Hepatopancreas Transcriptome Analysis of Spinibarbus sinensis to Reveal Different Growth-Related Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement, Experimental Animals, and Sample Collection
2.2. Date Measurement and Sample Collection
2.3. RNA Isolation, Transcriptome Library Construction, and Sequencing
2.4. Data Processing, Assembly, and Functional Annotation
2.5. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis of DEGs
2.6. Validation of the RNA-seq Analysis by RT-qPCR
2.7. Statistical Analysis
3. Results
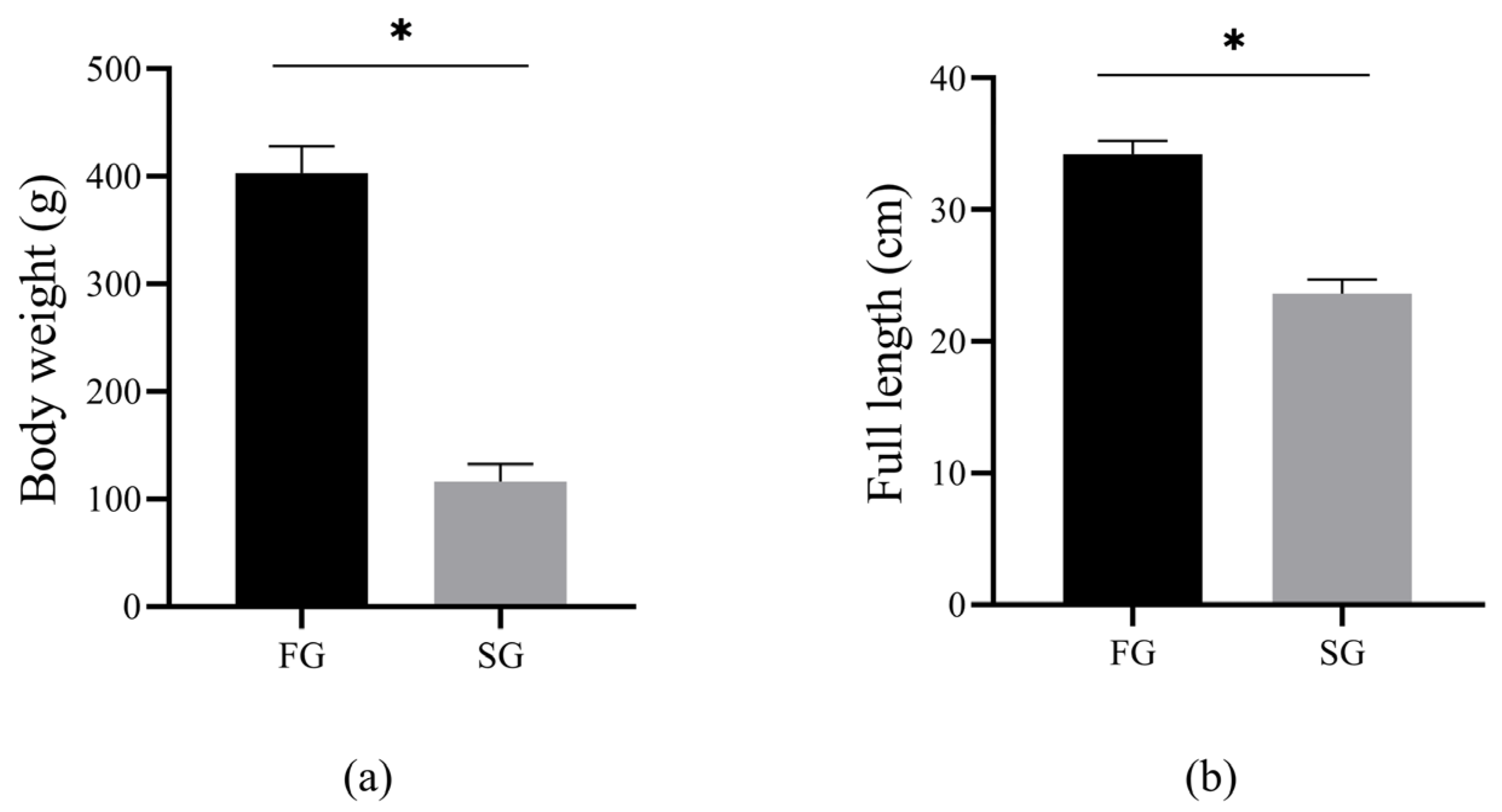
3.1. Analysis of Growth Data between FG and SG
3.2. Preliminary Analysis of the Transcriptome Sequences
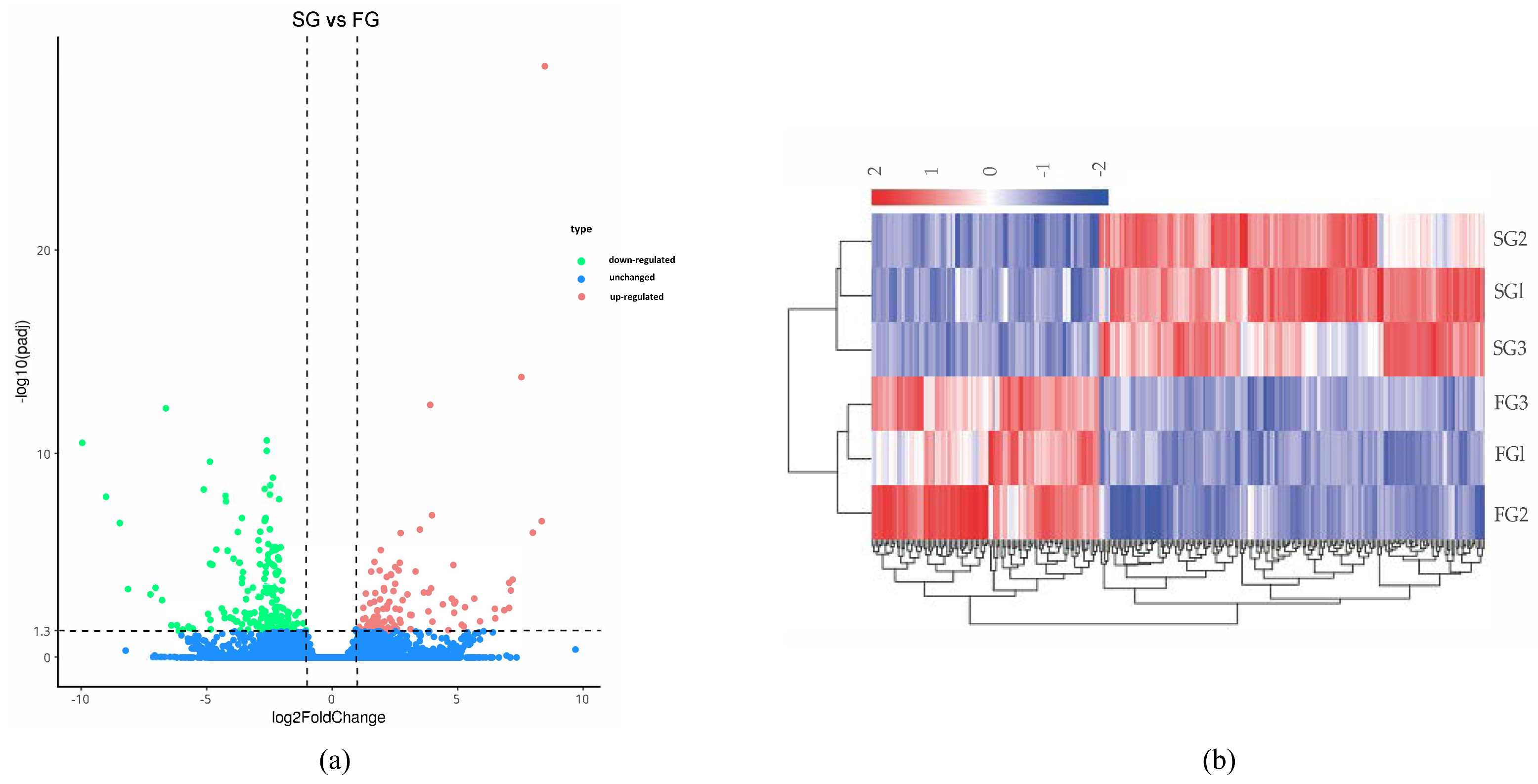
3.3. DEGs Analysis
3.4. Enrichment and Pathway Analysis of GO and KEGG
3.5. Verification of DEGs with qRT-PCR
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cai, Y.Z.; He, C.R.; Cai, Y.Q.; Jiang, J. Preliminary study on biology of Spinibarbus sinensis. Freshw. Fish. 2003, 33, 16–18. [Google Scholar]
- Li, P.; Wang, B.S. Spinibarbus sinensis. Bull. Biol. 2008, 43, 17–18. [Google Scholar]
- Chen, Z.Z.; Zhou, Z.M.; Huang, X.H.; Hu, J.J. Research advances of Spinibarbus sinensis Bleeker aquaculture technology. J. Anhui Agric. Sci. 2014, 42, 7043–7046. [Google Scholar]
- Xu, J.J.; Fu, S.J.; Fu, C. Physiological and behavioral stress responses to predators are Altered by prior predator experience in juvenile Qingbo (Spinibarbus sinensis). Biol. Open 2019, 8, bio041012. [Google Scholar] [CrossRef]
- Wang, L.; Tang, J.Y.; Qin, Y.L.; Zeng, L.Q.; Peng, J.L.; Fu, S.J. Effect of starvation on energy metabolism, fish behavior, and schooling behavior of Spinibarbus sinensis. Acta Ecol. Sin. 2019, 39, 1095–1104. [Google Scholar]
- Qiu, H.X.; Liu, W.M.; Yan, Y.L.; Long, J.; Xie, X.J. Effects of waterborne cadmium exposure on Spinibarbus sinensis hepatopancreas and kidney: Mitochondrial cadmium accumulation and respiratory metabolism. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2021, 248, 109115. [Google Scholar] [CrossRef]
- Tang, Z.H.; Fu, S.J. Qingbo (Spinibarbus sinensis) personalities and their effect on shoaling behavior. Acta Ethologica 2019, 22, 135–144. [Google Scholar] [CrossRef]
- Yang, C.G.; Wen, H.; Jiang, M. The complete mitochondrial genome of Spinibarbus sinensis (Bleeker). Mitochondrial DNA 2015, 26, 276–277. [Google Scholar] [CrossRef]
- Wu, P.; Yang, W.G.; Dong, Y.Y.; Wang, Y.L.; Zhang, Y.; Zou, X.J.; Ge, H.; Hu, D.X.; Cui, Y.B.; Chen, Z.B. Feasibility of cultivation of Spinibarbus sinensis with coconut oil and its effect on disease resistance (nonspecific immunity, antioxidation and mTOR and NF-kB signaling pathways). Fish Shellfish Immunol. 2019, 93, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.H.; Xu, W.F.; Yuan, J.J. Effects of immunopolysaccharide on nonspecific immune function and disease resistance of Spinibarbus sinensis. Jiangsu Agric. Sci. 2013, 41, 184–188. [Google Scholar]
- Li, Z.; Du, X.S.; Wen, L.T.; Li, Y.; Qin, J.Q.; Chen, Z.; Huang, Y.; Wu, X.; Luo, H.; Lin, Y.; et al. Transcriptome analysis reveals the involvement of ubiquitin-proteasome pathway in the regulation of muscle growth of rice flower carp. Comp. Biochem. Physiol. Part D Genom. Proteom. 2022, 41, 100948. [Google Scholar] [CrossRef]
- Zhang, J.H.; Shen, Y.B.; Xu, X.Y.; Dai, Y.F.; Li, J.L. Transcriptome analysis of the liver and muscle tissues of black carp (Mylopharyngodon Piceus) of different growth rates. Mar. Biotechnol. 2020, 22, 706–716. [Google Scholar] [CrossRef]
- Liu, X.G.; Zeng, S.; Liu, S.; Wang, G.P.; Lai, H.; Zhao, X.P.; Bi, S.; Guo, D.L.; Chen, X.L.; Yi, H.D.; et al. Identifying the related genes of muscle growth and exploring the functions by compensatory growth in mandarin fish (Siniperca chuatsi). Front. Physiol. 2020, 11, 553563. [Google Scholar] [CrossRef]
- Wang, W.; Wu, X.G.; Liu, Z.J.; Zheng, H.J.; Cheng, Y.X. Insights into hepatopancreatic functions for nutrition metabolism and ovarian development in the Crab Portunus trituberculatus: Gene discovery in the comparative transcriptome of different hepatopancreas stages. PLoS ONE 2014, 9, e84921. [Google Scholar] [CrossRef]
- Huang, S.; Wang, J.; Yue, W.C.; Chen, J.; Gaughan, S.; Lu, W.Q.; Lu, G.Q.; Wang, C.H. Transcriptomic variation of hepatopancreas reveals the energy metabolism and biological processes associated with molting in Chinese mitten crab, Eriocheir sinensis. Sci. Rep. 2015, 5, 14015. [Google Scholar] [CrossRef]
- Fang, F.; Yuan, Y.; Jin, M.; Shi, B.; Zhu, T.T.; Luo, J.X.; Lu, J.J.; Wang, X.X.; Jiao, L.F.; Zhou, Q.C. Hepatopancreas transcriptome analysis reveals the molecular responses to different dietary n-3 PUFA lipid sources in the swimming crab Portunus trituberculatus. Aquaculture 2021, 543, 737016. [Google Scholar] [CrossRef]
- Jin, H.H.; Li, Y.; Xiao, C.B.; Sun, W.B.; Liu, F.; Ke, Z.L.; Zhao, S.F.; Qin, F.; Lei, K.; Wu, J.Q.; et al. Transcriptome analysis reveals the effects of dietary protein level on growth performance and metabolism in adult procambarus clarkii farming in rice field. Aquac. Rep. 2024, 35, 101949. [Google Scholar] [CrossRef]
- Ye, H.; Xiao, S.J.; Wang, X.Q.; Wang, Z.Y.; Zhang, Z.S.; Zhu, C.K.; Hu, B.J.; Lv, C.H.; Zheng, S.M.; Luo, H. Characterization of spleen transcriptome of Schizothorax prenanti during Aeromonas hydrophila infection. Mar. Biotechnol. 2018, 20, 246–256. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, X.L.; Wei, X.K.; Li, Y.; Liu, W.; Gan, G.C.; Xiao, L.L.; Wang, X.Y.; Luo, H. Gonadal transcriptome analysis of paradise fish Macropodus opercularis to reveal sex-related genes. Comp. Biochem. Physiol. Part D Genom. Proteom. 2023, 48, 101125. [Google Scholar] [CrossRef]
- Luo, H.; Liu, H.P.; Zhang, J.; Hu, B.J.; Zhou, C.W.; Xiang, M.B.; Yang, Y.J.; Zhou, M.R.; Jing, T.S.; Li, Z.; et al. Full-length transcript sequencing accelerates the transcriptome research of Gymnocypris namensis, an iconic fish of the Tibetan Plateau. Sci. Rep. 2020, 10, 9668. [Google Scholar] [CrossRef]
- Jiang, S.F.; Zhang, W.Y.; Xiong, Y.W.; Cheng, D.; Wang, J.S.; Jin, S.B.; Gong, Y.S.; Wu, Y.; Qiao, H.; Fu, H.T. Hepatopancreas transcriptome analyses provide new insights into the molecular regulatory mechanism of fast ovary maturation in Macrobrachium nipponense. BMC Genom. 2022, 23, 625. [Google Scholar] [CrossRef] [PubMed]
- Ahmadifar, E.; Pourmohammadi Fallah, H.; Yousefi, M.; Dawood, M.A.O.; Hoseinifar, S.H.; Adineh, H.; Yilmaz, S.; Paolucci, M.; Doan, H.V. The gene regulatory roles of herbal extracts on the growth, immune system, and reproduction of fish. Animals 2021, 11, 2167. [Google Scholar] [CrossRef]
- Assan, D.; Huang, Y.L.; Mustapha, U.F.; Addah, M.N.; Li, G.L.P.; Chen, H. Fish feed intake, feeding behavior, and the physiological response of apelin to fasting and refeeding. Front. Endocrinol. 2021, 12, 798903. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, H.R.; Clark, T.D. Why do some fish grow faster than others? Fish Fish. 2023, 24, 796–811. [Google Scholar] [CrossRef]
- Qi, M.; Wu, Q.Q.; Liu, T.; Hou, Y.L.; Miao, Y.X.; Hu, M.H.; Liu, Q.G. Hepatopancreas transcriptome profiling analysis reveals physiological responses to acute hypoxia and reoxygenation in juvenile Qingtian paddy field carp Cyprinus carpio var qingtianensis. Front. Physiol. 2020, 11, 1110. [Google Scholar] [CrossRef] [PubMed]
- Vogt, G. Life-cycle and functional cytology of the hepatopancreatic cells of Astacus astacus (Crustacea, Decapoda). Zoomorphology 1994, 114, 83–101. [Google Scholar] [CrossRef]
- Wang, J.; Xue, M.; Wu, X.F.; Han, F.; Zheng, Y. Regulation mechanism of selective feed intake of fish when fed different protein source diets: A review. Chin. J. Anim. Nutr. 2014, 26, 833–842. [Google Scholar]
- Zhan, Q.; Wang, J.; Han, T. Research progress of fish feeding regulation. Feed. Ind. 2020, 41, 41–47. [Google Scholar]
- Crawley, J.N.; Beinfeld, M.C. Rapid development of tolerance to the behavioural actions of cholecystokinin. Nature 1983, 302, 703–706. [Google Scholar] [CrossRef]
- Xu, S.H. The Mechanisms of Cholecystokinin-Regulated Feeding and Reproduction in Grass Carp. Master’s Thesis, Huazhong Agriculture University, Wuhan, China, 2021. [Google Scholar]
- Nelson, L.E.; Sheridan, M.A. Regulation of somatostatins and their receptors in fish. Gen. Comp. Endocrinol. 2005, 142, 117–133. [Google Scholar] [CrossRef]
- Fabián, C.L.; Chang, J.P.; Richard, P.E. Neuroendocrine control of growth hormone in fish. Gen. Comp. Endocrinol. 2007, 151, 1–26. [Google Scholar]
- Özel, O.T.; Öztürk, E.; Garipoğlu, A.V. Lipid Metabolism in Fish. Aquac. Stud. 2017, 17, 303–317. [Google Scholar]
- Li, Y.; Zou, X.X.; Jin, H.H.; Zhou, B.; Zhou, J.; Zhang, L.; Li, Z.; Ling, L.Y.; Liu, F.; Gao, Y.; et al. Identification of genes related to growth from transcriptome profiles of the muscle and liver of Chinese longsnout catfish (Leiocassis longirostris). Comp. Biochem. Physiol. Part D Genom. Proteom. 2024, 49, 101180. [Google Scholar] [CrossRef] [PubMed]
- Masset, T.; Ferrari, B.J.D.; Dudefoi, W.; Schirmer, K.; Bergmann, A.; Vermeirssen, E.; Grandjean, D.; Harris, L.C.; Breider, F. Bioaccessibility of organic compounds associated with tire particles using a fish in vitro digestive model: Solubilization kinetics and effects of food coingestion. Environ. Sci. Technol. 2022, 56, 15607–15616. [Google Scholar] [CrossRef] [PubMed]
- Parlapani, F.F.; Anagnostopoulos, D.A.; Karamani, E.; Mallouchos, A.; Haroutounian, S.A.; Boziaris, I.S. Growth and volatile organic compound production of pseudomonas fish spoiler strains on fish juice agar model substrate at different temperatures. Microorganisms 2023, 11, 189. [Google Scholar] [CrossRef]
- Tinoco, A.B.; Näslund, J.; Delgado, M.J.; Pedro, N.D.; Johnsson, J.I.; Jönsson, E. Ghrelin increases food intake, swimming activity and growth in juvenile brown trout (Salmo trutta). Physiol. Behav. 2014, 124, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Frühbeck, G.; Catalán, V.; Valentí, V.; Moncada, R.; Gómez-Ambrosi, J.; Becerril, S.; Silva, C.; Portincasa, P.; Escalada, J.; Rodríguez, A. FNDC4 and FNDC5 reduce SARS-CoV-2 entry points and spike glycoprotein S1-induced pyroptosis, apoptosis, and necroptos is in human adipocytes. Cell. Mol. Immunol. 2021, 18, 2457–2459. [Google Scholar] [CrossRef]
- Li, J.W.; Wang, Y.S.; Wang, Y.; Yan, Y.Q.; Tong, H.L.; Li, S.F. Fibronectin type III domain containing four promotes differentiation of C2C12 through the Wnt/β-catenin signaling pathway. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 7759–7772. [Google Scholar] [CrossRef] [PubMed]
- Katsuhito, M.; Masanori, E.; Masaaki, I. Fetuin-A: A multifunctional protein. Recent Pat. Endocr. Metab. Immune Drug Discov. 2011, 5, 124–146. [Google Scholar]
- Shulman, G.I. Cellular mechanisms of insulin resistance. J. Clin. Investig. 2000, 106, 171–176. [Google Scholar] [CrossRef]
- Chen, T.; Bi, J.; Cao, C. Structure and biological functions of fetuin-A/α2-HS glycoprotein. Chin. J. Biochem. Mol. Biol. 2014, 30, 636–641. [Google Scholar]


| Gene ID | Annotation | Sequence |
|---|---|---|
| Cluster-30550.6193 | Serine/Threonine-Protein Kinase 31 (stk 31) | F:AATTGCCAATGATCTATCC R:TCCTCCCACTCAACTCTT |
| Cluster-30550.62137 | Probable G-Protein Coupled Receptor 149 (gpr 149) | F:GCATTCGCCACTGCCACTT R:GGTTCGCATCTCCGCTCCT |
| Cluster-35286.0 | Angiopoietin-related protein 1 (angptl 1) | F: TGGGAATCACAGACGAGA R: GGAGAAACGAGGCACATA |
| Cluster-30550.68321 | Follistatin-related protein 1 (fstl 1) | F: TGGCAGTAATGGGAAGACA R:CGGTCCGTAAGGTAACAAA |
| Cluster-30550.18115 | Serine/Threonine-Protein Kinase SIK1 (sik 1) | F: TTCAGGAGGGAAGGAGAT R:ATACGGCTTTGTGGATTG |
| Cluster-30550.104720 | Receptor Tyrosine Kinase Like Orphan Receptor 2 (ror 2) | F: ATCTGGCTGCTCGTAACATTCT R:TTGTAGTAGTCGGCTGAGTAGACC |
| Cluster-30550.59282 | NLR Family CARD Domain Containing 3 (nlrc 3) | F: CACGCTCTGCCTGTTCTTA R: TCCATTTCCACGCTGTTAT |
| Cluster-30550.58994 | PDZ And LIM Domain 2 (pdlim 2) | F:CGGACTCCAACTCCACCT R:GCACGGGATTCTTTGTTC |
| Cluster-30550.64357 | Neuron Navigator 2 (nav 2) | F:GTTACCTGGCATCCTCTG R: TCGCTTCCTCCATTTACT |
| Cluster-30550.65732 | Bone Morphogenetic Protein 1 (bmp 1) | F:AGAGGAGGCAGAGGACAG R: AAGAGCAGATTGCACCAG |
| Cluster-30550.15737 | G-Patch Domain Containing 8 (gpatch 8) | F:CACCGATGATGAGATTGAGA R: GAGAAGAGGGCTGGAGTATG |
| Cluster-30550.7707 | Somatostatin 2 (sst 2) | F:AAGGTCAGACAAGCCACA R:CCAAACAGAACAGGGAGA |
| Cluster-34620.0 | Kruppel-Like Factor 6 (klf 6) | F:GGTGTTCGTGGGATGGCTGTG R:TGGCTGCATTTGAAAGGTTTGG |
| Cluster-36962.2 | Cholecystokinin A Receptor (cckar) | F:TGCTGTAGGGATGATTTG R:CAGTTAGCCTTGTAAGTGTT |
| Cluster-30550.43338 | Syncoilin, Intermediate Filament Protein (sync 1) | F: CCCAGATGTTTGAGCATAGCA R:TACCCTTCCTTCTTGGATTTA |
| Cluster-30550.53986 | Basic Helix–Loop–Helix Family Member A15 (bhlha 15) | F:AGAGGCGTCTGTGAGGGTG R:AGGCGTTGTTGAGTTTGTG |
| Cluster-30550.15279 | Stanniocalcin 1 (stc 1) | F:GAGCCATTCTCGACACTA R: CCCAAGAAGCACTTACAG |
| Cluster-34288.0 | S100 Calcium-Binding Protein A1 (s100a1) | F: TGGTATTCCACCGTTATGC R: TCCTCAAAGTTCACCTCCC |
| Cluster-30550.58298 | β-actin (β-actin) | F:TTCTTGGGTATGGAGTCTTG R:AGGTCCTTACGGATGTCG |
| Sample | Raw Reads | Clean Reads | Clean Bases (%) | Error Rate | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|
| FG1 | 23,283,837 | 22,964,984 | 6.89 | 0.03 | 94.14 | 45.79 |
| FG2 | 22,613,144 | 22,367,822 | 6.71 | 0.03 | 94.03 | 46.32 |
| FG3 | 21,152,663 | 20,972,103 | 6.29 | 0.03 | 93.57 | 46.24 |
| SG1 | 22,624,801 | 22,377,351 | 6.71 | 0.03 | 94.05 | 46.09 |
| SG2 | 23,510,788 | 23,244,189 | 6.97 | 0.03 | 93.89 | 47.18 |
| SG3 | 23,310,396 | 23,118,045 | 6.94 | 0.02 | 94.22 | 46.04 |
| Assembly statistics (all the clean reads from the 6 libraries were assembled together) | ||||||
| Term | Unigene | |||||
| N50 | 1635 | |||||
| Total Length (bp) | 157,542,467 | |||||
| Max Length (bp) | 51,530 | |||||
| Min Length (bp) | 301 | |||||
| Average Length (bp) | 1050 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, B.; Ling, L.; Wang, B.; Yang, F.; Hou, M.; Liu, F.; Li, Y.; Luo, H.; He, W.; Ye, H. Hepatopancreas Transcriptome Analysis of Spinibarbus sinensis to Reveal Different Growth-Related Genes. Genes 2024, 15, 949. https://doi.org/10.3390/genes15070949
Zhou B, Ling L, Wang B, Yang F, Hou M, Liu F, Li Y, Luo H, He W, Ye H. Hepatopancreas Transcriptome Analysis of Spinibarbus sinensis to Reveal Different Growth-Related Genes. Genes. 2024; 15(7):949. https://doi.org/10.3390/genes15070949
Chicago/Turabian StyleZhou, Bo, Leyan Ling, Bin Wang, Fei Yang, Mengdan Hou, Fan Liu, Yu Li, Hui Luo, Wenping He, and Hua Ye. 2024. "Hepatopancreas Transcriptome Analysis of Spinibarbus sinensis to Reveal Different Growth-Related Genes" Genes 15, no. 7: 949. https://doi.org/10.3390/genes15070949
APA StyleZhou, B., Ling, L., Wang, B., Yang, F., Hou, M., Liu, F., Li, Y., Luo, H., He, W., & Ye, H. (2024). Hepatopancreas Transcriptome Analysis of Spinibarbus sinensis to Reveal Different Growth-Related Genes. Genes, 15(7), 949. https://doi.org/10.3390/genes15070949







