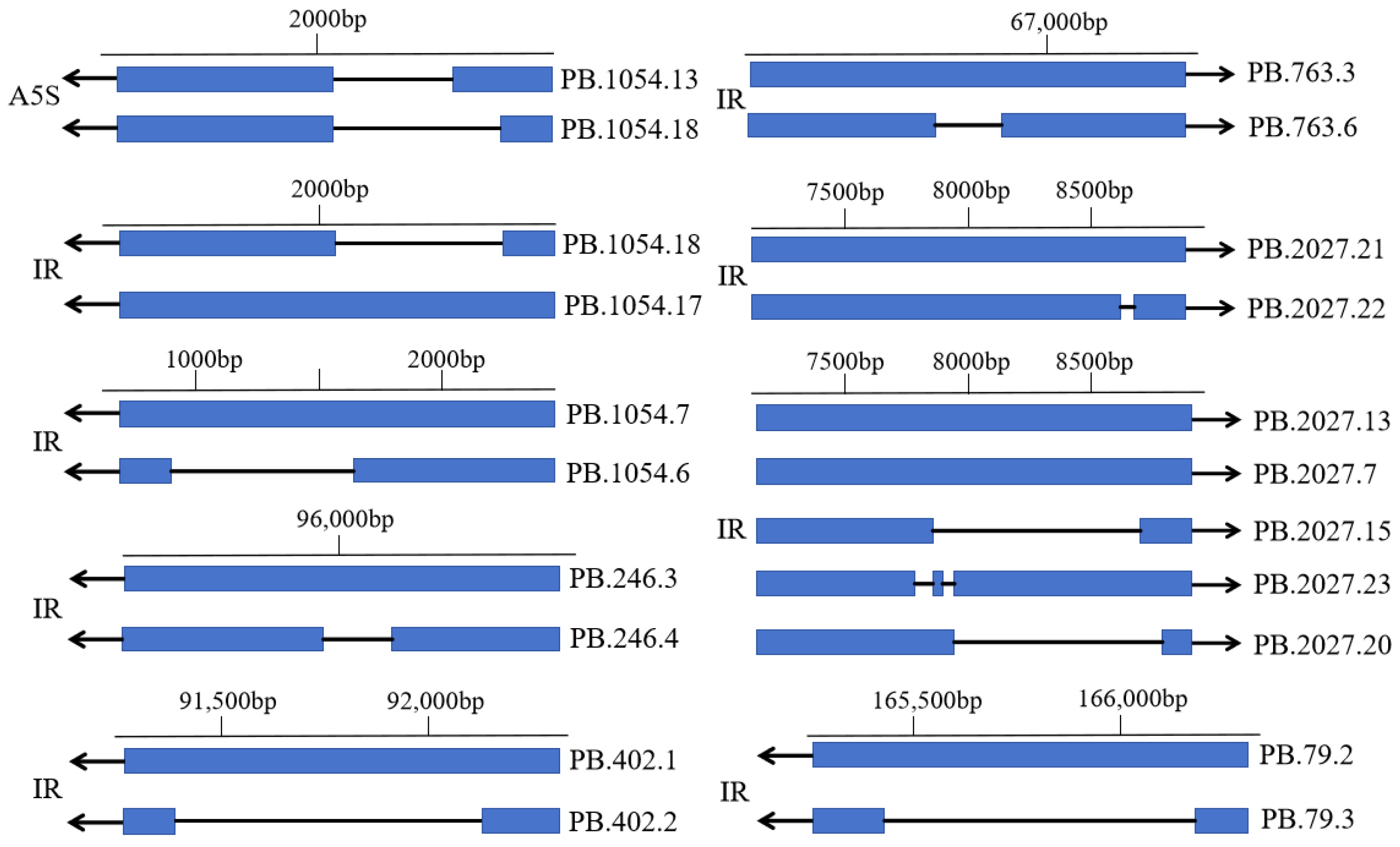
3.6. Analysis of AS Events of V. ceranae Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fries, I.; Feng, F.; Silva, A.D.; Slemenda, S.B.; Pieniazek, N.J. Nosema ceranae n. sp. (microspora, nosematidae), morphological and molecular characterization of a microsporidian parasite of the asian honey bee apis cerana (hymenoptera, apidae). J. Invertebr. Pathol. 1996, 32, 356–365. [Google Scholar] [CrossRef]
- Higes, M.; Martín, R.; Meana, A. Nosema ceranae, a new microsporidian parasite in honeybees in Europe. J. Invertebr. Pathol. 2006, 92, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.P.; Evans, J.D.; Murphy, C.; Gutell, R.; Zuker, M.; Gundensen-Rindal, D.; Pettis, J.S. Morphological, molecular, and phylogenetic characterization of Nosema ceranae, a microsporidian parasite isolated from the European honey bee, Apis mellifera. J. Eukaryot. Microbiol. 2009, 56, 142–147. [Google Scholar] [CrossRef]
- Smith, M.L. The honey bee parasite Nosema ceranae: Transmissible via food exchange? PLoS ONE 2012, 7, e43319. [Google Scholar] [CrossRef]
- Liang, Q.; Chen, D.F. Honey Bee Protection Studies; China Agriculture Press: Beijing, China, 2009; pp. 108–110. [Google Scholar]
- Cornman, R.S.; Chen, Y.; Schatz, M.C.; Street, C.; Zhao, Y.; Desany, B.; Egholm, M.; Hutchison, S.; Pettis, J.S.; Lipkin, W.I.; et al. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. PLoS Pathog. 2009, 5, e1000466. [Google Scholar] [CrossRef] [PubMed]
- Wilkes, R.P. Next-Generation Diagnostics for Pathogens. Vet. Clin. N. Am. Food Anim. Pract. 2023, 39, 165–173. [Google Scholar] [CrossRef]
- Kikuchi, T.; Eves-van den Akker, S.; Jones, J.T. Genome Evolution of Plant-Parasitic Nematodes. Annu. Rev. Phytopathol. 2017, 55, 333–354. [Google Scholar] [CrossRef]
- Zahedi, A.; Greay, T.L.; Paparini, A.; Linge, K.L.; Joll, C.A.; Ryan, U.M. Identification of eukaryotic microorganisms with 18S rRNA next-generation sequencing in wastewater treatment plants, with a more targeted NGS approach required for Cryptosporidium detection. Water Res. 2019, 158, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Chen, Y.; Wang, R.W.; Schwarz, R.S.; Evans, J.D. Honey bee microRNAs respond to infection by the microsporidian parasite Nosema ceranae. Sci. Rep. 2015, 5, 17494. [Google Scholar] [CrossRef]
- Garrido, P.M.; Porrini, M.P.; Antúnez, K.; Branchiccela, B.; Martínez-Noël, G.M.; Zunino, P.; Salerno, G.; Eguaras, M.J.; Ieno, E. Sublethal effects of acaricides and Nosema ceranae infection on immune related gene expression in honeybees. Vet. Res. 2016, 47, 51. [Google Scholar] [CrossRef]
- Badaoui, B.; Fougeroux, A.; Petit, F.; Anselmo, A.; Gorni, C.; Cucurachi, M.; Cersini, A.; Granato, A.; Cardeti, G.; Formato, G.; et al. RNA-sequence analysis of gene expression from honeybees (Apis mellifera) infected with Nosema ceranae. PLoS ONE 2017, 12, e0173438. [Google Scholar] [CrossRef]
- Li, Y.H.; Chang, Z.T.; Yen, M.R.; Huang, Y.F.; Chen, T.H.; Chang, J.C.; Wu, M.C.; Yang, Y.L.; Chen, Y.W.; Nai, Y.S. Transcriptome of Nosema ceranae and Upregulated Microsporidia Genes during Its Infection of Western Honey Bee (Apis mellifera). Insects 2022, 13, 716. [Google Scholar] [CrossRef] [PubMed]
- Pelin, A.; Selman, M.; Aris-Brosou, S.; Farinelli, L.; Corradi, N. Genome analyses suggest the presence of polyploidy and recent human-driven expansions in eight global populations of the honeybee pathogen Nosema ceranae. Environ. Microbiol. 2015, 17, 4443–4458. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.; Bahar, R.; Johan, M.F.; Mohamed Hashim, E.K.; Abdullah, W.Z.; Esa, E.; Abdul Hamid, F.S.; Zulkafli, Z. Next-Generation Sequencing (NGS) and Third-Generation Sequencing (TGS) for the Diagnosis of Thalassemia. Diagnostics 2023, 13, 373. [Google Scholar] [CrossRef]
- Lu, F.H.; McKenzie, N.; Kettleborough, G.; Heavens, D.; Clark, M.D.; Bevan, M.W. Independent assessment and improvement of wheat genome sequence assemblies using Fosill jumping libraries. Gigascience 2018, 7, giy053. [Google Scholar] [CrossRef]
- Magrini, V.; Gao, X.; Rosa, B.A.; McGrath, S.; Zhang, X.; Hallsworth-Pepin, K.; Martin, J.; Hawdon, J.; Wilson, R.K.; Mitreva, M. Improving eukaryotic genome annotation using single molecule mRNA sequencing. BMC Genom. 2018, 19, 172. [Google Scholar] [CrossRef]
- Chao, Y.; Yuan, J.; Li, S.; Jia, S.; Han, L.; Xu, L. Analysis of transcripts and splice isoforms in red clover (Trifolium pratense L.) by single-molecule long-read sequencing. BMC Plant Biol. 2018, 18, 300. [Google Scholar] [CrossRef]
- Li, Y.; Fang, C.; Fu, Y.; Hu, A.; Li, C.; Zou, C.; Li, X.; Zhao, S.; Zhang, C.; Li, C. A survey of transcriptome complexity in Sus scrofa using single-molecule long-read sequencing. DNA Res. 2018, 25, 421–437. [Google Scholar] [CrossRef]
- Chen, D.; Du, Y.; Fan, X.; Zhu, Z.; Jiang, H.; Wang, J.; Fan, Y.; Chen, H.; Zhou, D.; Xiong, C.; et al. Reconstruction and functional annotation of Ascosphaera apis full-length transcriptome utilizing PacBio long reads combined with Illumina short reads. J. Invertebr. Pathol. 2020, 176, 107475. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.M.; Chen, H.Z.; Liu, S.Y.; Zhu, Z.W.; Fan, X.X.; Fan, Y.C.; Wan, J.Q.; Zhang, L.; Xiong, C.L.; Xu, G.J.; et al. Immune Responses of Apis mellifera ligustia to Nosema ceranae Stress. Sci. Agric. Sin. 2019, 52, 3069–3082. (In Chinese) [Google Scholar]
- Malone, L.A.; Giacon, H.A.; Newton, M.R. Comparison of the responses of some New Zealand and Australian honey bees (Apis mellifera L) to Nosema apis. Apidologie 1999, 26, 495–502. [Google Scholar] [CrossRef]
- Chen, D.; Chen, H.; Du, Y.; Zhou, D.; Geng, S.; Wang, H.; Wan, J.; Xiong, C.; Zheng, Y.; Guo, R. Genome-Wide Identification of Long Non-Coding RNAs and Their Regulatory Networks Involved in Apis mellifera ligustica Response to Nosema ceranae Infection. Insects 2019, 10, 245. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.P.; Tseng, E.; Salamov, A.; Zhang, J.; Meng, X.; Zhao, Z.; Kang, D.; Underwood, J.; Grigoriev, I.V.; Figueroa, M.; et al. Widespread Polycistronic Transcripts in Fungi Revealed by Single-Molecule mRNA Sequencing. PLoS ONE 2015, 10, e0132628. [Google Scholar] [CrossRef]
- Wu, T.D.; Watanabe, C.K. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 2005, 21, 1859–1875. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Foissac, S.; Sammeth, M. ASTALAVISTA: Dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res. 2007, 35, W297–W299. [Google Scholar] [CrossRef]
- Pertea, G.; Pertea, M. GFF Utilities: GffRead and GffCompare. F1000Res 2020, 9, ISCB Comm J-304. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Computational identification of functional RNA homologs in metagenomic data. RNA Biol. 2013, 10, 1170–1179. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Faino, L.; Seidl, M.F.; Datema, E.; van den Berg, G.C.; Janssen, A.; Wittenberg, A.H.; Thomma, B.P. Single-Molecule Real-Time Sequencing Combined with Optical Mapping Yields Completely Finished Fungal Genome. mBio 2015, 6, e00936-15. [Google Scholar] [CrossRef] [PubMed]
- Kingan, S.B.; Heaton, H.; Cudini, J.; Lambert, C.C.; Baybayan, P.; Galvin, B.D.; Durbin, R.; Korlach, J.; Lawniczak, M.K.N. A High-Quality De novo Genome Assembly from a Single Mosquito Using PacBio Sequencing. Genes 2019, 10, 62. [Google Scholar] [CrossRef] [PubMed]
- Wittner, M. Historic Perspective on the Microsporidia: Expanding Horizons. In The Microsporidia and Microsporidiosis; Wiley: Hoboken, NJ, USA, 1999; pp. 1–6. [Google Scholar] [CrossRef]
- Liu, T.; Shen, Y.; Liu, Q.; Liu, B. The unique Entner-Doudoroff (ED) glycolysis pathway of glucose in Archaea—A review. Acta Microbiol. 2008, 48, 1126–1131. [Google Scholar]
- Paldi, N.; Glick, E.; Oliva, M.; Zilberberg, Y.; Aubin, L.; Pettis, J.; Chen, Y.; Evans, J.D. Effective gene silencing in a microsporidian parasite associated with honeybee (Apis mellifera) colony declines. Appl. Environ. Microbiol. 2010, 76, 5960–5964. [Google Scholar] [CrossRef]
- St Laurent, G.; Wahlestedt, C.; Kapranov, P. The Landscape of long noncoding RNA classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef]
- Zhu, Q.H.; Wang, M.B. Molecular functions of long non-coding RNAs in plants. Genes 2012, 3, 176–190. [Google Scholar] [CrossRef]
- Guo, R.; Chen, D.F.; Xiong, C.L.; Hou, C.S.; Zheng, Y.Z.; Fu, Z.M.; Liang, Q.; Diao, Q.Y.; Zhang, L.; Wang, H.Q.; et al. First identification of long non-coding RNAs in fungal parasite Nosema ceranae. Apidologie 2018, 49, 660–670. [Google Scholar] [CrossRef]
- Huang, Q.; Wu, Z.H.; Li, W.F.; Guo, R.; Xu, J.S.; Dang, X.Q.; Ma, Z.G.; Chen, Y.P.; Evans, J.D. Genome and Evolutionary Analysis of Nosema ceranae: A Microsporidian Parasite of Honey Bees. Front. Microbiol. 2021, 12, 645353. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cai, K.; Zhang, Q.; Pei, X.; Chen, S.; Jiang, L.; Han, Z.; Zhao, M.; Li, Y.; Zhang, X.; et al. The Manchurian Walnut Genome: Insights into Juglone and Lipid Biosynthesis. GigaScience 2022, 11, giac057. [Google Scholar] [CrossRef] [PubMed]
- Lovci, M.T.; Ghanem, D.; Marr, H.; Arnold, J.; Gee, S.; Parra, M.; Liang, T.Y.; Stark, T.J.; Gehman, L.T.; Hoon, S.; et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat. Struct. Mol. Biol. 2013, 20, 1434–1442. [Google Scholar] [CrossRef]
- Mayr, C.; Bartel, D.P. Widespread shortening of 3′utrs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 2009, 138, 673–684. [Google Scholar] [CrossRef]
- Mironov, A.; Fickett, J.; Gelfand, M. Frequent alternative splicing of human genes. Genome Res. 1999, 9, 1288–1293. [Google Scholar] [CrossRef]
- Filichkin, S.A.; Hamilton, M.; Dharmawardhana, P.D.; Singh, S.K.; Sullivan, C.; Ben-Hur, A.; Reddy, A.S.N.; Jaiswal, P. Abiotic stresses modulate landscape of poplar transcriptome via alternative splicing, differential intron retention, and isoform ratio switching. Front. Plant Sci. 2018, 9, 5. [Google Scholar] [CrossRef]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Z.; Fan, X.X.; Fan, Y.C.; Wang, J.; Zhu, Z.W.; Jiang, H.B.; Zhang, W.D.; Long, Q.; Xiong, C.L.; Zheng, Y.Z.; et al. Analysis of alternative splicing and alternative polyadenylation of Nosema ceranae genes. Mycosystema 2021, 40, 161–173. (In Chinese) [Google Scholar]
- Cui, J.; Shen, N.; Lu, Z.; Xu, G.; Wang, Y.; Jin, B. Analysis and comprehensive comparison of PacBio and nanopore-based RNA sequencing of the Arabidopsis transcriptome. Plant Methods 2020, 16, 85. [Google Scholar] [CrossRef]
- Bruijnesteijn, J. HLA/MHC and KIR characterization in humans and non-human primates using Oxford Nanopore Technologies and Pacific Biosciences sequencing platforms. HLA 2023, 101, 205–221. [Google Scholar] [CrossRef]

| Full Names | Abbreviations |
|---|---|
| Single-molecule real-time | SMRT |
| Circular consensus | CCS |
| Full-length non-chimeric | FLNC |
| alternative splicing | AS |
| Next-generation sequencing | NGS |
| Third generation sequencing | TGS |
| Alternative polyadenylatio | APA |
| Full-length | FL |
| Non-full-length | nFL |
| Genomic Mapping and Alignment Program | GMAP |
| Exon skipping | ES |
| Intron retention | IR |
| Alternative 5′ splice site | A5S |
| Alternative 3′ splice site | A3S |
| Mutually exclusive exon | MEE |
| Potential Calculator | CPC |
| Coding-Non-Coding Index | CNCI |
| Coding-Potential Assessment Tool | CPAT |
| Number | |
|---|---|
| CCS | 41,950 |
| Read bases of CCS | 61,124,354 |
| Mean read length of CCS | 1457 |
| Mean number of passes | 95 |
| Full-length non-chimeric reads | 25,068 |
| Full-length non-chimeric percentage | 59.76% |
| Consensus isoforms | 10,900 |
| Average read length of consensus isoforms | 775 |
| Polished high-quality isoforms | 10,900 |
| Polished low-quality isoforms | 0 |
| Annotation | Transcript ID | |
|---|---|---|
| Virulence factors | ADP/ATP carrier protein | PB.150.1, PB.677.1, PB.1292.3, PB.1293.1 |
| Chitin synthase | PB.161.1, PB.1296.3 | |
| Iron-sulfur clusters transporter | PB.1602.1, PB.1302.1 | |
| ABC transporter | PB.54.5 | |
| Iron-sulfur clusters transporter, ABC transporter | PB.1301.8 | |
| Spore wall protein | PB.414.3 | |
| Energy metabolism | Glycolysis/Gluconeogenesis | PB.1959.3, PB.294.1, PB.293.1, PB.780.1, PB.1959.3 |
| Oxidative phosphorylation | PB.1107.1, PB.1797.6, PB.1608.1, PB.1608.2 | |
| Frataxin | PB.1632.4 |
| Gene ID | Locus | Strand | Site | Original Site | Optimize Site |
|---|---|---|---|---|---|
| gene-AAJ76_3600024363 | NW_020169331.1:24363–25655 | + | 3′ | 25,376 | 25,655 |
| gene-AAJ76_800027588 | NW_020169303.1:26663–27588 | − | 5′ | 26,860 | 26,663 |
| gene-AAJ76_1100030560 | NW_020169306.1:30560–32315 | + | 3’ | 31,945 | 32,315 |
| gene-AAJ76_1900029507 | NW_020169314.1:29503–30905 | + | 5’ | 29,507 | 29,503 |
| gene-AAJ76_1900029507 | NW_020169314.1:29503–30905 | + | 3′ | 30,883 | 30,905 |
| gene-AAJ76_1180002683 | NW_020169413.1:1478–3041 | − | 3′ | 2683 | 3041 |
| gene-AAJ76_2200042944 | NW_020169317.1:42944–44008 | + | 3′ | 43,930 | 44,008 |
| gene-AAJ76_1100054231 | NW_020169306.1:54231–55342 | + | 3′ | 54,851 | 55,342 |
| gene-AAJ76_200036678 | NW_020169297.1:35879–36711 | − | 5′ | 35,890 | 35,879 |
| gene-AAJ76_200036678 | NW_020169297.1:35879–36711 | − | 3′ | 36,706 | 36,711 |
| gene-AAJ76_100042163 | NW_020169296.1:41211–42484 | − | 5′ | 41,228 | 41,211 |
| gene-AAJ76_100042163 | NW_020169296.1:41211–42484 | − | 3′ | 42,163 | 42,484 |
| gene-AAJ76_580008628 | NW_020169353.1:7727–8628 | − | 5′ | 8332 | 7727 |
| Type of AS Event | Genomic Location | Gene ID | Transcript ID |
|---|---|---|---|
| Alternative 5′ splice site | NW_020169316.1:348-2409C | PB.1054 | PB.1054.18, PB.1054.13 |
| Intron retention | NW_020169316.1:348-2409C | PB.1054 | PB.1054.17, PB.1054.18 |
| Intron retention | NW_020169316.1:348-2409C | PB.1054 | PB.1054.7, PB.1054.6 |
| Intron retention | NW_020169298.1:95363-96278C | PB.246 | PB.246.3, PB.246.4 |
| Intron retention | NW_020169300.1:91266-92414C | PB.402 | PB.402.1, PB.402.2 |
| Intron retention | NW_020169306.1:66670-67167W | PB.763 | PB.763.3, PB.763.6 |
| Intron retention | NW_020169296.1:164677-166306C | PB.79 | PB.79.2, PB.79.3 |
| Intron retention | NW_020169415.1:7382-8798W | PB.2027 | PB.2027.21, PB.2027.22 |
| Intron retention | NW_020169415.1:7382-8798W | PB.2027 | PB.2027.13/PB.2027.7, PB.2027.15 |
| Intron retention | NW_020169415.1:7382-8798W | PB.2027 | PB.2027.13/PB.2027.7, PB.2027.23 |
| Intron retention | NW_020169415.1:7382-8798W | PB.2027 | PB.2027.13/PB.2027.7, PB.2027.20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, S.; Zang, H.; Liu, X.; Jing, X.; Liu, Z.; Zhang, W.; Wang, M.; Zheng, Y.; Li, Z.; Qiu, J.; et al. Full-Length Transcriptome Construction and Systematic Characterization of Virulence Factor-Associated Isoforms in Vairimorpha (Nosema) Ceranae. Genes 2024, 15, 1111. https://doi.org/10.3390/genes15091111
Guo S, Zang H, Liu X, Jing X, Liu Z, Zhang W, Wang M, Zheng Y, Li Z, Qiu J, et al. Full-Length Transcriptome Construction and Systematic Characterization of Virulence Factor-Associated Isoforms in Vairimorpha (Nosema) Ceranae. Genes. 2024; 15(9):1111. https://doi.org/10.3390/genes15091111
Chicago/Turabian StyleGuo, Sijia, He Zang, Xiaoyu Liu, Xin Jing, Zhitan Liu, Wende Zhang, Mengyi Wang, Yidi Zheng, Zhengyuan Li, Jianfeng Qiu, and et al. 2024. "Full-Length Transcriptome Construction and Systematic Characterization of Virulence Factor-Associated Isoforms in Vairimorpha (Nosema) Ceranae" Genes 15, no. 9: 1111. https://doi.org/10.3390/genes15091111
APA StyleGuo, S., Zang, H., Liu, X., Jing, X., Liu, Z., Zhang, W., Wang, M., Zheng, Y., Li, Z., Qiu, J., Chen, D., Yan, T., & Guo, R. (2024). Full-Length Transcriptome Construction and Systematic Characterization of Virulence Factor-Associated Isoforms in Vairimorpha (Nosema) Ceranae. Genes, 15(9), 1111. https://doi.org/10.3390/genes15091111








