Genome-Wide Identification of the PIP5K Gene Family in Camellia sinensis and Their Roles in Metabolic Regulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of the Tea CsPIP5K Family
2.2. Structural Analysis of the CsPIP5K Gene Family in Tea
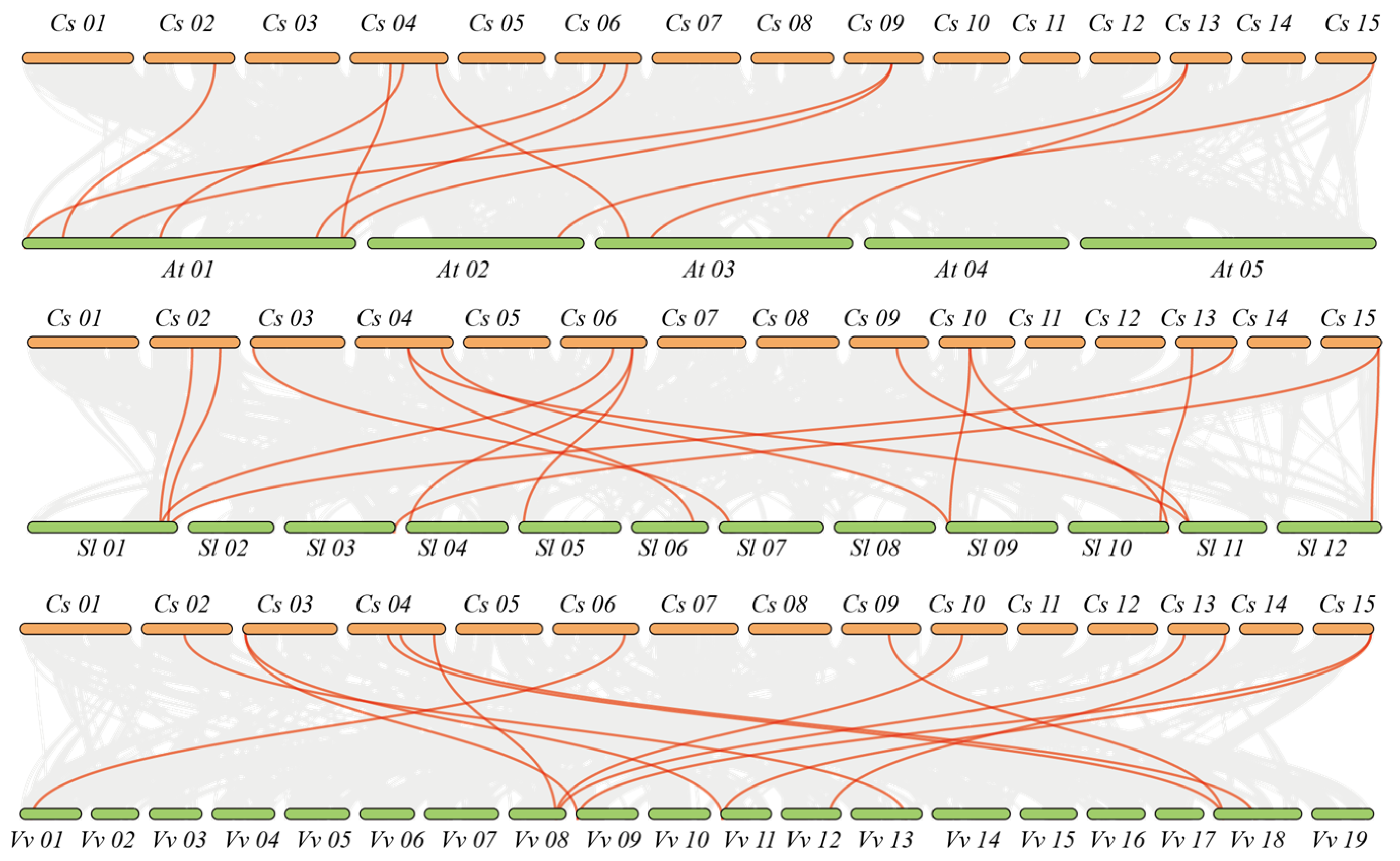
2.3. Chromosome Location and Collinearity Analysis of the CsPIP5K
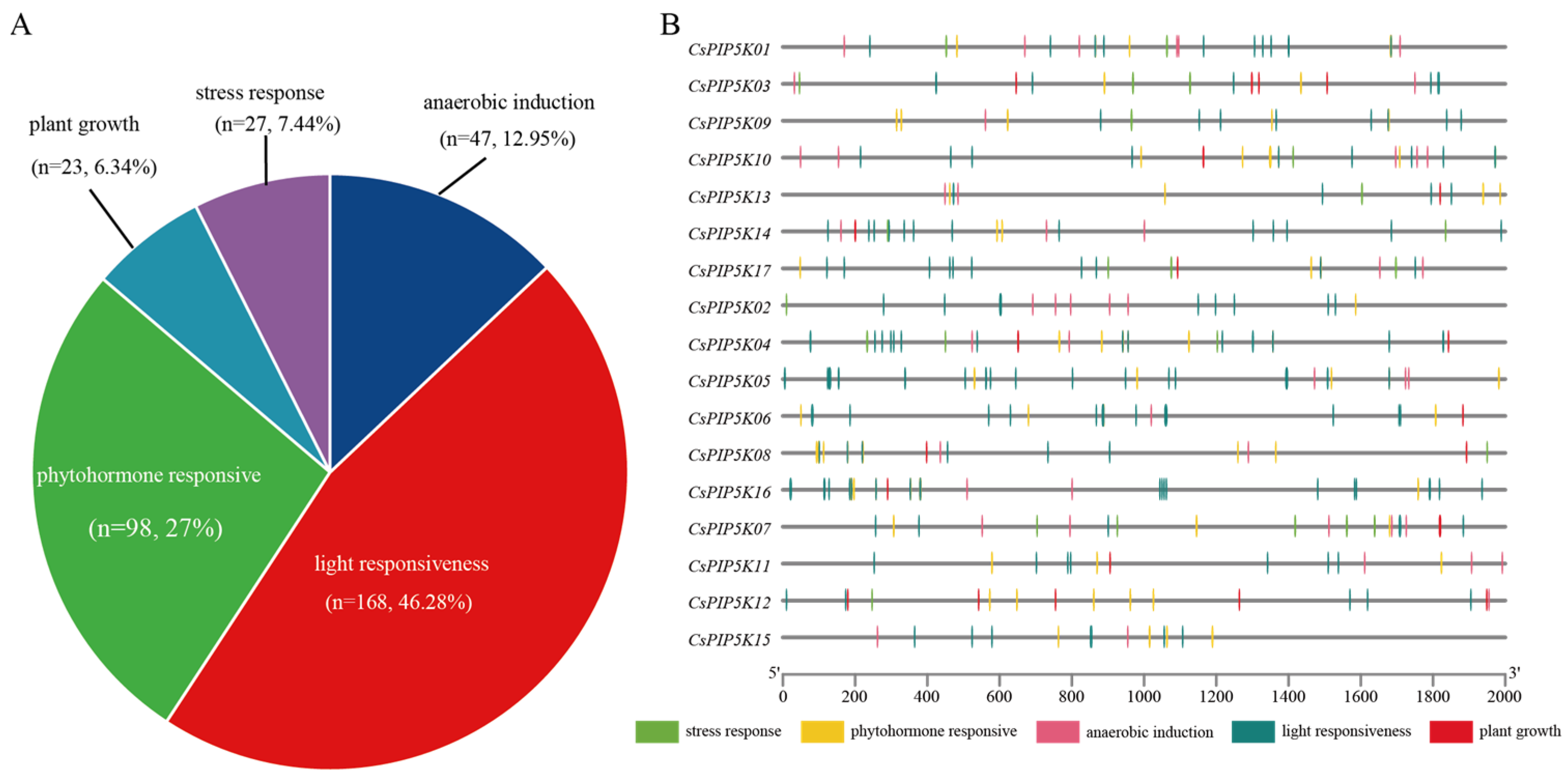
2.4. Cis Element Analysis of the CsPIP5K Gene in Tea
2.5. Analysis of the Expression Pattern of CsPIP5Ks under the Spider Mite Infestation
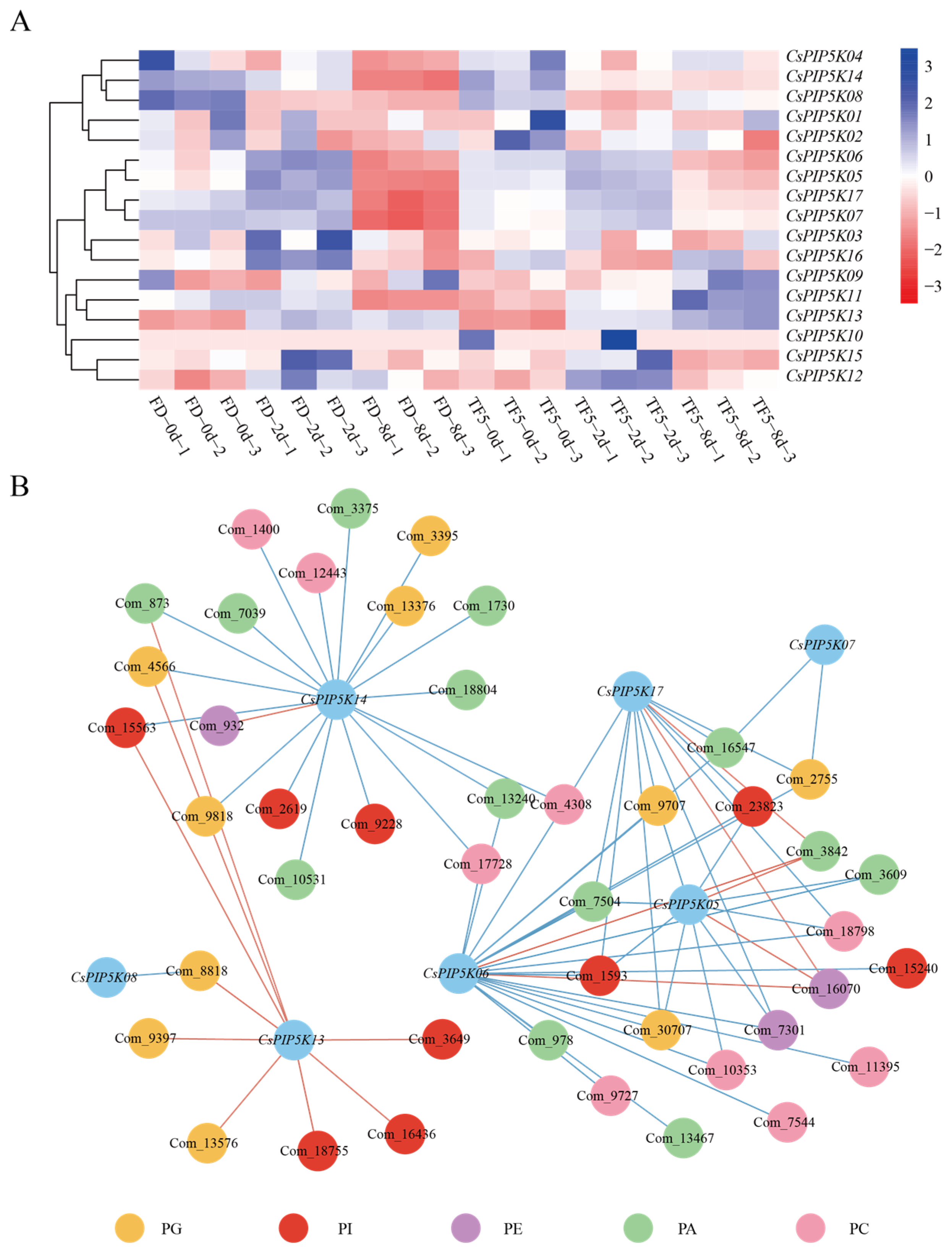
2.6. Integrated Analysis of Metabolomics and Transcriptomics
2.7. RNA Extraction
2.8. Real-Time Quantitative RT-PCR
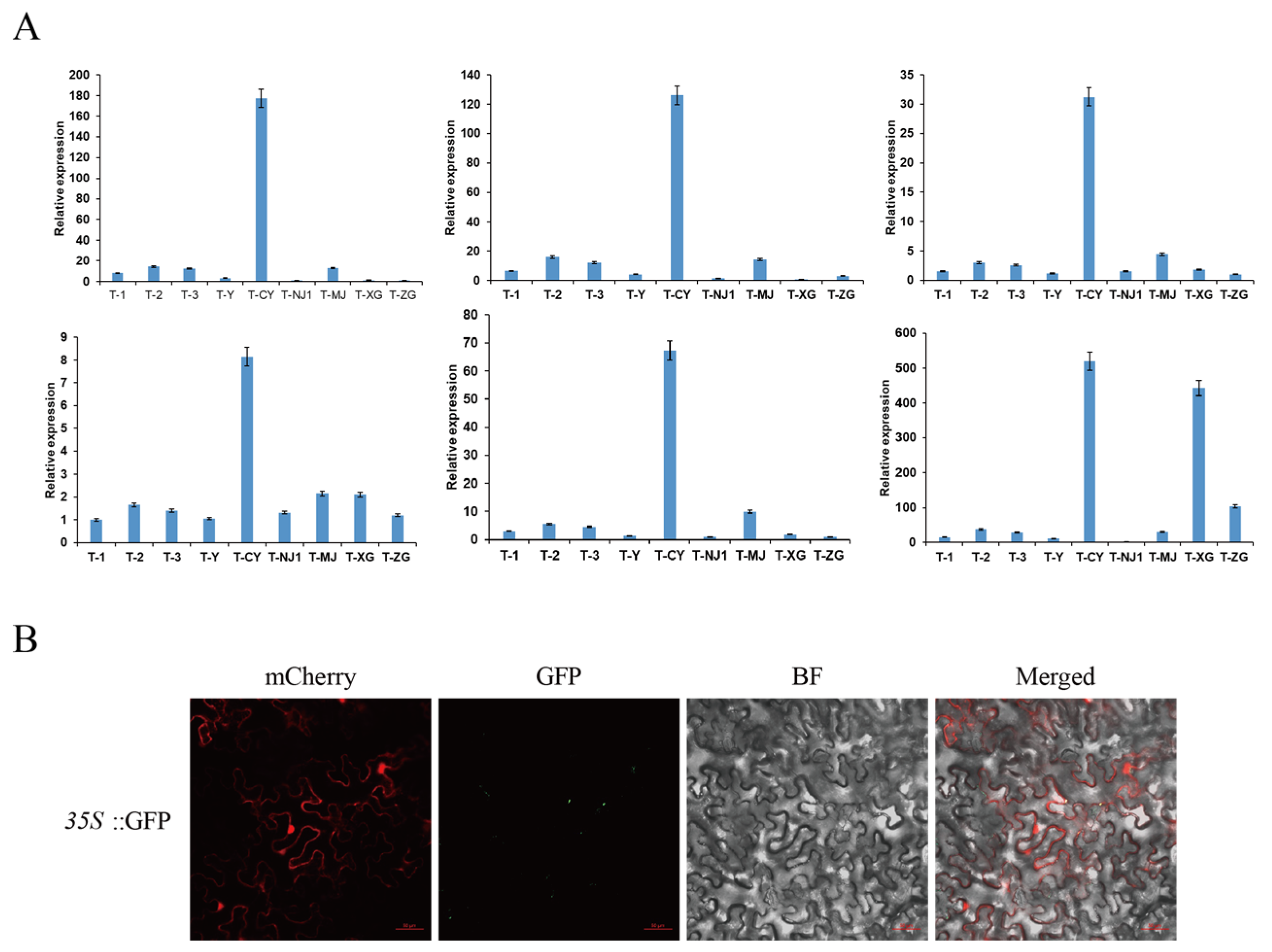
2.9. Subcellular Localization of CsPIP5K
3. Results
3.1. Characterization of Members of the CsPIP5K Gene Family in Camellia sinensis
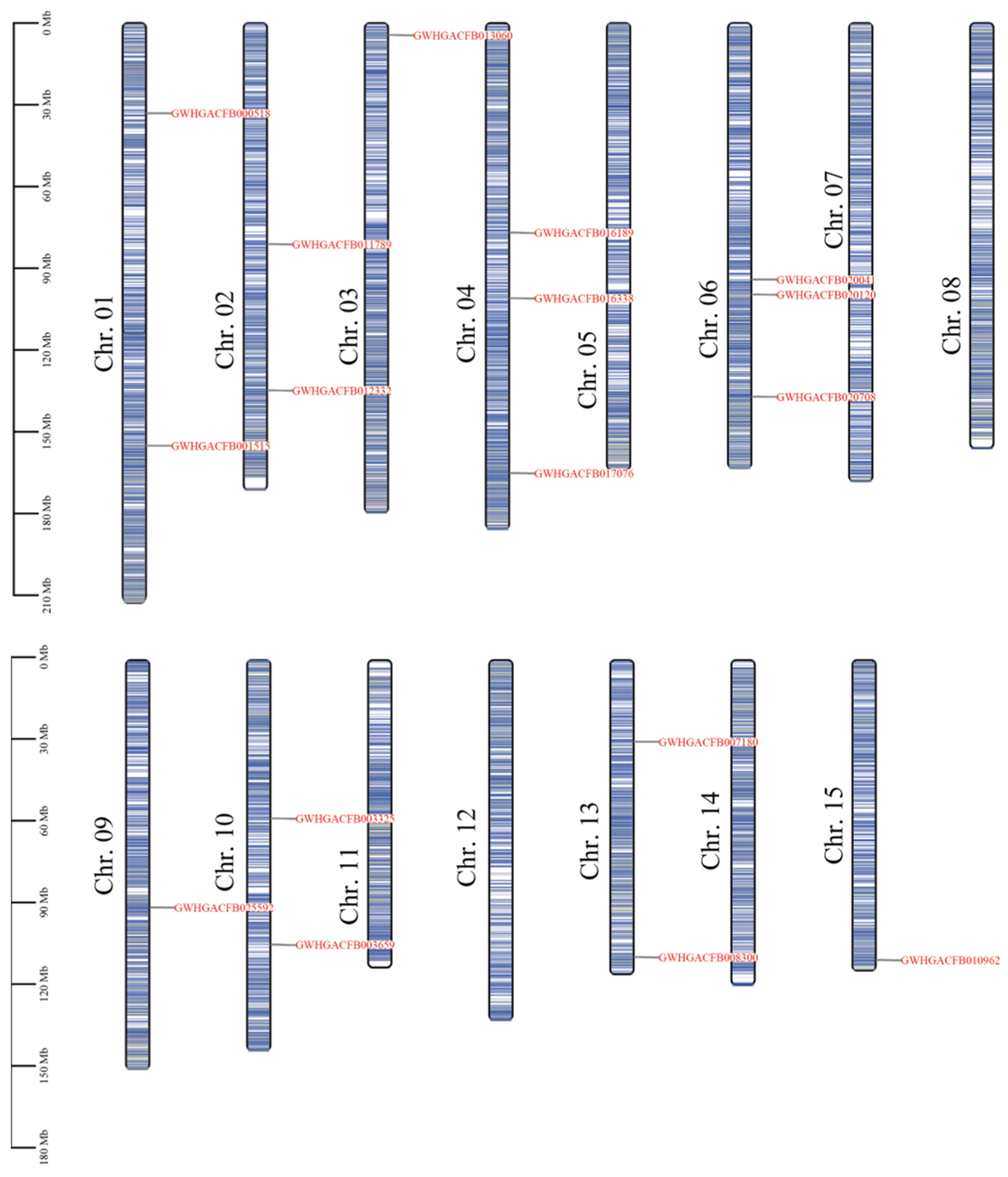
3.2. Chromosomal Localization of the CsPIP5K Gene Gamily
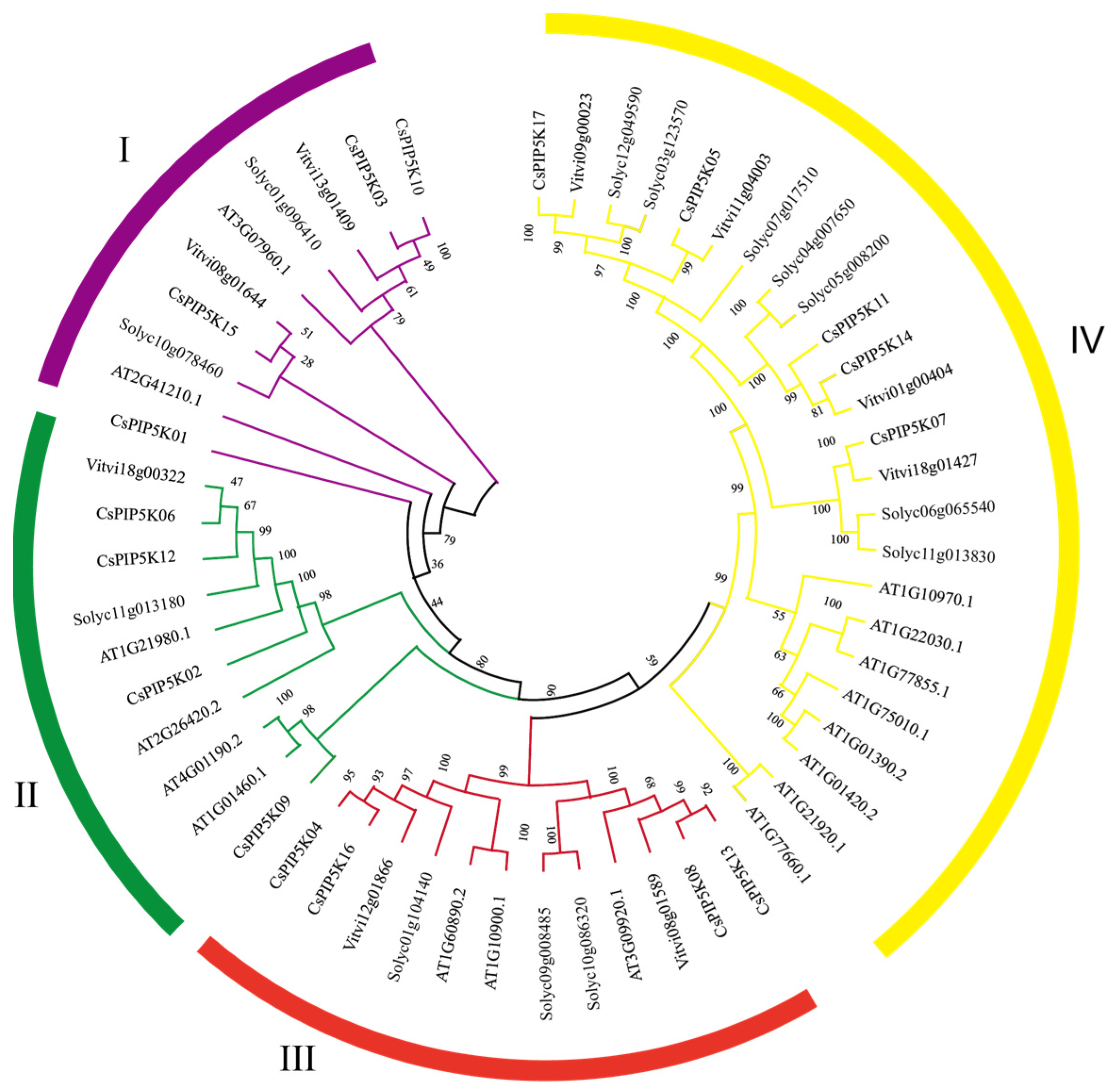
3.3. Phylogenetic Classification of the CsPIP5K Gene
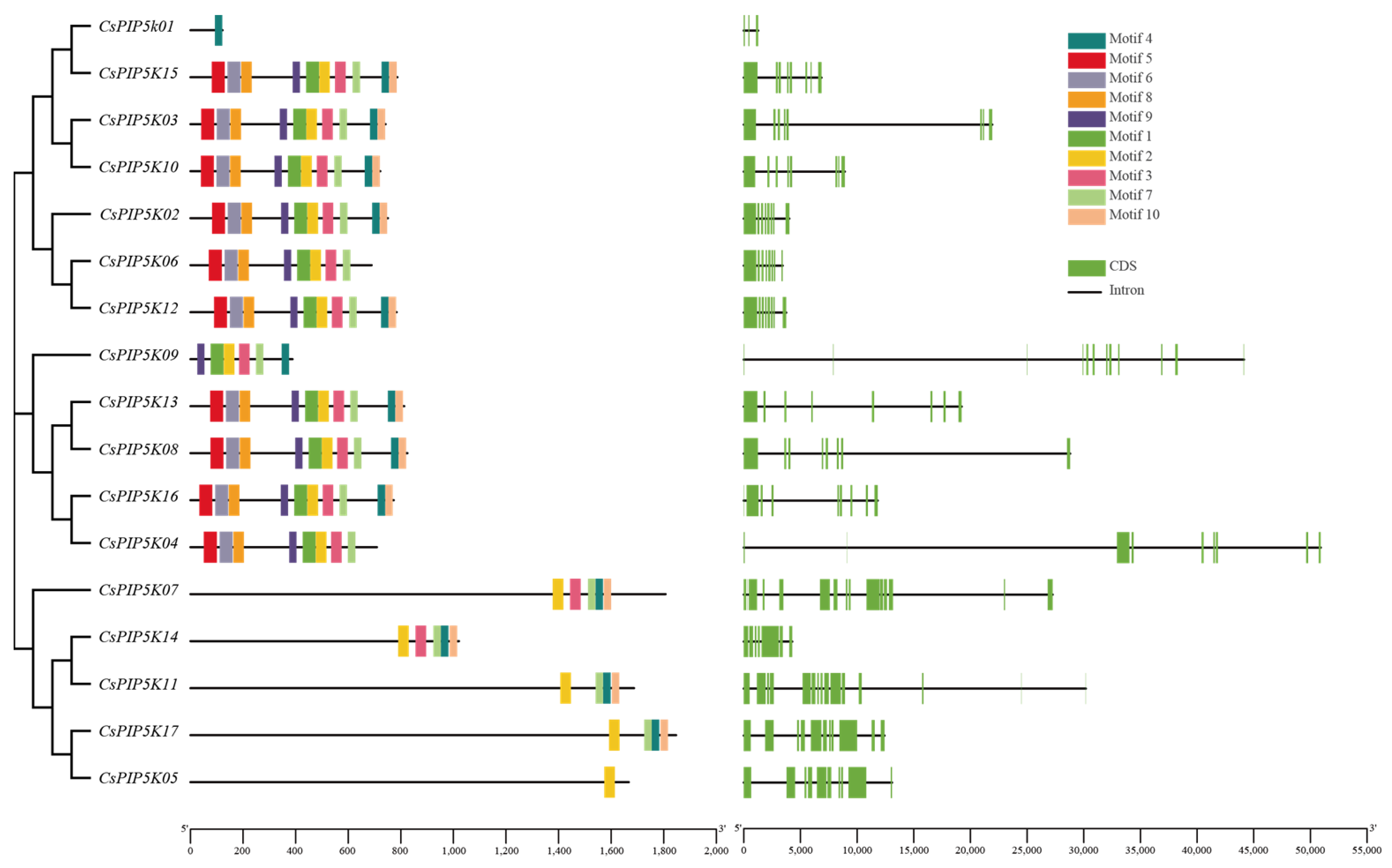
3.4. CsPIP5K Gene Family Member Motifs and Gene Structures
3.5. Analysis of Protein Structural Domains of Tea CsPIP5K Gene Family Members
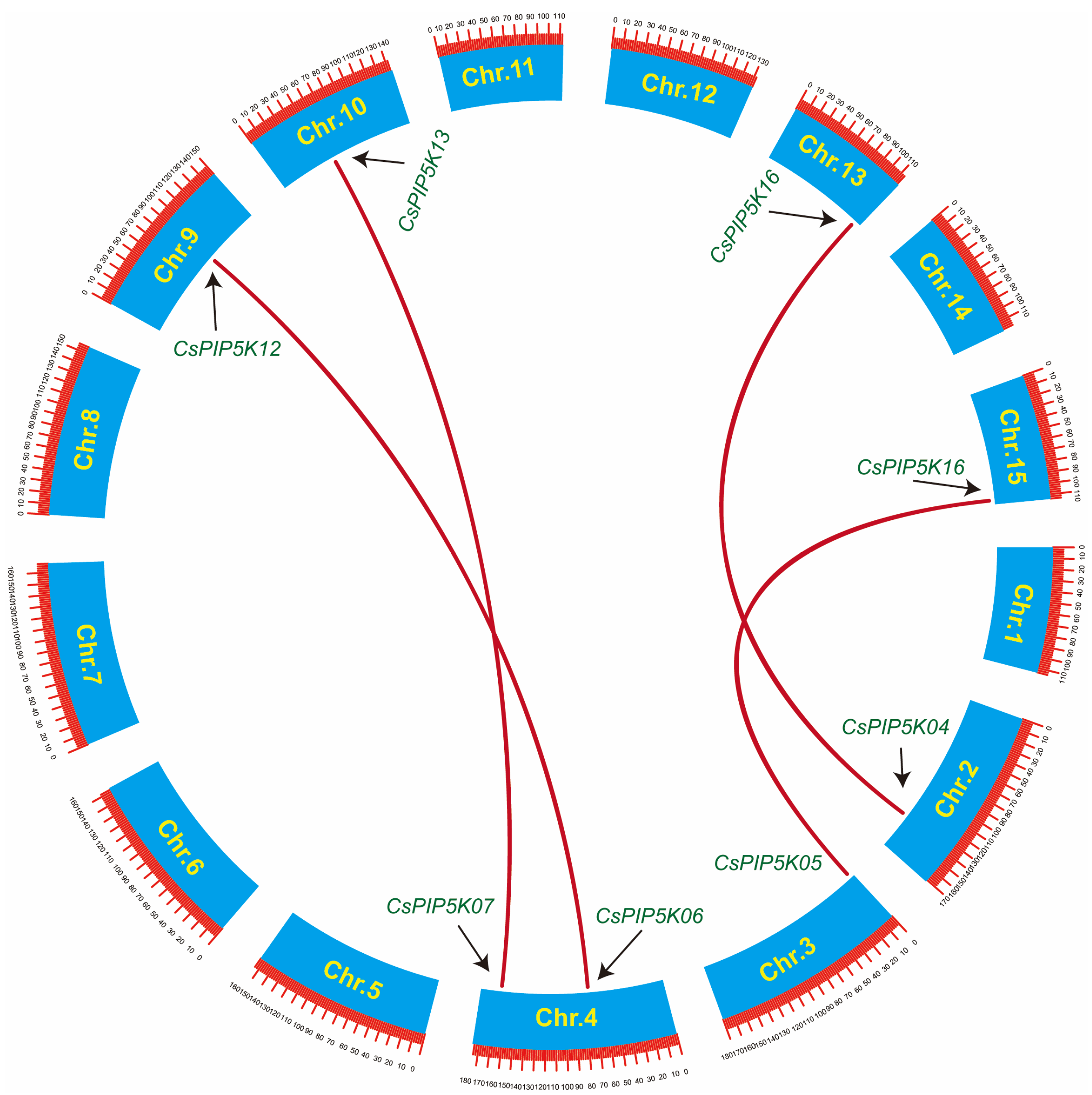
3.6. Analysis of Covariance and Evolutionary Pressure Analysis for Members of the CsPIP5Ks
3.7. Analysis of Cis-Acting Elements of CsPIP5K Promoter
3.8. Analysis of Expression Pattern of CsPIP5K under the Spider Mite Infestation
3.9. Metabolic Regulation of CsPIP5K Family Genes under Spider Mite Infection
3.10. Analysis of Multi-Tissue Expression Profile and Subcellular Localization of CsPIP5K06
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, X.-C.; Zhao, Q.-Y.; Ma, C.-L.; Zhang, Z.-H.; Cao, H.-L.; Kong, Y.-M.; Yue, C.; Hao, X.-Y.; Chen, L.; Ma, J.-Q.; et al. Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genom. 2013, 14, 415. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Zhang, J.; Yang, Y.; Yang, X.; Xu, B.; Yang, W.; Tong, T.; Jin, S.; Shen, C.; Rao, H.; et al. Earliest tea as evidence for one branch of the Silk Road across the Tibetan Plateau. Sci. Rep. 2016, 6, 18955. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Mukhopadhyay, A.; Gurusubramanian, G. Baseline susceptibility of Oligonychus coffeae (Acarina: Tetranychidae) to acaricides in North Bengal tea plantations, India. Int. J. Acarol. 2010, 36, 357–362. [Google Scholar] [CrossRef]
- Carafoli, E. Calcium signaling: A tale for all seasons. Proc. Natl. Acad. Sci. USA 2002, 99, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Sarwar, M.; Syed Khaja, A.S.; Aleskandarany, M.; Karlsson, R.; Althobiti, M.; Ødum, N.; Mongan, N.P.; Dizeyi, N.; Johnson, H.; Green, A.R.; et al. The role of PIP5K1α/pAKT and targeted inhibition of growth of subtypes of breast cancer using PIP5K1α inhibitor. Oncogene 2019, 38, 375–389. [Google Scholar] [CrossRef] [PubMed]
- van den Bout, I.; Divecha, N. PIP5K-driven PtdIns(4,5)P2 synthesis: Regulation and cellular functions. J. Cell Sci. 2009, 122, 3837–3850. [Google Scholar] [CrossRef] [PubMed]
- Katan, M.; Cockcroft, S. Phosphatidylinositol(4,5)bisphosphate: Diverse functions at the plasma membrane. Essays Biochem. 2020, 64, 513–531. [Google Scholar] [CrossRef]
- Li, S.; Huang, F.; Xia, T.; Shi, Y.; Yue, T. Phosphatidylinositol 4,5-bisphosphate sensing lipid raft via inter-leaflet coupling regulated by acyl chain length of sphingomyelin. Langmuir 2023, 39, 5995–6005. [Google Scholar] [CrossRef] [PubMed]
- Divecha, N.; Truong, O.; Hsuan, J.J.; Hinchliffe, K.A.; Irvine, R.F. The cloning and sequence of the C isoform of PtdIns4P 5-kinase. Biochem. J. 1995, 309 Pt 3, 715–719. [Google Scholar] [CrossRef]
- Khadka, B.; Gupta, R.S. Novel molecular signatures in the PIP4K/PIP5K family of proteins specific for different isozymes and subfamilies provide important insights into the evolutionary divergence of this protein family. Genes 2019, 10, 312. [Google Scholar] [CrossRef]
- Yang, X.; Kwon, H.; Kim, M.Y.; Lee, S.-H. RNA-seq profiling in leaf tissues of two soybean (Glycine max [L.] Merr.) cultivars that show contrasting responses to drought stress during early developmental stages. Mol. Breed. 2023, 43, 42. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Fang, T.; Du, Y.; Li, R.; Fan, M.; Ma, F.; Jin, P. miR-2013 negatively regulates phylogenetically conserved PIP5K involved in TLR4 mediated immune responses of amphioxus (Branchiostoma belcheri Tsingtaunese). Dev. Comp. Immunol. 2022, 133, 104430. [Google Scholar] [CrossRef] [PubMed]
- Doumane, M.; Lebecq, A.; Colin, L.; Fangain, A.; Stevens, F.D.; Bareille, J.; Hamant, O.; Belkhadir, Y.; Munnik, T.; Jaillais, Y.; et al. Inducible depletion of PI(4,5)P2 by the synthetic iDePP system in Arabidopsis. Nat. Plants 2021, 7, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Mikami, K.; Katagiri, T.; Iuchi, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J. 1998, 15, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, Y.; Huang, K.; Xu, W.; Zhang, C.; Yuan, H. Genome-wide systematic characterization and expression analysis of the phosphatidylinositol 4-phosphate 5-kinases in plants. Gene 2020, 756, 144915. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, R.; Kato, M.; Tsuge, T.; Aoyama, T. Arabidopsis phosphatidylinositol 4-phosphate 5-kinase genes PIP5K7, PIP5K8, and PIP5K9 are redundantly involved in root growth adaptation to osmotic stress. Plant J. 2021, 106, 913–927. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Sun, Z.; Hu, L.; Yue, Z. Genome-wide identification of PIP5K in wheat and its relationship with anther male sterility induced by high temperature. BMC Plant Biol. 2021, 21, 598. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-S.; Kidokoro, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulatory networks in plant responses to drought and cold stress. Plant Physiol. 2024, 195, 170–189. [Google Scholar] [CrossRef]
- Yang, Y.; Park, M.; Maekawa, M.; Fairn, G.D. Enforced expression of phosphatidylinositol 4-phosphate 5-kinase homolog alters PtdIns(4,5)P2 distribution and the localization of small G-proteins. Sci. Rep. 2019, 9, 14789. [Google Scholar] [CrossRef]
- Zhou, H.; Shi, H.; Yang, Y.; Feng, X.; Chen, X.; Xiao, F.; Lin, H.; Guo, Y. Insights into plant salt stress signaling and tolerance. J. Genet. Genom. 2024, 51, 16–34. [Google Scholar] [CrossRef]
- Hong, Y.; Zhang, W.; Wang, X. Phospholipase D and phosphatidic acid signalling in plant response to drought and salinity. Plant Cell Environ. 2010, 33, 627–635. [Google Scholar] [CrossRef]
- Elge, S.; Brearley, C.; Xia, H.-J.; Kehr, J.; Xue, H.-W.; Mueller-Roeber, B. An Arabidopsis inositol phospholipid kinase strongly expressed in procambial cells: Synthesis of PtdIns(4,5)P2 and PtdIns(3,4,5)P3 in insect cells by 5-phosphorylation of precursors. Plant J. 2001, 26, 561–571. [Google Scholar] [CrossRef]
- Amsalingam, R.; Gajjeraman, P.; Sam, N.; Rahman, V.J.; Azariah, B. Mechanism of fenpropathrin resistance in red spider mite, oligonychus coffeae (Acarina: Tetranychidae), infesting tea [Camellia sinensis L. (O. Kuntze)]. Appl. Biochem. Biotechnol. 2017, 181, 548–561. [Google Scholar] [CrossRef]
- Rahman, V.J.; Babu, A.; Roobakkumar, A.; Perumalsamy, K. Functional and numerical responses of the predatory mite, neoseiulus longispinosus, to the red spider mite, Oligonychus coffeae, infesting tea. J. Insect Sci. 2012, 12, 125. [Google Scholar] [CrossRef]
- Wang, X.; Feng, H.; Chang, Y.; Ma, C.; Wang, L.; Hao, X.; Li, A.L.; Cheng, H.; Wang, L.; Cui, P.; et al. Population sequencing enhances understanding of tea plant evolution. Nat. Commun. 2020, 11, 4447. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Cell-Ploc 2.0: An improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat. Sci. 2010, 02, 1090–1103. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. Mega11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Letunic, i.; Bork, P. Interactive Tree of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. the meme suite. Nucleic Acids Res. 2015, 43, w39–w49. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Rombauts, S.; Déhais, P.; Van Montagu, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xiang, Y.; Sun, M.; Xiong, Y.; Li, C.; Zhang, T.; Ma, W.; Wang, Y.; Liu, X. Transcriptomic and metabolomic analyses reveals keys genes and metabolic pathways in tea (Camellia sinensis) against six-spotted spider mite (Eotetranychus sexmaculatus). BMC Plant Biol. 2023, 23, 638. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- William Roy, S.; Gilbert, W. The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat. Rev. Genet. 2006, 7, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, M.B.; Junior, M.L.; Grossi-de-Sá, M.F.; Petrofeza, S. Exogenous application of methyl jasmonate induces a defense response and resistance against Sclerotinia sclerotiorum in dry bean plants. J. Plant Physiol. 2015, 182, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.-C. Effects of Exogenous Methyl Jasmonate on Induced Physical Defensive Response of Prunus Mongolica. Design 2013, 36. Available online: https://www.alljournals.cn/view_abstract.aspx?pcid=03F54A49DE00578AA0E5DDF5BC021AA7&cid=336C48A7E1AE975A&jid=3734DD28E23CEFA6E34CAF0531DD05A3&aid=5A0D620F4831EB6757620B7D7AC3D11B&yid=FF7AA908D58E97FA&vid=&iid=&sid=&eid=&from_absract=1 (accessed on 10 July 2024).
- Gomes, G.L.B.; Scortecci, K.C. Auxin and its role in plant development: Structure, signalling, regulation and response mechanisms. Plant Biol. 2021, 23, 894–904. [Google Scholar] [CrossRef] [PubMed]
- Martínez, C.; Espinosa-Ruiz, A.; Prat, S. Gibberellins and plant vegetative growth. In Annual Plant Reviews Online; Wiley: Hoboken, NJ, USA, 2018; pp. 285–322. [Google Scholar] [CrossRef]
- Lundquist, M.R.; Goncalves, M.D.; Loughran, R.M.; Possik, E.; Vijayaraghavan, T.; Yang, A.; Pauli, C.; Ravi, A.; Verma, A.; Yang, Z.; et al. Phosphatidylinositol-5-Phosphate 4-Kinases regulate cellular lipid metabolism by facilitating autophagy. Mol. Cell 2018, 70, 531–544.e9. [Google Scholar] [CrossRef]
- Zhang, Q.; Cai, M.; Yu, X.; Wang, L.; Guo, C.; Ming, R.; Zhang, J. Transcriptome dynamics of Camellia sinensis in response to continuous salinity and drought stress. Tree Genet. Genomes 2017, 13, 78. [Google Scholar] [CrossRef]
- Laxalt, A.M.; Munnik, T. Phospholipid signalling in plant defence. Curr. Opin. Plant Biol. 2002, 5, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Kutateladze, T.G. Phosphatidylinositol 3-phosphate recognition and membrane docking by the FYVE domain. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2006, 1761, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Sankaran, V.G.; Klein, D.E.; Sachdeva, M.M.; Lemmon, M.A. High-affinity binding of a FYVE domain to phosphatidylinositol 3-phosphate requires intact phospholipid but not FYVE domain oligomerization. Biochemistry 2001, 40, 8581–8587. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Z.; Li, X.; Chen, Z.; Liu, Y.; Zhang, F.; Dai, Q.; Yu, Q.; Li, N. Identification and analysis of the expression of the PIP5K gene family in tomatoes. Int. J. Mol. Sci. 2024, 25, 159. [Google Scholar] [CrossRef] [PubMed]
- Lyu, R.; Singh, S.K.; Liu, Y.; Patra, B.; Zhou, Y.; Wang, B.; Pattanaik, S.; Yuan, L. Reprogramming plant specialized metabolism by manipulating protein kinases. aBIOTECH 2021, 2, 226–239. [Google Scholar] [CrossRef] [PubMed]
- Kerchev, P.I.; Fenton, B.; Foyer, C.H.; Hancock, R.D. Plant responses to insect herbivory: Interactions between photosynthesis, reactive oxygen species and hormonal signalling pathways. Plant Cell Environ. 2012, 35, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Ma, T.; Song, S.; Li, X.; Fu, P.; Wu, W.; Liu, J.; Gao, Y.; Ye, W.; Dry, I.B.; et al. Arabidopsis downy mildew effector HaRxLL470 suppresses plant immunity by attenuating the DNA-binding activity of bZIP transcription factor HY5. New Phytol. 2021, 230, 1562–1577. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Wang, H.; Yang, X.-M.; Hu, Z.-W.; Zhou, X.-H.; Xiang, L.; Xiong, X.-Y.; He, X.-R.; Zhu, Y.; Li, G.-B.; et al. Osa-miR160a confers broad-spectrum resistance to fungal and bacterial pathogens in rice. New Phytol. 2022, 236, 2216–2232. [Google Scholar] [CrossRef]
- Cao, Y.; Wu, Y.; Zheng, Z.; Song, F. Overexpression of the rice EREBP-like gene OsBIERF3 enhances disease resistance and salt tolerance in transgenic tobacco. Physiol. Mol. Plant Pathol. 2005, 67, 202–211. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Xiong, Y.; Tang, X.; Zhang, T.; Ma, W.; Wang, Y.; Li, C. Genome-Wide Identification of the PIP5K Gene Family in Camellia sinensis and Their Roles in Metabolic Regulation. Genes 2024, 15, 932. https://doi.org/10.3390/genes15070932
Wang X, Xiong Y, Tang X, Zhang T, Ma W, Wang Y, Li C. Genome-Wide Identification of the PIP5K Gene Family in Camellia sinensis and Their Roles in Metabolic Regulation. Genes. 2024; 15(7):932. https://doi.org/10.3390/genes15070932
Chicago/Turabian StyleWang, Xiaoping, Yuanyuan Xiong, Xiaobo Tang, Ting Zhang, Weiwei Ma, Yun Wang, and Chunhua Li. 2024. "Genome-Wide Identification of the PIP5K Gene Family in Camellia sinensis and Their Roles in Metabolic Regulation" Genes 15, no. 7: 932. https://doi.org/10.3390/genes15070932
APA StyleWang, X., Xiong, Y., Tang, X., Zhang, T., Ma, W., Wang, Y., & Li, C. (2024). Genome-Wide Identification of the PIP5K Gene Family in Camellia sinensis and Their Roles in Metabolic Regulation. Genes, 15(7), 932. https://doi.org/10.3390/genes15070932




