Optimizing Genomic Parental Selection for Categorical and Continuous–Categorical Multi-Trait Mixtures
Abstract
1. Introduction
2. Materials and Methods
2.1. General Structure of Phenotypic and Genomic Data
2.2. General Model Formulation
2.3. Categorical Multi-Trait (CM)
2.4. Continuous–Categorical Multi-Trait Mixtures (CCMM)
2.4.1. Posterior Predictive Distribution and Posterior Expected Loss
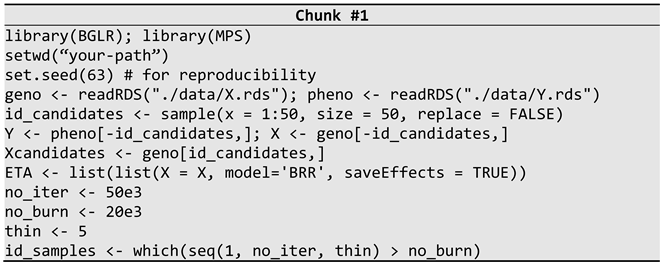
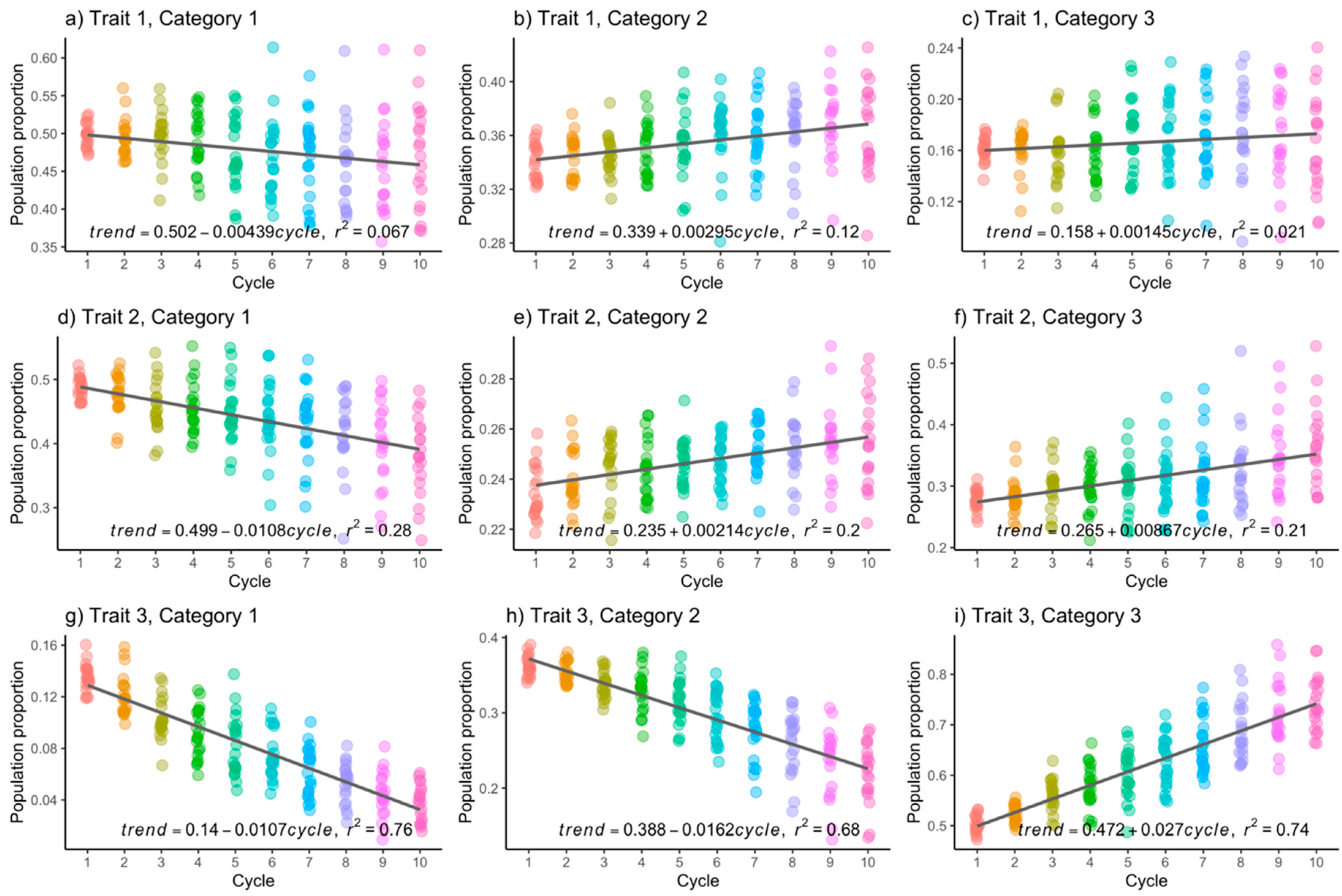
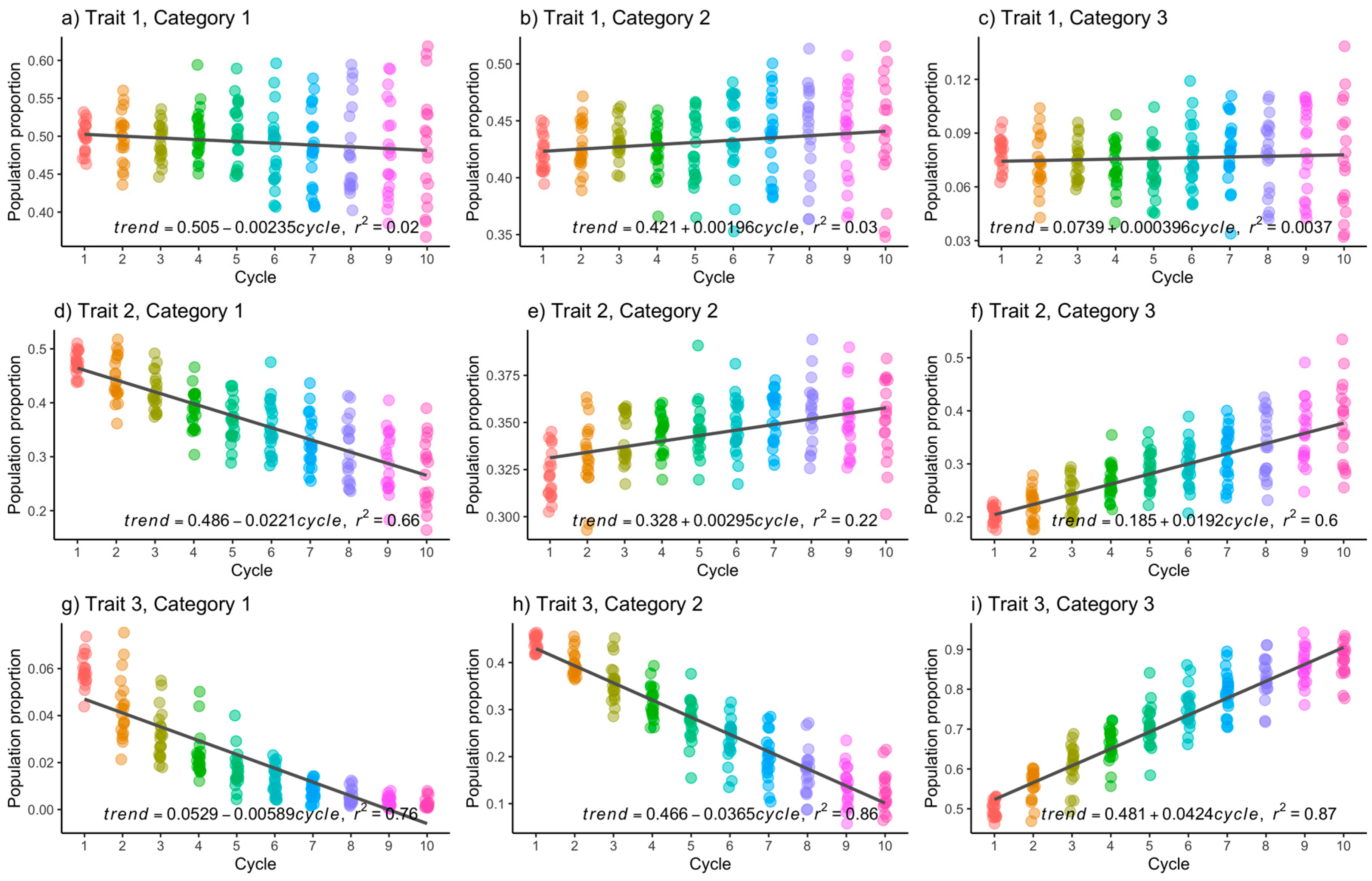
2.4.2. Simulation Study
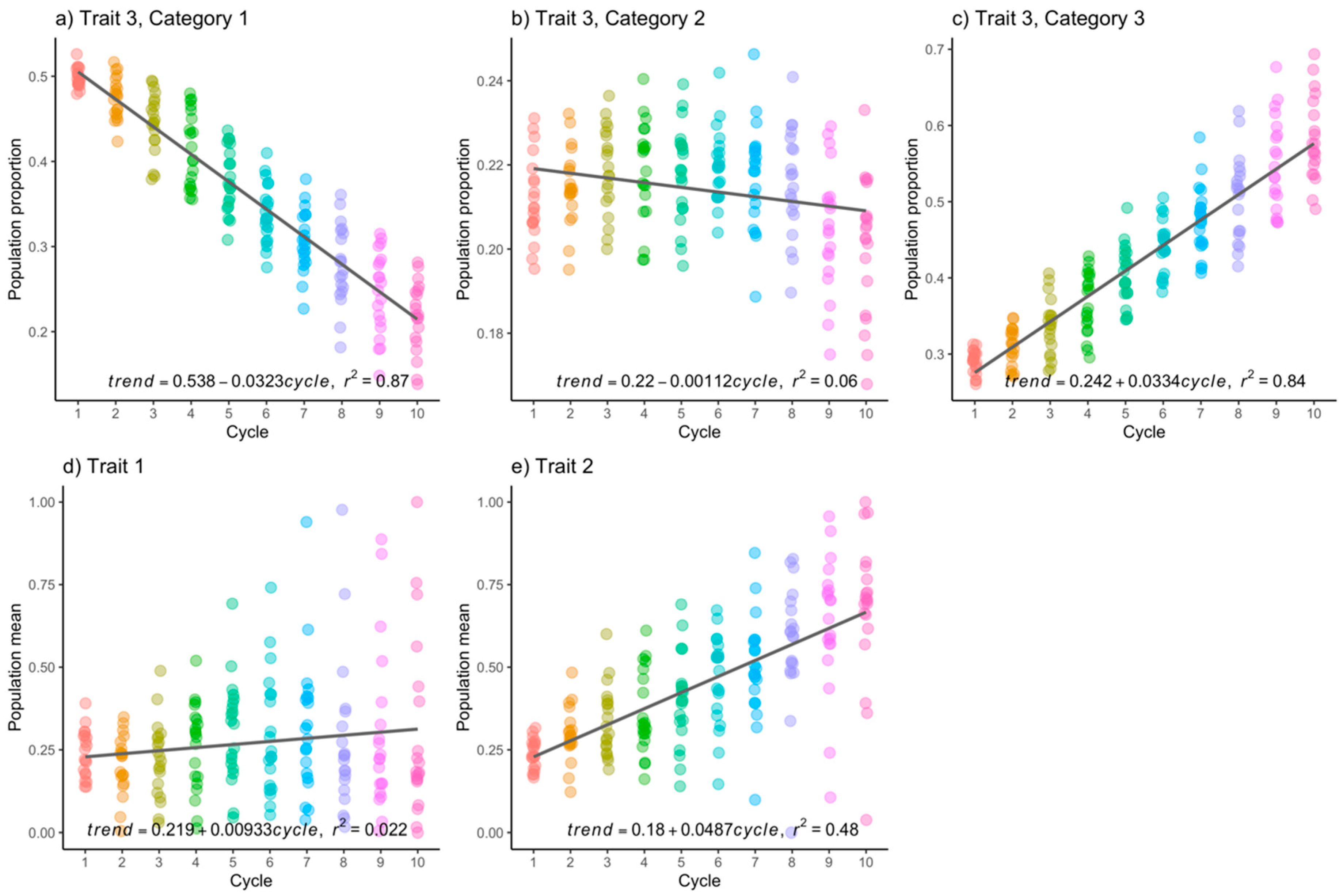
2.4.3. Experimental Data
3. Results
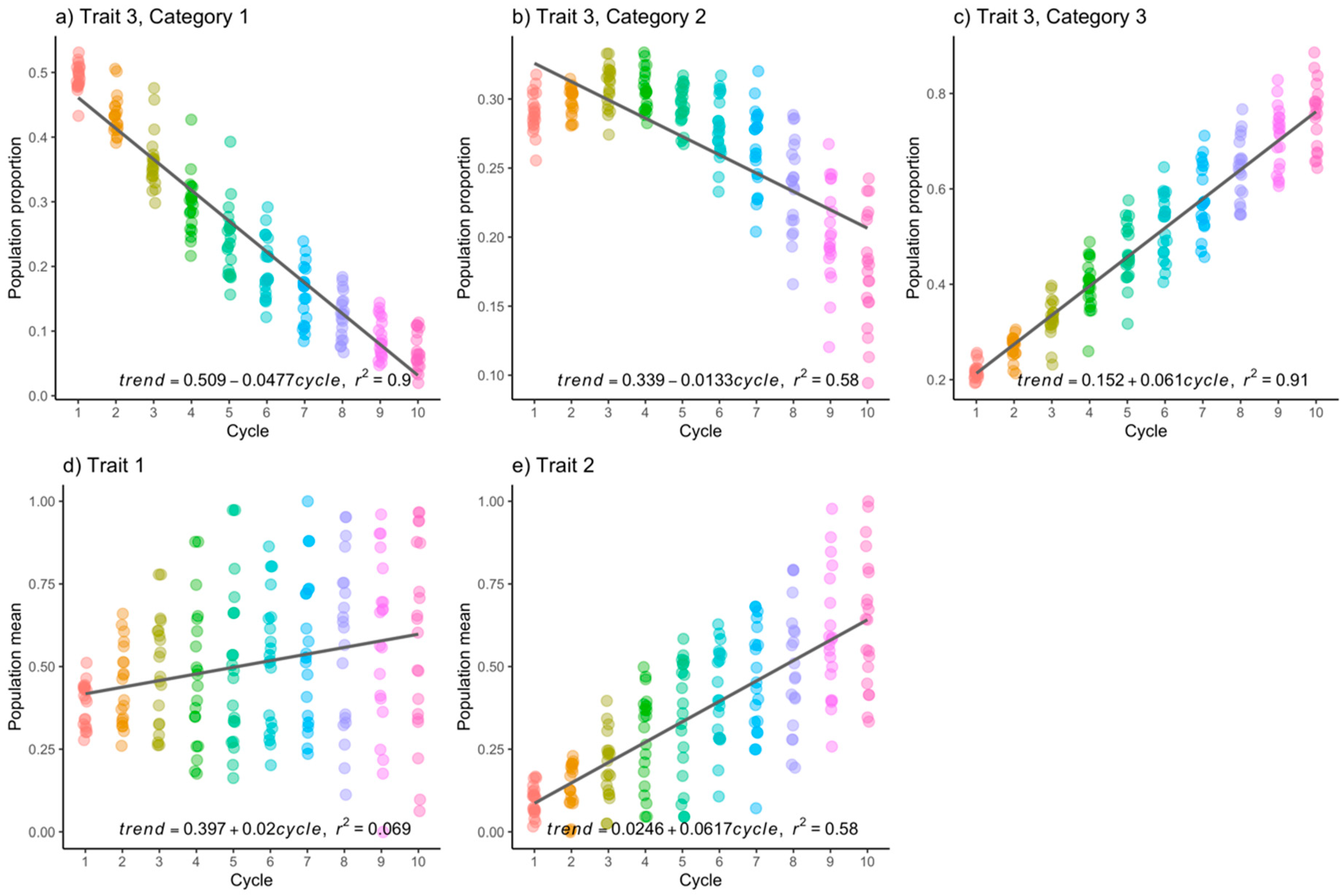
3.1. Simulated Data
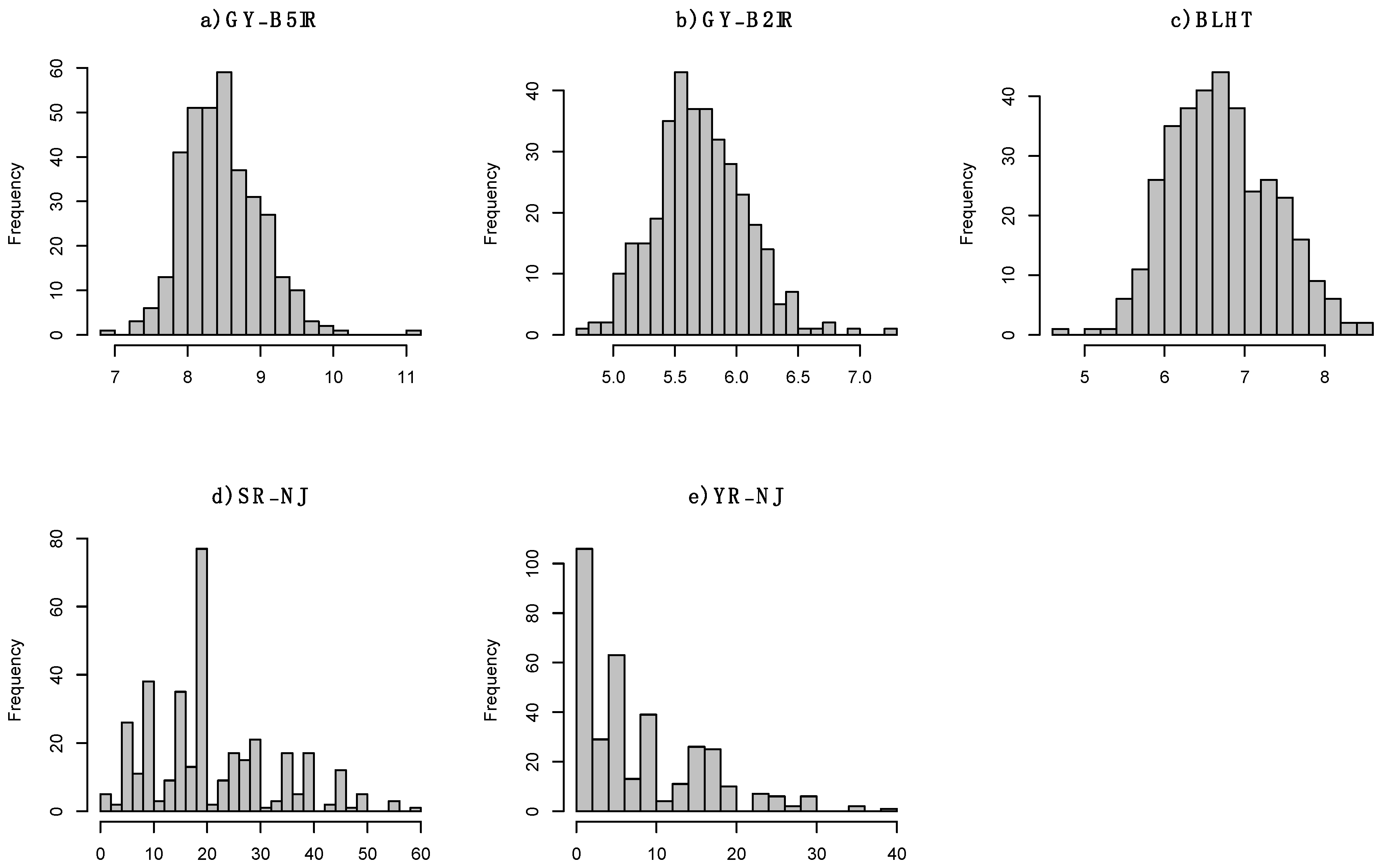
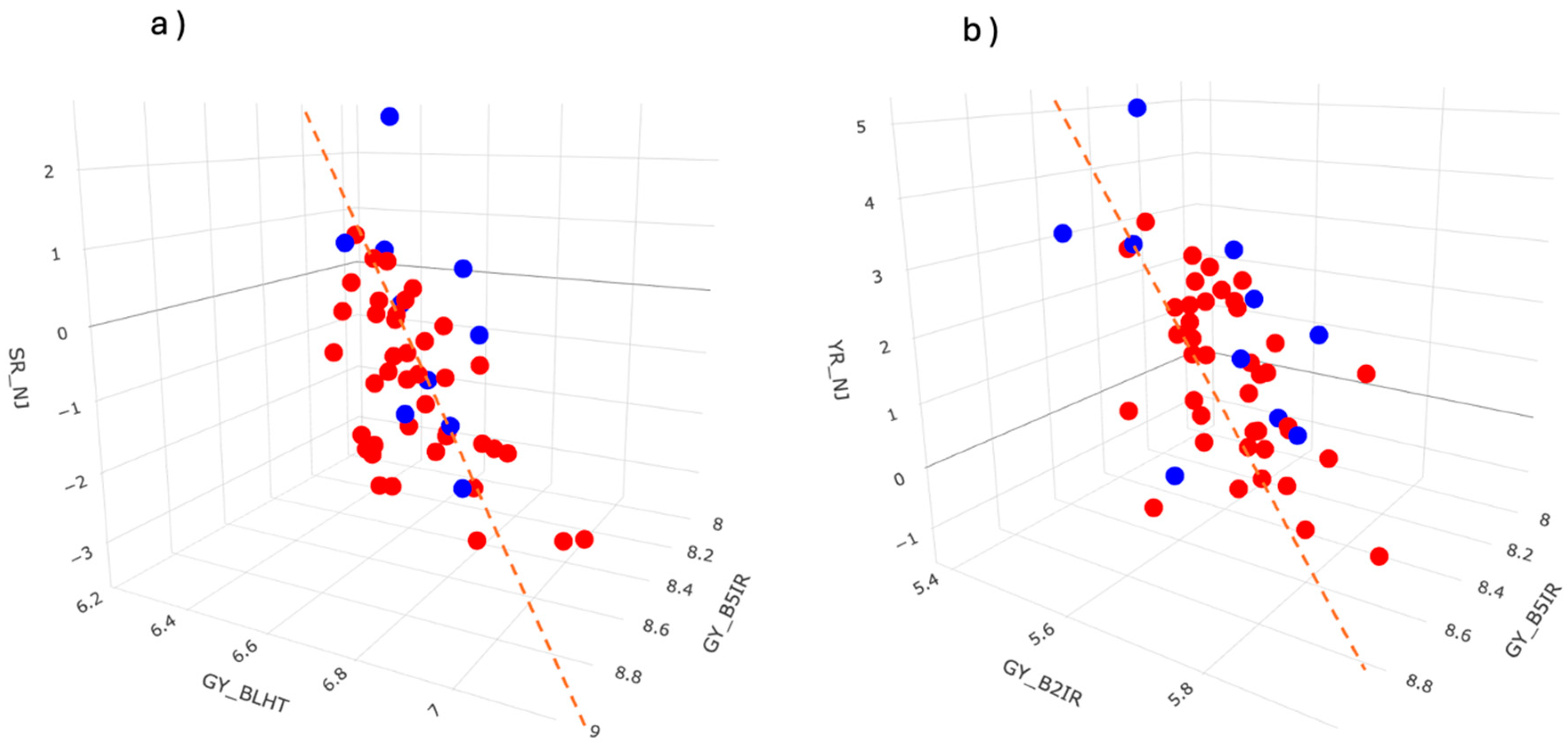
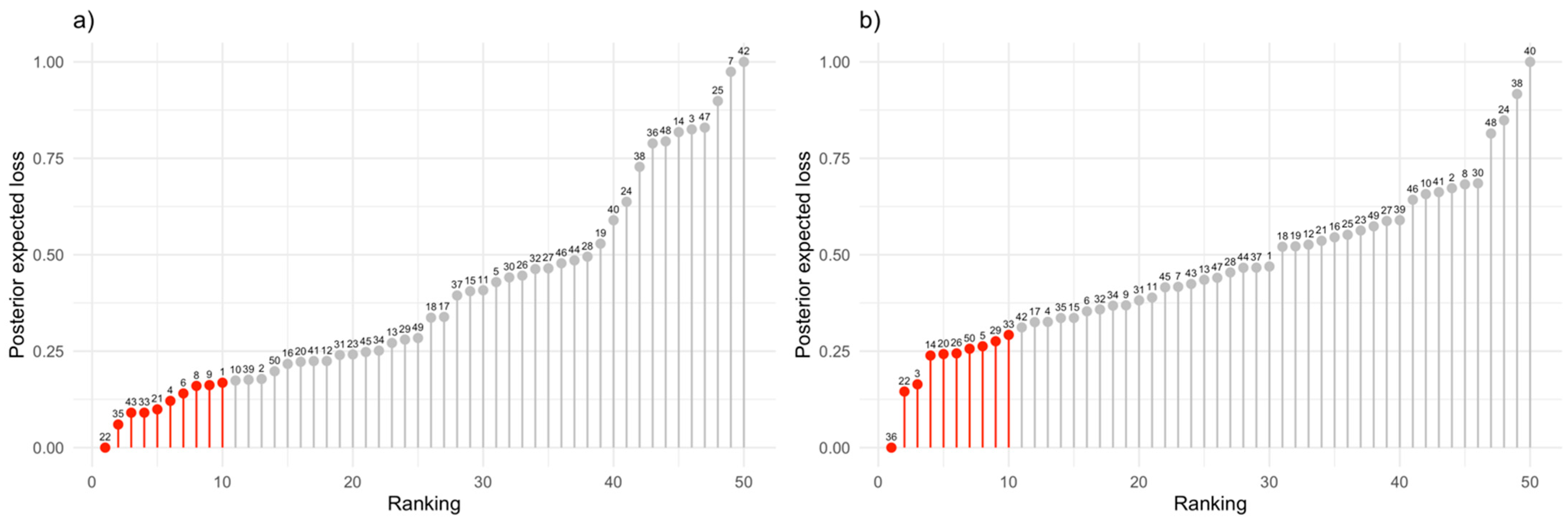
3.2. Experimental Data
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A


References
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Beyene, Y.; Semagn, K.; Mugo, S.; Tarekegne, A.; Babu, R.; Meisel, B.; Sehabiague, P.; Makumbi, D.; Magorokosho, C.; Oikeh, S.; et al. Genetic gains in grain yield through genomic selection in eight bi-parental maize populations under drought stress. Crop Sci. 2015, 55, 154–163. [Google Scholar] [CrossRef]
- Crossa, J.; Perez, P.; Hickey, J.; Burgueno, J.; Ornella, L.; Cerón-Rojas, J.; Zhang, X.; Dreisigacker, S.; Babu, R.; Li, Y.; et al. Genomic prediction in CIMMYT maize and wheat breeding programs. Heredity 2014, 112, 48–60. [Google Scholar] [CrossRef]
- Heffner, E.L.; Lorenz, A.J.; Jannink, J.-L.; Sorrells, M.E. Plant breeding with genomic selection: Gain per unit time and cost. Crop Sci. 2010, 50, 1681–1690. [Google Scholar] [CrossRef]
- Poland, J.; Endelman, J.; Dawson, J.; Rutkoski, J.; Wu, S.; Manes, Y.; Dreisigacker, S.; Crossa, J.; Sánchez-Villeda, H.; Sorrells, M.; et al. Genomic selection in wheat breeding using genotyping-by-sequencing. Plant Genome 2012, 5, 103–113. [Google Scholar] [CrossRef]
- Spindel, J.; Begum, H.; Akdemir, D.; Collard, B.; Redoña, E.; Jannink, J.; McCouch, S. Genome-wide prediction models that incorporate de novo GWAS are a powerful new tool for tropical rice improvement. Heredity 2016, 116, 395–408. [Google Scholar] [CrossRef]
- Calus, M.; Meuwissen, T.; De Roos, A.; Veerkamp, R. Accuracy of genomic selection using different methods to define haplotypes. Genetics 2008, 178, 553–561. [Google Scholar] [CrossRef]
- Dassonneville, R.; Brøndum, R.; Druet, T.; Fritz, S.; Guillaume, F.; Guldbrandtsen, B.; Lund, M.; Ducrocq, V.; Su, G. Effect of imputing markers from a low-density chip on the reliability of genomic breeding values in holstein populations. J. Dairy Sci. 2011, 94, 3679–3686. [Google Scholar] [CrossRef]
- Erbe, M.; Hayes, B.; Matukumalli, L.; Goswami, S.; Bowman, P.; Reich, C.; Mason, B.; Goddard, M. Improving accuracy of genomic predictions within and between dairy cattle breeds with imputed high-density single nucleotide polymorphism panels. J. Dairy Sci. 2012, 95, 4114–4129. [Google Scholar] [CrossRef]
- Hayes, B.J.; Bowman, P.J.; Chamberlain, A.J.; Goddard, M.E. Genomic selection in dairy cattle: Progress and challenges. J. Dairy Sci. 2009, 92, 433–443. [Google Scholar] [CrossRef]
- Laverdière, J.-P.; Lenz, P.; Nadeau, S.; Depardieu, C.; Isabel, N.; Perron, M.; Beaulieu, J.; Bousquet, J. Breeding for adaptation to climate change: Genomic selection for drought response in a white spruce multi-site polycross test. Evol. Appl. 2022, 15, 383–402. [Google Scholar] [CrossRef]
- Lenz, P.R.; Nadeau, S.; Mottet, M.-J.; Perron, M.; Isabel, N.; Beaulieu, J.; Bousquet, J. Multi-trait genomic selection for weevil resistance, growth, and wood quality in norway spruce. Evol. Appl. 2020, 13, 76–94. [Google Scholar] [CrossRef]
- Pérez, R.; de los Campos, G. Genome-wide regression and prediction with the BGLR statistical package. Genetics 2014, 198, 483–495. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, P.; Montesinos-López, O.A.; Montesinos-López, A.; Crossa, J. Bayesian regularized quantile regression: A robust alternative for genome-based prediction of skewed data. Crop J. 2020, 8, 713–722. [Google Scholar] [CrossRef]
- Xu, Z.; Kurek, A.; Cannon, S.; Beavis, W. Predictions from algorithmic modeling result in better decisions than from data modeling for soybean iron deficiency chlorosis. PLoS ONE 2021, 16, e0240948. [Google Scholar] [CrossRef]
- Cerón-Rojas, J.J.; Crossa, J. Combined multistage linear genomic selection indices to predict the net genetic merit in plant breeding. G3 Genes Genomes Genet. 2020, 10, 2087–2101. [Google Scholar] [CrossRef]
- Villar-Hernández, B.d.J.; Pérez-Elizalde, S.; Martini, J.W.; Toledo, F.; Pérez-Rodríguez, P.; Krause, M.; García-Calvillo, I.D.; Covarrubias-Pazaran, G.; Crossa, J. Application of multi-trait bayesian decision theory for parental genomic selection. G3 Genes Genomes Genet. 2021, 11, jkab012. [Google Scholar] [CrossRef]
- Villar-Hernández, B.d.J.; Pérez-Elizalde, S.; Crossa, J.; Pérez-Rodríguez, P.; Toledo, F.H.; Burgueño, J. A bayesian decision theory approach for genomic selection. G3 Genes Genomes Genet. 2018, 8, 3019–3037. [Google Scholar] [CrossRef]
- Villar-Hernández, B.d.J.; Dreisigacker, S.; Crespo, L.; Pérez-Rodríguez, P.; Pérez-Elizalde, S.; Toledo, F.; Crossa, J. A Bayesian optimization R package for multitrait parental selection. Plant Genome 2024, 17, e20433. [Google Scholar] [CrossRef]
- Pérez, R.; de los Campos, G. Multitrait Bayesian shrinkage and variable selection models with the BGLR-R package. Genetics 2022, 222, iyac112. [Google Scholar] [CrossRef]
- McCullagh, P. Regression models for ordinal data. J. R. Stat. Soc. Ser. B Methodol. 1980, 42, 109–127. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, X.; Ding, X.; Li, J.; Zhang, Q. GPOPSIM: A simulation tool for whole-genome genetic data. BMC Genet. 2015, 16, 10. [Google Scholar] [CrossRef] [PubMed]
| CM: Categorical Multi-Trait | ||||||||
|---|---|---|---|---|---|---|---|---|
| Trait | Type | Category | Notation | Goal | Achieved | Average % Change | Achieved | Average % Change |
| 1 | Categorical | 1 | T1C1 | Decrease | Yes | −5.36 | Yes | −2.95 |
| 1 | Categorical | 2 | T1C2 | Increase/decrease | Yes | 6.65 | No | 4.39 |
| 1 | Categorical | 3 | T1C3 | Increase | Yes | 2.47 | Yes | −4.72 |
| 2 | Categorical | 1 | T2C1 | Decrease | Yes | −21.93 | Yes | −42.62 |
| 2 | Categorical | 2 | T2C2 | Increase/decrease | Yes | 8.72 | Yes | 9.09 |
| 2 | Categorical | 3 | T2C3 | Increase | Yes | 31.06 | Yes | 84.84 |
| 3 | Categorical | 1 | T3C1 | Decrease | Yes | −71.97 | Yes | −95.29 |
| 3 | Categorical | 2 | T3C2 | Increase/decrease | Yes | −38.96 | Yes | −71.58 |
| 3 | Categorical | 3 | T3C3 | Increase | Yes | 47.47 | Yes | 74.45 |
| CCMM: Continuous–Categorical Multi-trait Mixture | ||||||||
| Trait | Type | Category | Notation | Goal | Achieved | Average % change | Achieved | Average % change |
| 1 | Continuous | - | T1 | Increase | Yes | 23.96 | Yes | 50.77 |
| 2 | Continuous | - | T2 | Increase | Yes | 185.26 | Yes | 572.10 |
| 3 | Categorical | 1 | T3C1 | Decrease | Yes | −56.32 | Yes | −85.88 |
| 3 | Categorical | 2 | T3C2 | Increase/Decrease | Yes | −5.19 | Yes | −39.03 |
| 3 | Categorical | 3 | T3C3 | Increase | Yes | 100.61 | Yes | 246.36 |
| Trait 2 Category 3, = 0.3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| p-Value = 2.14 × 10−6 from the Kruskal–Wallis Test | |||||||||
| p-Values from the Mann–Whitney U Test Using the Bonferroni Correction | |||||||||
| Cycle 1 | Cycle 2 | Cycle 3 | Cycle 4 | Cycle 5 | Cycle 6 | Cycle 7 | Cycle 8 | Cycle 9 | |
| Cycle 2 | NS | ||||||||
| Cycle 3 | NS | NS | |||||||
| Cycle 4 | NS | NS | NS | ||||||
| Cycle 5 | NS | NS | NS | NS | |||||
| Cycle 6 | NS | NS | NS | NS | NS | ||||
| Cycle 7 | NS | NS | NS | NS | NS | NS | |||
| Cycle 8 | NS | NS | NS | NS | NS | NS | NS | ||
| Cycle 9 | S | S | NS | NS | NS | NS | NS | NS | |
| Cycle 10 | S | S | NS | S | NS | NS | NS | NS | NS |
| Trait 3 Category 3, = 0.3 | |||||||||
| p-value = 7.5510−29 from the Kruskal–Wallis test | |||||||||
| p-values from the Mann–Whitney U test using the Bonferroni correction | |||||||||
| Cycle 1 | Cycle 2 | Cycle 3 | Cycle 4 | Cycle 5 | Cycle 6 | Cycle 7 | Cycle 8 | Cycle 9 | |
| Cycle 2 | S | ||||||||
| Cycle 3 | S | S | |||||||
| Cycle 4 | S | S | NS | ||||||
| Cycle 5 | S | S | NS | NS | |||||
| Cycle 6 | S | S | S | NS | NS | ||||
| Cycle 7 | S | S | S | S | NS | NS | |||
| Cycle 8 | S | S | S | S | S | NS | NS | ||
| Cycle 9 | S | S | S | S | S | S | NS | NS | |
| Cycle 10 | S | S | S | S | S | S | S | NS | NS |
| Trait 2 Category 3, = 0.6 | |||||||||
| p-Value = 3.90 × 10−23 from the Kruskal–Wallis Test | |||||||||
| p-Values from the Mann–Whitney U Test Using the Bonferroni Correction | |||||||||
| Cycle 1 | Cycle 2 | Cycle 3 | Cycle 4 | Cycle 5 | Cycle 6 | Cycle 7 | Cycle 8 | Cycle 9 | |
| Cycle 2 | NS | ||||||||
| Cycle 3 | S | NS | |||||||
| Cycle 4 | S | S | NS | ||||||
| Cycle 5 | S | S | S | NS | |||||
| Cycle 6 | S | S | S | NS | NS | ||||
| Cycle 7 | S | S | S | NS | NS | NS | |||
| Cycle 8 | S | S | S | S | NS | NS | NS | ||
| Cycle 9 | S | S | S | S | S | S | NS | NS | |
| Cycle 10 | S | S | S | S | S | NS | NS | NS | NS |
| Trait 3 Category 3, = 0.6 | |||||||||
| p-value = 2.74 × 10−32 from the Kruskal–Wallis test | |||||||||
| p-values from the Mann–Whitney U test using the Bonferroni correction | |||||||||
| Cycle 1 | Cycle 2 | Cycle 3 | Cycle 4 | Cycle 5 | Cycle 6 | Cycle 7 | Cycle 8 | Cycle 9 | |
| Cycle 2 | S | ||||||||
| Cycle 3 | S | S | |||||||
| Cycle 4 | S | S | NS | ||||||
| Cycle 5 | S | S | S | NS | |||||
| Cycle 6 | S | S | S | S | NS | ||||
| Cycle 7 | S | S | S | S | S | NS | |||
| Cycle 8 | S | S | S | S | S | S | NS | ||
| Cycle 9 | S | S | S | S | S | S | S | NS | |
| Cycle 10 | S | S | S | S | S | S | S | NS | NS |
| Trait 2 Continuous, = 0.3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| p-Value = 8.46 × 10−22 from the Kruskal–Wallis Test | |||||||||
| p-Values from the Mann–Whitney U Test Using the Bonferroni Correction | |||||||||
| Cycle 1 | Cycle 2 | Cycle 3 | Cycle 4 | Cycle 5 | Cycle 6 | Cycle 7 | Cycle 8 | Cycle 9 | |
| Cycle 2 | NS | ||||||||
| Cycle 3 | NS | NS | |||||||
| Cycle 4 | S | NS | NS | ||||||
| Cycle 5 | S | NS | NS | NS | |||||
| Cycle 6 | S | S | NS | NS | NS | ||||
| Cycle 7 | S | S | S | NS | NS | NS | |||
| Cycle 8 | S | S | S | S | S | NS | NS | ||
| Cycle 9 | S | S | S | S | S | S | NS | NS | |
| Cycle 10 | S | S | S | S | S | S | NS | NS | NS |
| Cycle 10 | NS | NS | S | NS | S | S | S | NS | NS |
| Trait 3 Category 3, = 0.3 | |||||||||
| p-value = 1.79 × 10−34 from the Kruskal–Wallis test | |||||||||
| p-values from the Mann–Whitney U test using the Bonferroni correction | |||||||||
| Cycle 1 | Cycle 2 | Cycle 3 | Cycle 4 | Cycle 5 | Cycle 6 | Cycle 7 | Cycle 8 | Cycle 9 | |
| Cycle 2 | NS | ||||||||
| Cycle 3 | S | NS | |||||||
| Cycle 4 | S | S | NS | ||||||
| Cycle 5 | S | S | S | NS | |||||
| Cycle 6 | S | S | S | S | NS | ||||
| Cycle 7 | S | S | S | S | S | NS | |||
| Cycle 8 | S | S | S | S | S | S | NS | ||
| Cycle 9 | S | S | S | S | S | S | S | NS | |
| Cycle 10 | S | S | S | S | S | S | S | S | NS |
| Trait 2 Continuous, = 0.6 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| p-Value = 8.46 × 10−22 from the Kruskal–Wallis Test | |||||||||
| p-Values from the Mann–Whitney U Test Using the Bonferroni Correction | |||||||||
| Cycle 1 | Cycle 2 | Cycle 3 | Cycle 4 | Cycle 5 | Cycle 6 | Cycle 7 | Cycle 8 | Cycle 9 | |
| Cycle 2 | NS | ||||||||
| Cycle 3 | S | NS | |||||||
| Cycle 4 | S | NS | NS | ||||||
| Cycle 5 | S | S | NS | NS | |||||
| Cycle 6 | S | S | S | NS | NS | ||||
| Cycle 7 | S | S | S | NS | NS | NS | |||
| Cycle 8 | S | S | S | S | NS | NS | NS | ||
| Cycle 9 | S | S | S | S | S | NS | NS | NS | |
| Cycle 10 | S | S | S | S | S | S | NS | NS | NS |
| p-value = 1.79 × 10−34 from the Kruskal–Wallis test | |||||||||
| p-values from the Mann–Whitney U test using the Bonferroni correction | |||||||||
| Cycle 1 | Cycle 2 | Cycle 3 | Cycle 4 | Cycle 5 | Cycle 6 | Cycle 7 | Cycle 8 | Cycle 9 | |
| Cycle 2 | S | ||||||||
| Cycle 3 | S | S | |||||||
| Cycle 4 | S | S | S | ||||||
| Cycle 5 | S | S | S | S | |||||
| Cycle 6 | S | S | S | S | NS | ||||
| Cycle 7 | S | S | S | S | S | NS | |||
| Cycle 8 | S | S | S | S | S | S | NS | ||
| Cycle 9 | S | S | S | S | S | S | S | NS | |
| Cycle 10 | S | S | S | S | S | S | S | S | NS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villar-Hernández, B.d.J.; Pérez-Rodríguez, P.; Vitale, P.; Gerard, G.; Montesinos-Lopez, O.A.; Saint Pierre, C.; Crossa, J.; Dreisigacker, S. Optimizing Genomic Parental Selection for Categorical and Continuous–Categorical Multi-Trait Mixtures. Genes 2024, 15, 995. https://doi.org/10.3390/genes15080995
Villar-Hernández BdJ, Pérez-Rodríguez P, Vitale P, Gerard G, Montesinos-Lopez OA, Saint Pierre C, Crossa J, Dreisigacker S. Optimizing Genomic Parental Selection for Categorical and Continuous–Categorical Multi-Trait Mixtures. Genes. 2024; 15(8):995. https://doi.org/10.3390/genes15080995
Chicago/Turabian StyleVillar-Hernández, Bartolo de Jesús, Paulino Pérez-Rodríguez, Paolo Vitale, Guillermo Gerard, Osval A. Montesinos-Lopez, Carolina Saint Pierre, José Crossa, and Susanne Dreisigacker. 2024. "Optimizing Genomic Parental Selection for Categorical and Continuous–Categorical Multi-Trait Mixtures" Genes 15, no. 8: 995. https://doi.org/10.3390/genes15080995
APA StyleVillar-Hernández, B. d. J., Pérez-Rodríguez, P., Vitale, P., Gerard, G., Montesinos-Lopez, O. A., Saint Pierre, C., Crossa, J., & Dreisigacker, S. (2024). Optimizing Genomic Parental Selection for Categorical and Continuous–Categorical Multi-Trait Mixtures. Genes, 15(8), 995. https://doi.org/10.3390/genes15080995






