Splice Variant of Spalax Heparanase Skipping Exon 12
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. RNA and cDNA Preparation
2.3. Cloning of Spalax Heparanase Splice Variants
2.4. Cells and Transfections
2.5. Western Blot Analysis
2.6. Heparanase Activity
3. Results
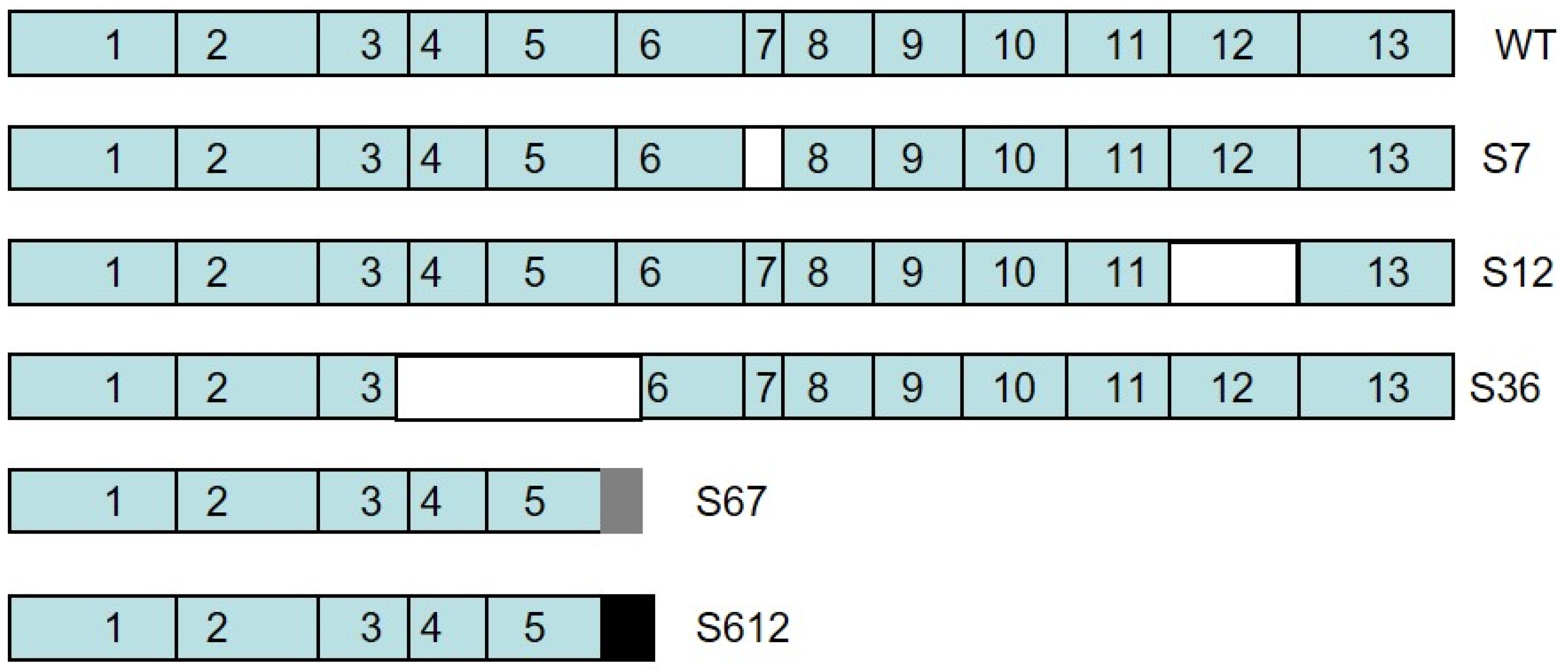
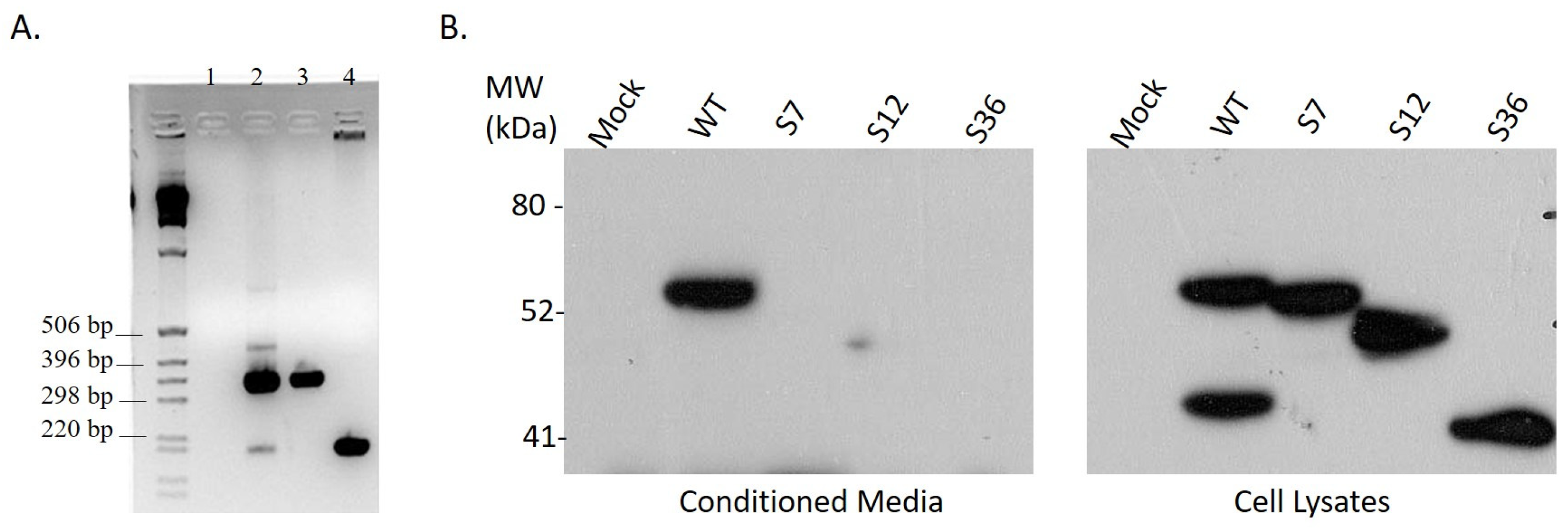
3.1. Cloning of Splice Variant #12 of Spalax Heparanase
3.2. Expression of Spalax Heparanase Splice Variants in U87 Cells
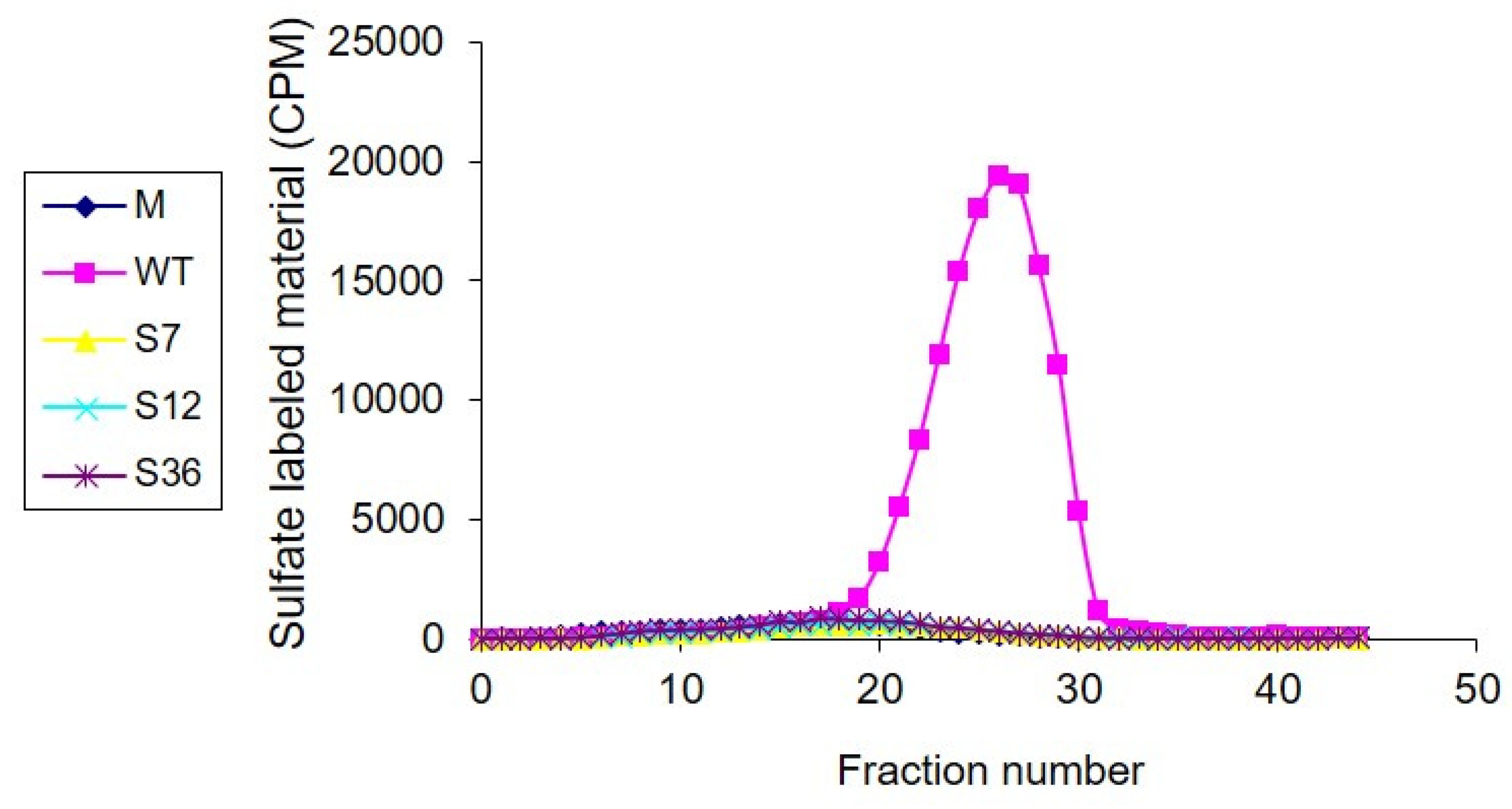
3.3. Heparanase Enzymatic Activity
4. Discussion
5. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fang, X.; Nevo, E.; Han, L.; Levanon, E.Y.; Zhao, J.; Avivi, A.; Larkin, D.; Jiang, X.; Feranchuk, S.; Zhu, Y.; et al. Genome-wide adaptive complexes to underground stresses in blind mole rats Spalax. Nat. Commun. 2014, 5, 3966. [Google Scholar] [CrossRef] [PubMed]
- Nevo, E. Mosaic Evolution of Subterranean Mammals: Regression, Progression, and Global Convergence; Oxford University Press: Oxford, UK, 1999. [Google Scholar]
- Avivi, A.; Albrecht, U.; Oster, H.; Joel, A.; Beiles, A.; Nevo, E. Biological clock in total darkness: The Clock/MOP3 circadian system of the blind subterranean mole rat. Proc. Natl. Acad. Sci. USA 2001, 98, 13751–13756. [Google Scholar] [CrossRef] [PubMed]
- Avivi, A.; Shams, I.; Joel, A.; Lache, O.; Levy, A.P.; Nevo, E. Increased blood vessel density provides the mole rat physiological tolerance to its hypoxic subterranean habitat. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2005, 19, 1314–1316. [Google Scholar] [CrossRef] [PubMed]
- Kimchi, T.; Terkel, J. Magnetic compass orientation in the blind mole rat Spalax ehrenbergi. J. Exp. Biol. 2001, 204, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Malewski, S.; Begall, S.; Schleich, C.E.; Antenucci, C.D.; Burda, H. Do subterranean mammals use the Earth’s magnetic field as a heading indicator to dig straight tunnels? PeerJ 2018, 6, e5819. [Google Scholar] [CrossRef] [PubMed]
- Nevo, E.; Heth, G.; Pratt, H. Seismic communication in a blind subterranean mammal: A major somatosensory mechanism in adaptive evolution underground. Proc. Natl. Acad. Sci. USA 1991, 88, 1256–1260. [Google Scholar] [CrossRef] [PubMed]
- Adwan Shekhidem, H.; Sharvit, L.; Huffman, D.M.; Manov, I.; Atzmon, G.; Shams, I. Damage-Free Shortening of Telomeres Is a Potential Strategy Supporting Blind Mole-Rat Longevity. Genes 2023, 14, 845. [Google Scholar] [CrossRef] [PubMed]
- Nevo, E.; Ivanitskaia, E.; Beiles, A. Adaptive Radiation of Blind Subterranean Mole Rats; Backhuys Publishers: Oegstgeest, The Netherlands, 2001. [Google Scholar]
- Widmer, H.R.; Hoppeler, H.; Nevo, E.; Taylor, C.R.; Weibel, E.R. Working underground: Respiratory adaptations in the blind mole rat. Proc. Natl. Acad. Sci. USA 1997, 94, 2062–2067. [Google Scholar] [CrossRef] [PubMed]
- Edoute, Y.; Arieli, R.; Nevo, E. Evidence for improved myocardial oxygen delivery and function during hypoxia in the mole rat. J. Comp. Physiol. B 1988, 158, 575–582. [Google Scholar] [CrossRef]
- Nasser, N.J.; Kaplan, M.; Nevo, E.; Aviram, M. Lipid profile and serum characteristics of the blind subterranean mole rat, Spalax. PLoS ONE 2009, 4, e4528. [Google Scholar] [CrossRef]
- Sandwall, E.; Bodevin, S.; Nasser, N.J.; Nevo, E.; Avivi, A.; Vlodavsky, I.; Li, J.P. Molecular structure of heparan sulfate from Spalax. Implications of heparanase and hypoxia. J. Biol. Chem. 2009, 284, 3814–3822. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xiong, M.; Chen, Z.; Seabra, G.; Liu, J.; Li, C.; Cui, L. Design Principle of Heparanase Inhibitors: A Combined In Vitro and In Silico Study. ACS Med. Chem. Lett. 2024, 15, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Nasser, N.J. Heparanase involvement in physiology and disease. Cell Mol. Life Sci. 2008, 65, 1706–1715. [Google Scholar] [CrossRef] [PubMed]
- Nasser, N.J.; Fox, J.; Agbarya, A. Potential Mechanisms of Cancer-Related Hypercoagulability. Cancers 2020, 12, 566. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Friedmann, Y.; Elkin, M.; Aingorn, H.; Atzmon, R.; Ishai-Michaeli, R.; Bitan, M.; Pappo, O.; Peretz, T.; Michal, I.; et al. Mammalian heparanase: Gene cloning, expression and function in tumor progression and metastasis. Nat. Med. 1999, 5, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Jayatilleke, K.M.; Hulett, M.D. Heparanase and the hallmarks of cancer. J. Transl. Med. 2020, 18, 453. [Google Scholar] [CrossRef] [PubMed]
- Koganti, R.; Suryawanshi, R.; Shukla, D. Heparanase, cell signaling, and viral infections. Cell. Mol. Life Sci. 2020, 77, 5059–5077. [Google Scholar] [CrossRef] [PubMed]
- Mayfosh, A.J.; Nguyen, T.K.; Hulett, M.D. The heparanase regulatory network in health and disease. Int. J. Mol. Sci. 2021, 22, 11096. [Google Scholar] [CrossRef]
- Lerner, I.; Baraz, L.; Pikarsky, E.; Meirovitz, A.; Edovitsky, E.; Peretz, T.; Vlodavsky, I.; Elkin, M. Function of heparanase in prostate tumorigenesis: Potential for therapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 668–676. [Google Scholar] [CrossRef]
- Nasser, N.J.; Klein, J.; Agbarya, A. Markers of Toxicity and Response to Radiation Therapy in Patients With Prostate Cancer. Adv. Radiat. Oncol. 2021, 6, 100603. [Google Scholar] [CrossRef]
- Zhang, Q.; Ming, J.; Li, Y.; Zhang, S.; Li, B.; Qiu, X.; Wang, E. Heparanase expression correlates with angiogenesis and lymphangiogenesis in human lung cancer. Chin. J. Lung Cancer 2009, 12, 864–867. [Google Scholar]
- Vornicova, O.; Naroditsky, I.; Boyango, I.; Shachar, S.S.; Mashiach, T.; Ilan, N.; Vlodavsky, I.; Bar-Sela, G. Prognostic significance of heparanase expression in primary and metastatic breast carcinoma. Oncotarget 2018, 9, 6238–6244. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Friedmann, Y.; Vlodavsky, I.; Aingorn, H.; Aviv, A.; Peretz, T.; Pecker, I.; Pappo, O. Expression of heparanase in normal, dysplastic, and neoplastic human colonic mucosa and stroma. Evidence for its role in colonic tumorigenesis. Am. J. Pathol. 2000, 157, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Mikami, S.; Oya, M.; Shimoda, M.; Mizuno, R.; Ishida, M.; Kosaka, T.; Mukai, M.; Nakajima, M.; Okada, Y. Expression of heparanase in renal cell carcinomas: Implications for tumor invasion and prognosis. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 6055–6061. [Google Scholar] [CrossRef] [PubMed][Green Version]
- McKenzie, E.; Tyson, K.; Stamps, A.; Smith, P.; Turner, P.; Barry, R.; Hircock, M.; Patel, S.; Barry, E.; Stubberfield, C. Cloning and expression profiling of Hpa2, a novel mammalian heparanase family member. Biochem. Biophys. Res. Commun. 2000, 276, 1170–1177. [Google Scholar] [CrossRef] [PubMed]
- Gross-Cohen, M.; Yanku, Y.; Kessler, O.; Barash, U.; Boyango, I.; Cid-Arregui, A.; Neufeld, G.; Ilan, N.; Vlodavsky, I. Heparanase 2 (Hpa2) attenuates tumor growth by inducing Sox2 expression. Matrix Biol. J. Int. Soc. Matrix Biol. 2021, 99, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Hilwi, M.; Kayal, Y.; Soboh, S.; Ilan, N. Impact of heparanase-2 (Hpa2) on cancer and inflammation: Advances and paradigms. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2024, 38, e23670. [Google Scholar] [CrossRef] [PubMed]
- Kayal, Y.; Barash, U.; Naroditsky, I.; Ilan, N.; Vlodavsky, I. Heparanase 2 (Hpa2)- a new player essential for pancreatic acinar cell differentiation. Cell Death Dis. 2023, 14, 465. [Google Scholar] [CrossRef] [PubMed]
- Lopes, F.M.; Grenier, C.; Jarvis, B.W.; Al Mahdy, S.; Lène-McKay, A.; Gurney, A.M.; Newman, W.G.; Waddington, S.N.; Woolf, A.S.; Roberts, N.A. Human HPSE2 gene transfer ameliorates bladder pathophysiology in a mutant mouse model of urofacial syndrome. eLife 2024, 13, RP91828. [Google Scholar] [CrossRef]
- Daly, S.B.; Urquhart, J.E.; Hilton, E.; McKenzie, E.A.; Kammerer, R.A.; Lewis, M.; Kerr, B.; Stuart, H.; Donnai, D.; Long, D.A.; et al. Mutations in HPSE2 cause urofacial syndrome. Am. J. Hum. Genet. 2010, 86, 963–969. [Google Scholar] [CrossRef]
- Nasser, N.J.; Nevo, E.; Shafat, I.; Ilan, N.; Vlodavsky, I.; Avivi, A. Adaptive evolution of heparanase in hypoxia-tolerant Spalax: Gene cloning and identification of a unique splice variant. Proc. Natl. Acad. Sci. USA 2005, 102, 15161–15166. [Google Scholar] [CrossRef] [PubMed]
- Nasser, N.J.; Avivi, A.; Shafat, I.; Edovitsky, E.; Zcharia, E.; Ilan, N.; Vlodavsky, I.; Nevo, E. Alternatively spliced Spalax heparanase inhibits extracellular matrix degradation, tumor growth, and metastasis. Proc. Natl. Acad. Sci. USA 2009, 106, 2253–2258. [Google Scholar] [CrossRef] [PubMed]
- Nasser, N.J.; Avivi, A.; Vlodavsky, I.; Nevo, E. Cloning of two splice variants of Spalax heparanase encoding for truncated proteins. Anti-Cancer Drugs 2020, 31, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Nasser, N.J.; Avivi, A.; Shushy, M.; Vlodavsky, I.; Nevo, E. Cloning, expression, and characterization of an alternatively spliced variant of human heparanase. Biochem. Biophys. Res. Commun. 2007, 354, 33–38. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Weissmann, M.; Arvatz, G.; Horowitz, N.; Feld, S.; Naroditsky, I.; Zhang, Y.; Ng, M.; Hammond, E.; Nevo, E.; Vlodavsky, I.; et al. Heparanase-neutralizing antibodies attenuate lymphoma tumor growth and metastasis. Proc. Natl. Acad. Sci. USA 2016, 113, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Zetser, A.; Levy-Adam, F.; Kaplan, V.; Gingis-Velitski, S.; Bashenko, Y.; Schubert, S.; Flugelman, M.Y.; Vlodavsky, I.; Ilan, N. Processing and activation of latent heparanase occurs in lysosomes. J. Cell Sci. 2004, 117, 2249–2258. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Kukula, A.K.; Toyoshima, M.; Nakajima, M. Genomic organization and chromosome localization of the newly identified human heparanase gene. Gene 2000, 253, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Cassinelli, G.; Torri, G.; Naggi, A. Non-anticoagulant heparins as heparanase inhibitors. In Heparanase: From Basic Research to Clinical Applications; Springer: Cham, Switzerland, 2020; pp. 493–522. [Google Scholar]
- de Boer, C.; Armstrong, Z.; Lit, V.A.; Barash, U.; Ruijgrok, G.; Boyango, I.; Weitzenberg, M.M.; Schröder, S.P.; Sarris, A.J.; Meeuwenoord, N.J. Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo. Proc. Natl. Acad. Sci. USA 2022, 119, e2203167119. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wei, Q.; Guo, C.; Dong, G.; Liu, Y.; Tang, C.; Dong, Z. Hypoxia, HIF, and associated signaling networks in chronic kidney disease. Int. J. Mol. Sci. 2017, 18, 950. [Google Scholar] [CrossRef]
- Kanbay, M.; Altıntas, A.; Yavuz, F.; Copur, S.; Sanchez-Lozada, L.G.; Lanaspa, M.A.; Johnson, R.J. Responses to Hypoxia: How Fructose Metabolism and Hypoxia-Inducible Factor-1a Pathways Converge in Health and Disease. Curr. Nutr. Rep. 2023, 12, 181–190. [Google Scholar] [CrossRef]
- Arieli, R.; Nevo, E. Hypoxic survival differs between two mole rat species (Spalax ehrenbergi) of humid and arid habitats. Comp. Biochem. Physiol. A Comp. Physiol. 1991, 100, 543–545. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Ye, K.; Liu, D.; Pan, D.; Gu, S.; Wang, Z. Evolution of hemoglobin genes in a subterranean rodent species (Lasiopodomys mandarinus). Biology 2020, 9, 106. [Google Scholar] [CrossRef] [PubMed]
- Šumbera, R.; Lovy, M.; Nevo, E.; Okrouhlík, J. Thermal Biology in the Upper Galili Mountain Blind Mole Rat (Nannospalax galili) and an Overview of Spalacine Energetics. J. Therm. Biol. 2023, 115, 103618. [Google Scholar] [CrossRef] [PubMed]
- Izraelson, M.; Metsger, M.; Davydov, A.; Shagina, I.; Dronina, M.; Obraztsova, A.; Miskevich, D.; Mamedov, I.; Volchkova, L.; Nakonechnaya, T. Distinct organization of adaptive immunity in the long-lived rodent Spalax galili. Nat. Aging 2021, 1, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Oltean, S.; Bates, D.O. Hallmarks of alternative splicing in cancer. Oncogene 2014, 33, 5311–5318. [Google Scholar] [CrossRef] [PubMed]
- Reixachs-Solé, M.; Eyras, E. Uncovering the impacts of alternative splicing on the proteome with current omics techniques. Wiley Interdiscip. Rev. RNA 2022, 13, e1707. [Google Scholar] [CrossRef] [PubMed]
- Fux, L.; Ilan, N.; Sanderson, R.D.; Vlodavsky, I. Heparanase: Busy at the cell surface. Trends Biochem. Sci. 2009, 34, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Lai, N.S.; Simizu, S.; Morisaki, D.; Muroi, M.; Osada, H. Requirement of the conserved, hydrophobic C-terminus region for the activation of heparanase. Exp. Cell Res. 2008, 314, 2834–2845. [Google Scholar] [CrossRef] [PubMed]
- Levy-Adam, F.; Abboud-Jarrous, G.; Guerrini, M.; Beccati, D.; Vlodavsky, I.; Ilan, N. Identification and characterization of heparin/heparan sulfate binding domains of the endoglycosidase heparanase. J. Biol. Chem. 2005, 280, 20457–20466. [Google Scholar] [CrossRef]
- Levy-Adam, F.; Miao, H.-Q.; Heinrikson, R.L.; Vlodavsky, I.; Ilan, N. Heterodimer formation is essential for heparanase enzymatic activity. Biochem. Biophys. Res. Commun. 2003, 308, 885–891. [Google Scholar] [CrossRef]
- Fux, L.; Feibish, N.; Cohen-Kaplan, V.; Gingis-Velitski, S.; Feld, S.; Geffen, C.; Vlodavsky, I.; Ilan, N. Structure-function approach identifies a COOH-terminal domain that mediates heparanase signaling. Cancer Res. 2009, 69, 1758–1767. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yuan, F.; Zhou, H.; Quan, J.; Liu, C.; Wang, Y.; Xiao, F.; Liu, Q.; Liu, J.; Zhang, Y.; et al. Potential roles of heparanase in cancer therapy: Current trends and future direction. J. Cell. Physiol. 2023, 238, 896–917. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasser, N.J.; Nevo, E.; Avivi, A. Splice Variant of Spalax Heparanase Skipping Exon 12. Genes 2024, 15, 1039. https://doi.org/10.3390/genes15081039
Nasser NJ, Nevo E, Avivi A. Splice Variant of Spalax Heparanase Skipping Exon 12. Genes. 2024; 15(8):1039. https://doi.org/10.3390/genes15081039
Chicago/Turabian StyleNasser, Nicola J., Eviatar Nevo, and Aaron Avivi. 2024. "Splice Variant of Spalax Heparanase Skipping Exon 12" Genes 15, no. 8: 1039. https://doi.org/10.3390/genes15081039
APA StyleNasser, N. J., Nevo, E., & Avivi, A. (2024). Splice Variant of Spalax Heparanase Skipping Exon 12. Genes, 15(8), 1039. https://doi.org/10.3390/genes15081039





