The Potential Links between lncRNAs and Drug Tolerance in Lung Adenocarcinoma
Abstract
1. Introduction
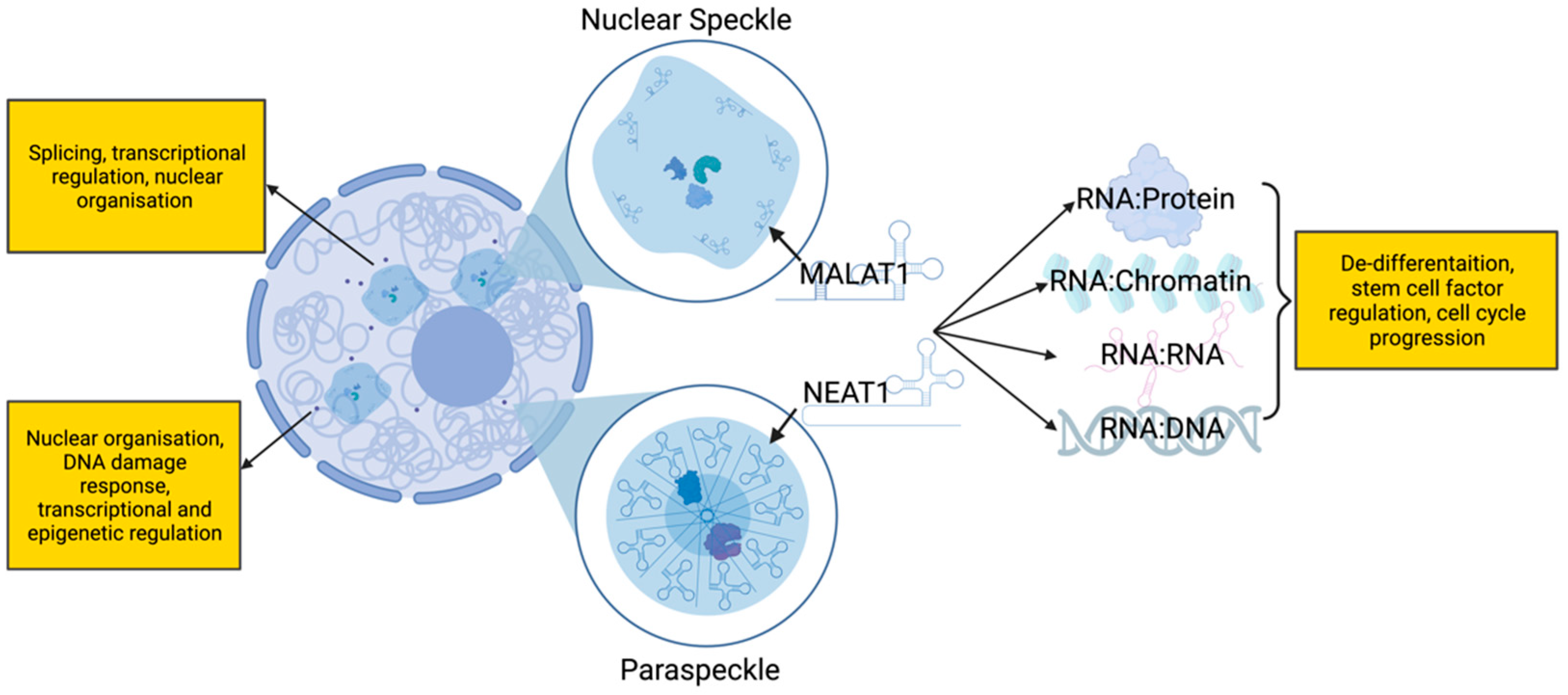
2. Long Noncoding RNAs
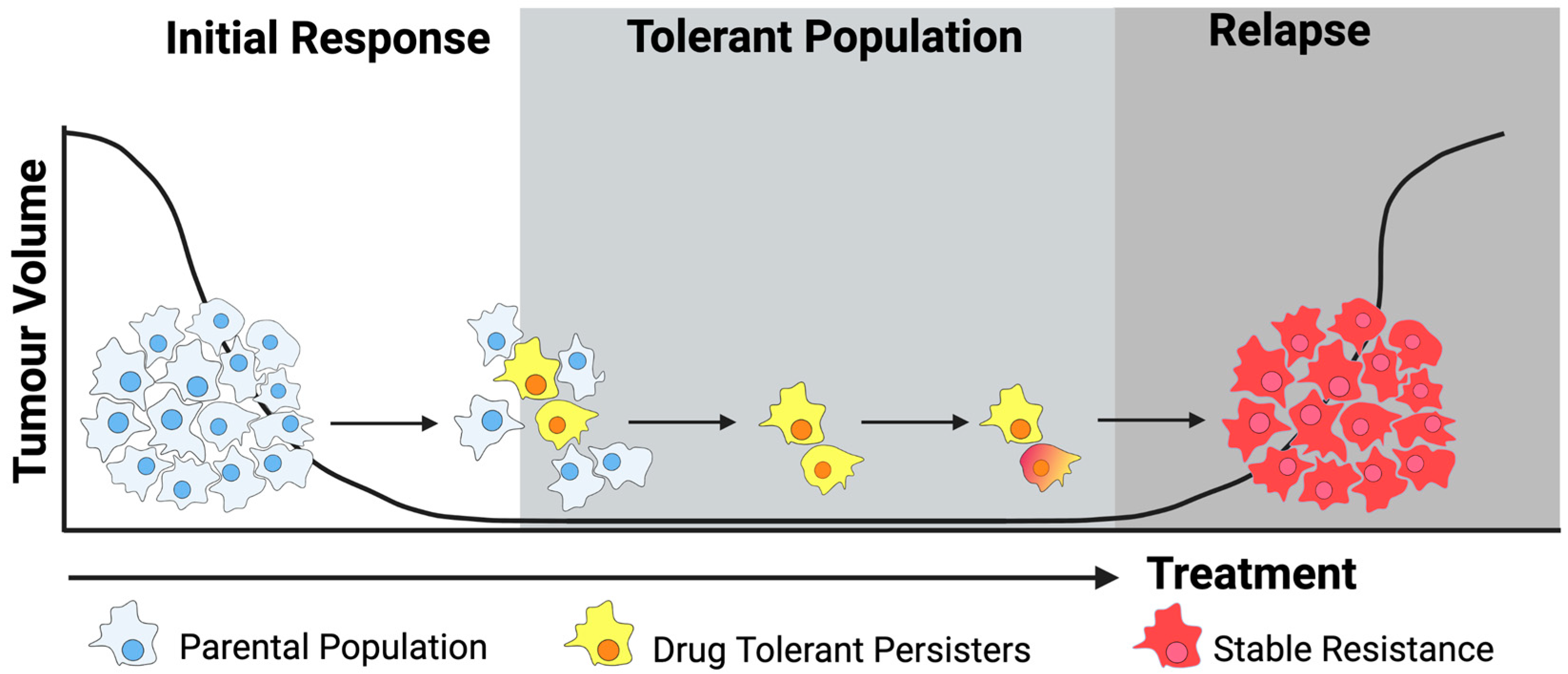
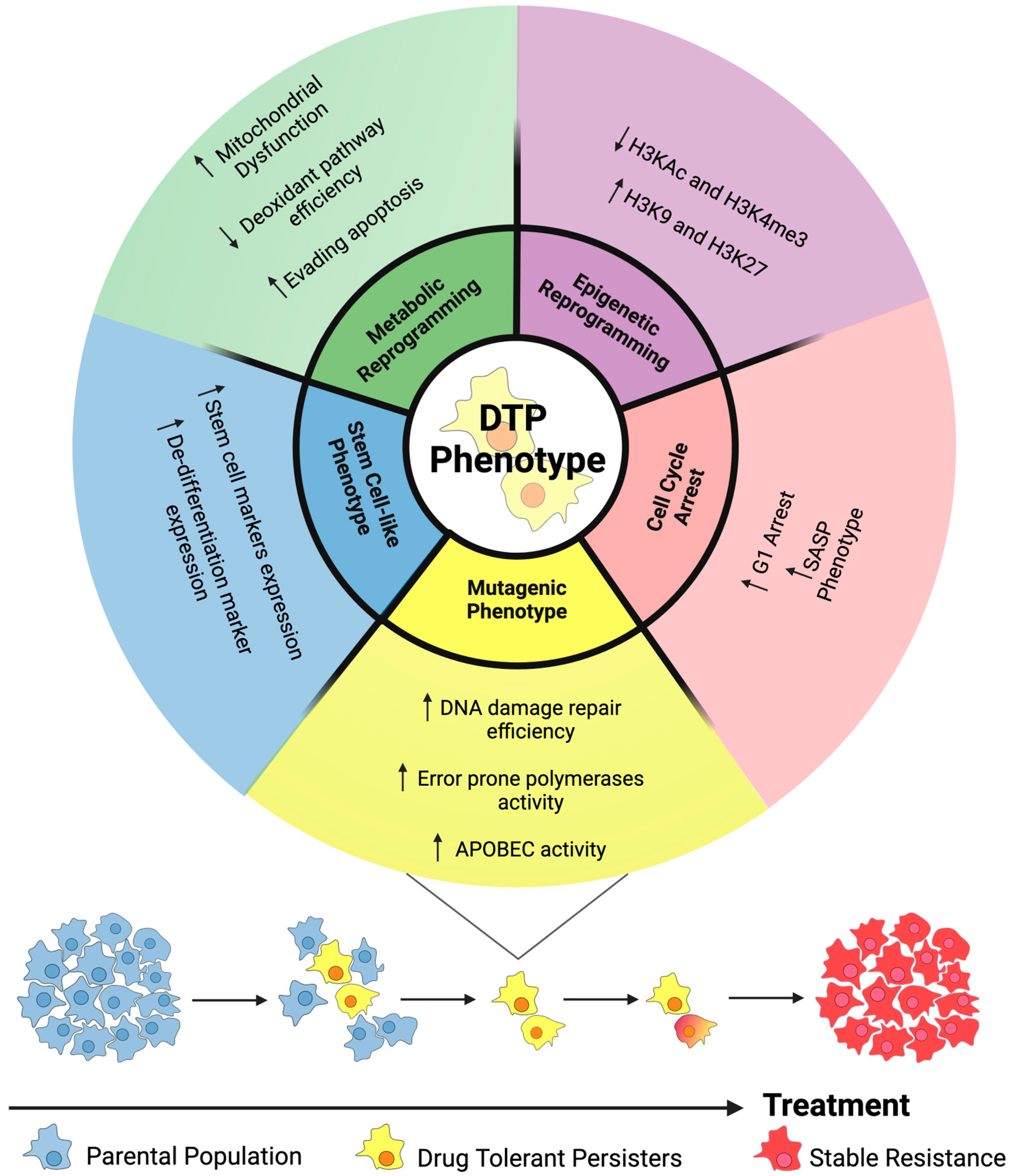
3. Drug Tolerance
3.1. Epigenetic Reprogramming
3.2. Slow Cycling Phenotype
3.3. Regulation of a SASP-like Phenotype
3.4. Stem and Developmental Cell-like Features in DTPs
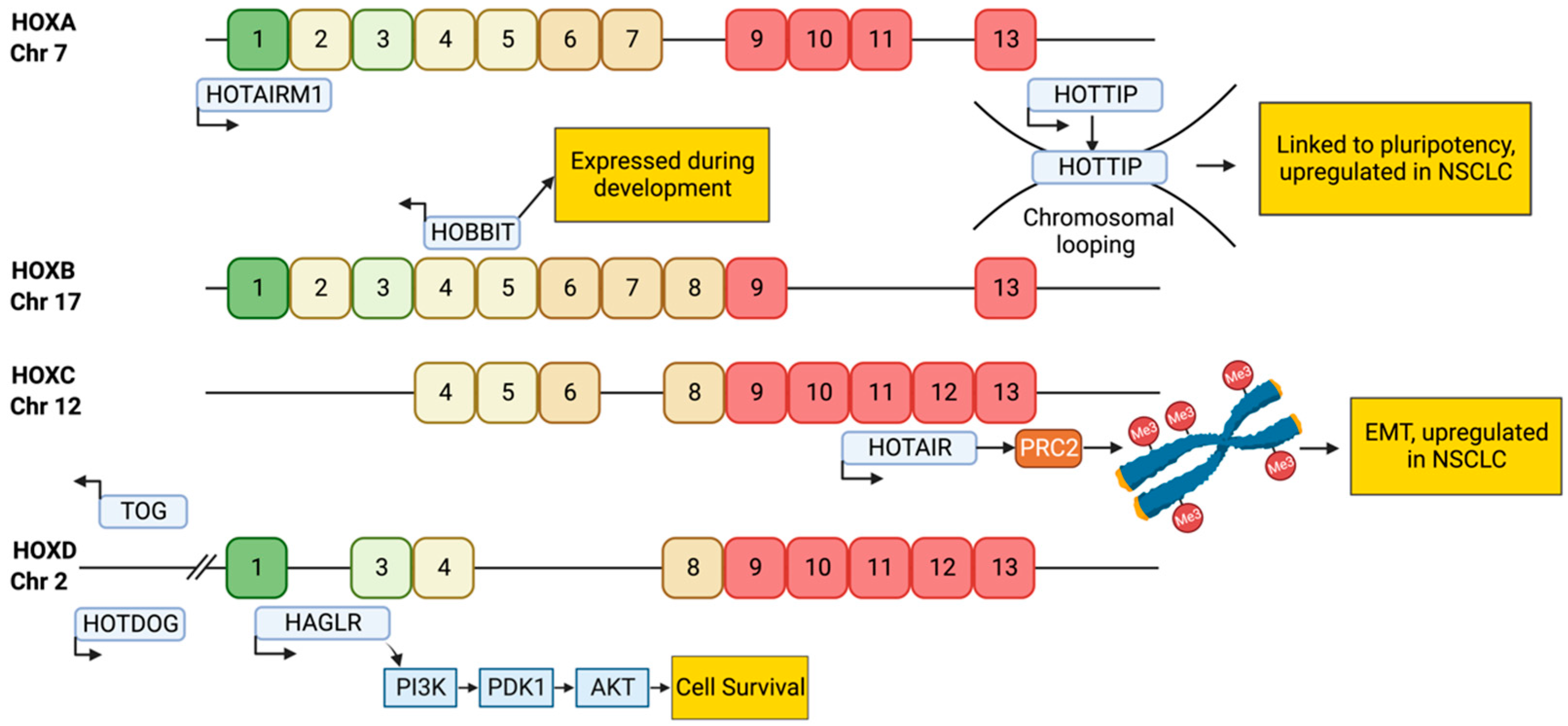
Developmental Lung Genes Co-Ordinated by lncRNAs
3.5. Metabolic Reprogramming
3.6. Adaptive Mutability
4. Therapeutic Challenges and Potential
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rotow, J.; Bivona, T.G. Understanding and targeting resistance mechanisms in NSCLC. Nat. Rev. Cancer 2017, 17, 637–658. [Google Scholar] [CrossRef] [PubMed]
- Pu, Y.; Li, L.; Peng, H.; Liu, L.; Heymann, D.; Robert, C.; Vallette, F.; Shen, S. Drug-tolerant persister cells in cancer: The cutting edges and future directions. Nat. Rev. Clin. Oncol. 2023, 20, 799–813. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.V.; Lee, D.Y.; Li, B.; Quinlan, M.P.; Takahashi, F.; Maheswaran, S.; McDermott, U.; Azizian, N.; Zou, L.; Fischbach, M.A.; et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010, 141, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Madhusoodanan, J. How persister bacteria evade antibiotics, prolong infections. Proc. Natl. Acad. Sci. USA 2022, 119, e2215617119. [Google Scholar] [CrossRef] [PubMed]
- Oren, Y.; Tsabar, M.; Cuoco, M.S.; Amir-Zilberstein, L.; Cabanos, H.F.; Hütter, J.-C.; Hu, B.; Thakore, P.I.; Tabaka, M.; Fulco, C.P.; et al. Cycling cancer persister cells arise from lineages with distinct programs. Nature 2021, 596, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.; Crisafulli, G.; Sogari, A.; Reilly, N.M.; Arena, S.; Lamba, S.; Bartolini, A.; Amodio, V.; Magrì, A.; Novara, L.; et al. Adaptive mutability of colorectal cancers in response to targeted therapies. Science 2019, 366, 1473–1480. [Google Scholar] [CrossRef]
- Noronha, A.; Nataraj, N.B.; Lee, J.S.; Zhitomirsky, B.; Oren, Y.; Oster, S.; Lindzen, M.; Mukherjee, S.; Will, R.; Ghosh, S.; et al. AXL and error-prone DNA replication confer drug resistance and offer strategies to treat EGFR-mutant lung cancer. Cancer Discov. 2022, 12, 2666–2683. [Google Scholar] [CrossRef] [PubMed]
- Rambow, F.; Marine, J.C.; Goding, C.R. Melanoma plasticity and phenotypic diversity: Therapeutic barriers and opportunities. Genes Dev. 2019, 33, 1295–1318. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.A.; Jen, J.; Jiang, S.; Sayad, A.; Mer, A.S.; Brown, K.R.; Nixon, A.M.; Dhabaria, A.; Tang, K.H.; Venet, D.; et al. Ontogeny and Vulnerabilities of Drug-Tolerant Persisters in HER2+ Breast Cancer. Cancer Discov. 2022, 12, 1022–1045. [Google Scholar] [CrossRef]
- Liau, B.B.; Sievers, C.; Donohue, L.K.; Gillespie, S.M.; Flavahan, W.A.; Miller, T.E.; Venteicher, A.S.; Hebert, C.H.; Carey, C.D.; Rodig, S.J.; et al. Adaptive Chromatin Remodeling Drives Glioblastoma Stem Cell Plasticity and Drug Tolerance. Cell Stem Cell 2017, 20, 233–246.e7. [Google Scholar] [CrossRef]
- Maynard, A.; McCoach, C.E.; Rotow, J.K.; Harris, L.; Haderk, F.; Kerr, D.L.; Yu, E.A.; Schenk, E.L.; Tan, W.; Zee, A.; et al. Therapy-Induced Evolution of Human Lung Cancer Revealed by Single-Cell RNA Sequencing. Cell 2020, 182, 1232–1251.e22. [Google Scholar] [CrossRef] [PubMed]
- Aissa, A.F.; Islam, A.B.M.M.K.; Ariss, M.M.; Go, C.C.; Rader, A.E.; Conrardy, R.D.; Gajda, A.M.; Rubio-Perez, C.; Valyi-Nagy, K.; Pasquinelli, M.; et al. Single-cell transcriptional changes associated with drug tolerance and response to combination therapies in cancer. Nat. Commun. 2021, 12, 1628. [Google Scholar] [CrossRef] [PubMed]
- Mikubo, M.; Inoue, Y.; Liu, G.; Tsao, M.-S. Mechanism of Drug Tolerant Persister Cancer Cells: The Landscape and Clinical Implication for Therapy. J. Thorac. Oncol. 2021, 16, 1798–1809. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437. [Google Scholar] [CrossRef] [PubMed]
- van Bakel, H.; Nislow, C.; Blencowe, B.J.; Hughes, T.R. Most “Dark Matter” Transcripts Are Associated with Known Genes. PLoS Biol. 2010, 8, e1000371. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Gencel-Augusto, J.; Wu, W.; Bivona, T.G. Long Non-Coding RNAs as Emerging Targets in Lung Cancer. Cancers 2023, 15, 3135. [Google Scholar] [CrossRef]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Kitagawa, M.; Kitagawa, K.; Kotake, Y.; Niida, H.; Ohhata, T. Cell cycle regulation by long non-coding RNAs. Cell. Mol. Life Sci. 2013, 70, 4785–4794. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Shoorei, H.; Anamag, F.T.; Taheri, M. The Role of Non-Coding RNAs in Controlling Cell Cycle Related Proteins in Cancer Cells. Front. Oncol. 2020, 10, 608975. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, D.; Choi, Y.E.; Brault, M.E. Charity begins at home: Non-coding RNA functions in DNA repair. Nat. Rev. Mol. Cell Biol. 2013, 14, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Taiana, E.; Ronchetti, D.; Todoerti, K.; Nobili, L.; Tassone, P.; Amodio, N.; Neri, A. LncRNA NEAT1 in Paraspeckles: A Structural Scaffold for Cellular DNA Damage Response Systems? Noncoding RNA 2020, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Guler, G.D.; Tindell, C.A.; Pitti, R.; Wilson, C.; Nichols, K.; Cheung, T.K.; Kim, H.-J.; Wongchenko, M.; Yan, Y.; Haley, B.; et al. Repression of Stress-Induced LINE-1 Expression Protects Cancer Cell Subpopulations from Lethal Drug Exposure. Cancer Cell 2017, 32, 221–237.e13. [Google Scholar] [CrossRef] [PubMed]
- Vinogradova, M.; Gehling, V.S.; Gustafson, A.; Arora, S.; A Tindell, C.; Wilson, C.; E Williamson, K.; Guler, G.D.; Gangurde, P.; Manieri, W.; et al. An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat. Chem. Biol. 2016, 12, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Huang, P.; Zhang, J. Hypermethylation of MEG3 promoter correlates with inactivation of MEG3 and poor prognosis in patients with retinoblastoma. J. Transl. Med. 2017, 15, 268. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, Z.; Duan, Y. LncRNA MEG3 inhibits non-small cell lung cancer via interaction with DKC1 protein. Oncol. Lett. 2020, 20, 2183–2190. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhong, Y.; Wang, Y.; Zhang, X.; Batista, D.L.; Gejman, R.; Ansell, P.J.; Zhao, J.; Weng, C.; Klibanski, A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007, 282, 24731–24742. [Google Scholar] [CrossRef] [PubMed]
- Uroda, T.; Anastasakou, E.; Rossi, A.; Teulon, J.-M.; Pellequer, J.-L.; Annibale, P.; Pessey, O.; Inga, A.; Chillón, I.; Marcia, M. Conserved Pseudoknots in lncRNA MEG3 Are Essential for Stimulation of the p53 Pathway. Mol. Cell 2019, 75, 982–995.e9. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Fu, X.-D. Chromatin-associated RNAs as facilitators of functional genomic interactions. Nat. Rev. Genet. 2019, 20, 503–519. [Google Scholar] [CrossRef] [PubMed]
- Limouse, C.; Smith, O.K.; Jukam, D.; Fryer, K.A.; Greenleaf, W.J.; Straight, A.F. Global mapping of RNA-chromatin contacts reveals a proximity-dominated connectivity model for ncRNA-gene interactions. Nat. Commun. 2023, 14, 6073. [Google Scholar] [CrossRef]
- Dueva, R.; Akopyan, K.; Pederiva, C.; Trevisan, D.; Dhanjal, S.; Lindqvist, A.; Farnebo, M. Neutralization of the Positive Charges on Histone Tails by RNA Promotes an Open Chromatin Structure. Cell Chem. Biol. 2019, 26, 1436–1449.e5. [Google Scholar] [CrossRef]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef]
- Oh, H.J.; Aguilar, R.; Kesner, B.; Lee, H.-G.; Kriz, A.J.; Chu, H.-P.; Lee, J.T. Jpx RNA regulates CTCF anchor site selection and formation of chromosome loops. Cell 2021, 184, 6157–6173.e24. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Li, H.; Yu, Q.; Xiao, W.; Wang, D.O. LncRNA-mediated DNA methylation: An emerging mechanism in cancer and beyond. J. Exp. Clin. Cancer Res. 2022, 41, 100. [Google Scholar] [CrossRef] [PubMed]
- Merry, C.R.; Forrest, M.E.; Sabers, J.N.; Beard, L.; Gao, X.-H.; Hatzoglou, M.; Jackson, M.W.; Wang, Z.; Markowitz, S.D.; Khalil, A.M. DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer. Hum. Mol. Genet. 2015, 24, 6240–6253. [Google Scholar] [CrossRef]
- Long, Y.; Wang, X.; Youmans, D.T.; Cech, T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017, 3, eaao2110. [Google Scholar] [CrossRef]
- Bose, D.A.; Donahue, G.; Reinberg, D.; Shiekhattar, R.; Bonasio, R.; Berger, S.L. RNA Binding to CBP Stimulates Histone Acetylation and Transcription. Cell 2017, 168, 135–149.e22. [Google Scholar] [CrossRef]
- Piunti, A.; Shilatifard, A. The roles of Polycomb repressive complexes in mammalian development and cancer. Nat. Rev. Mol. Cell Biol. 2021, 22, 326–345. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Morales, D.R.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef]
- Davidovich, C.; Zheng, L.; Goodrich, K.J.; Cech, T.R. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013, 20, 1250–1257. [Google Scholar] [CrossRef]
- Long, Y.; Hwang, T.; Gooding, A.R.; Goodrich, K.J.; Rinn, J.L.; Cech, T.R. RNA is essential for PRC2 chromatin occupancy and function in human pluripotent stem cells. Nat. Genet. 2020, 52, 931–938. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- De Kumar, B.; Krumlauf, R. HOXs and lincRNAs: Two sides of the same coin. Sci. Adv. 2016, 2, e1501402. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Sukumar, S. The HOX genes and their roles in oncogenesis. Nat. Rev. Cancer 2010, 10, 361–371. [Google Scholar] [CrossRef]
- Li, L.; Zhang, X.; Liu, Q.; Yin, H.; Diao, Y.; Zhang, Z.; Wang, Y.; Gao, Y.; Ren, X.; Li, J.; et al. Emerging role of HOX genes and their related long noncoding RNAs in lung cancer. Crit. Rev. Oncol. Hematol. 2019, 139, 1–6. [Google Scholar] [CrossRef]
- Steens, J.; Klein, D. HOX genes in stem cells: Maintaining cellular identity and regulation of differentiation. Front. Cell Dev. Biol. 2022, 10, 1002909. [Google Scholar] [CrossRef]
- Battistelli, C.; Garbo, S.; Riccioni, V.; Montaldo, C.; Santangelo, L.; Vandelli, A.; Strippoli, R.; Tartaglia, G.G.; Tripodi, M.; Cicchini, C. Design and Functional Validation of a Mutant Variant of the LncRNA HOTAIR to Counteract Snail Function in Epithelial-to-Mesenchymal Transition. Cancer Res. 2021, 81, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.-C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Hajjari, M.; Salavaty, A. HOTAIR: An oncogenic long non-coding RNA in different cancers. Cancer Biol. Med. 2015, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Jarroux, J.; Foretek, D.; Bertrand, C.; Gabriel, M.; Szachnowski, U.; Saci, Z.; Guo, S.; Londoño-Vallejo, A.; Pinskaya, M.; Morillon, A. HOTAIR lncRNA promotes epithelial-mesenchymal transition by redistributing LSD1 at regulatory chromatin regions. EMBO Rep. 2021, 22, e50193. [Google Scholar] [CrossRef]
- Liu, X.H.; Liu, Z.L.; Sun, M.; Liu, J.; Wang, Z.X.; De, W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer 2013, 13, 464. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.Y.; Li, X.Q.; Gao, T.H.; Cui, Y.; Ma, N.; Zhou, Y.; Zhang, G.J. Elevated HOTAIR expression associated with cisplatin resistance in non-small cell lung cancer patients. J. Thorac. Dis. 2016, 8, 3314–3322. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Xu, Q.; Liu, F.; Ye, X.; Wang, J.; Meng, X. Identification and validation of long noncoding RNA biomarkers in human non-small-cell lung carcinomas. J. Thorac. Oncol. 2015, 10, 645–654. [Google Scholar] [CrossRef]
- Navarro, A.; Moises, J.; Santasusagna, S.; Marrades, R.M.; Viñolas, N.; Castellano, J.J.; Canals, J.; Muñoz, C.; Ramírez, J.; Molins, L.; et al. Clinical significance of long non-coding RNA HOTTIP in early-stage non-small-cell lung cancer. BMC Pulm. Med. 2019, 19, 55. [Google Scholar] [CrossRef]
- Hata, A.N.; Niederst, M.J.; Archibald, H.L.; Gomez-Caraballo, M.; Siddiqui, F.M.; Mulvey, H.E.; Maruvka, Y.E.; Ji, F.; Bhang, H.-E.C.; Radhakrishna, V.K.; et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat. Med. 2016, 22, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial persistence as a phenotypic switch. Science 2004, 305, 1622–1625. [Google Scholar] [CrossRef]
- Harms, A.; Maisonneuve, E.; Gerdes, K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 2016, 354, aaf4268. [Google Scholar] [CrossRef]
- Tripathi, V.; Shen, Z.; Chakraborty, A.; Giri, S.; Freier, S.M.; Wu, X.; Zhang, Y.; Gorospe, M.; Prasanth, S.G.; Lal, A.; et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013, 9, e1003368. [Google Scholar] [CrossRef]
- Marín-Béjar, O.; Marchese, F.P.; Athie, A.; Sánchez, Y.; González, J.; Segura, V.; Huang, L.; Moreno, I.; Navarro, A.; Monzó, M.; et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013, 14, R104. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, H.; Li, F.; Heindl, L.M.; He, X.; Yu, J.; Yang, J.; Ge, S.; Ruan, J.; Jia, R.; et al. Long Non-coding RNA LINC-PINT Suppresses Cell Proliferation and Migration of Melanoma via Recruiting EZH2. Front. Cell Dev. Biol. 2019, 7, 350. [Google Scholar] [CrossRef] [PubMed]
- Marín-Béjar, O.; Mas, A.M.; González, J.; Martinez, D.; Athie, A.; Morales, X.; Galduroz, M.; Raimondi, I.; Grossi, E.; Guo, S.; et al. The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element. Genome Biol. 2017, 18, 202. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.R.; Korolchuk, S.; Martin, M.P.; Stanley, W.A.; Moukhametzianov, R.; Noble, M.E.M.; Endicott, J.A. CDK1 structures reveal conserved and unique features of the essential cell cycle CDK. Nat. Commun. 2015, 6, 6769. [Google Scholar] [CrossRef]
- Zhang, E.-B.; Yin, D.-D.; Sun, M.; Kong, R.; Liu, X.-H.; You, L.-H.; Han, L.; Xia, R.; Wang, K.-M.; Yang, J.-S.; et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014, 5, e1243. [Google Scholar] [CrossRef]
- Zhang, E.; He, X.; Yin, D.; Han, L.; Qiu, M.; Xu, T.; Xia, R.; Xu, L.; Yin, R.; De, W. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis. 2016, 7, e2109. [Google Scholar] [CrossRef]
- Niu, Y.; Ma, F.; Huang, W.; Fang, S.; Li, M.; Wei, T.; Guo, L. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol. Cancer 2017, 16, 5. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.-T.; Jiang, D.; Yuan, J.; Cui, Y.-M.; Shi, X.-W.; Chen, C.-M.; Bian, X.-W.; Deng, Y.-J.; Ding, Y.-Q. HOXB7 as a Prognostic Factor and Mediator of Colorectal Cancer Progression. Clin. Cancer Res. 2011, 17, 3569–3578. [Google Scholar] [CrossRef] [PubMed]
- Kunimasa, K.; Nagano, T.; Shimono, Y.; Dokuni, R.; Kiriu, T.; Tokunaga, S.; Tamura, D.; Yamamoto, M.; Tachihara, M.; Kobayashi, K.; et al. Glucose metabolism-targeted therapy and withaferin A are effective for epidermal growth factor receptor tyrosine kinase inhibitor-induced drug-tolerant persisters. Cancer Sci. 2017, 108, 1368–1377. [Google Scholar] [CrossRef] [PubMed]
- Kurppa, K.J.; Liu, Y.; To, C.; Zhang, T.; Fan, M.; Vajdi, A.; Knelson, E.H.; Xie, Y.; Lim, K.; Cejas, P.; et al. Treatment-Induced Tumor Dormancy through YAP-Mediated Transcriptional Reprogramming of the Apoptotic Pathway. Cancer Cell 2020, 37, 104–122.e12. [Google Scholar] [CrossRef] [PubMed]
- Leonce, C.; Saintigny, P.; Ortiz-Cuaran, S. Cell-Intrinsic Mechanisms of Drug Tolerance to Systemic Therapies in Cancer. Mol. Cancer Res. 2022, 20, 11–29. [Google Scholar] [CrossRef] [PubMed]
- Montes, M.; Nielsen, M.M.; Maglieri, G.; Jacobsen, A.; Højfeldt, J.; Agrawal-Singh, S.; Hansen, K.; Helin, K.; van de Werken, H.J.G.; Pedersen, J.S.; et al. The lncRNA MIR31HG regulates p16INK4A expression to modulate senescence. Nat. Commun. 2015, 6, 6967. [Google Scholar] [CrossRef] [PubMed]
- Montes, M.; Lubas, M.; Arendrup, F.S.; Mentz, B.; Rohatgi, N.; Tumas, S.; Harder, L.M.; Skanderup, A.J.; Andersen, J.S.; Lund, A.H. The long non-coding RNA MIR31HG regulates the senescence associated secretory phenotype. Nat. Commun. 2021, 12, 2459. [Google Scholar] [CrossRef] [PubMed]
- Cipponi, A.; Goode, D.L.; Bedo, J.; McCabe, M.J.; Pajic, M.; Croucher, D.R.; Rajal, A.G.; Junankar, S.R.; Saunders, D.N.; Lobachevsky, P.; et al. MTOR signaling orchestrates stress-induced mutagenesis, facilitating adaptive evolution in cancer. Science 2020, 368, 1127–1131. [Google Scholar] [CrossRef] [PubMed]
- Faget, D.V.; Ren, Q.; Stewart, S.A. Unmasking senescence: Context-dependent effects of SASP in cancer. Nat. Rev. Cancer 2019, 19, 439–453. [Google Scholar] [CrossRef] [PubMed]
- Yap, K.L.; Li, S.; Muñoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.-M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 2010, 38, 662–674. [Google Scholar] [CrossRef]
- Muniz, L.; Lazorthes, S.; Delmas, M.; Ouvrard, J.; Aguirrebengoa, M.; Trouche, D.; Nicolas, E. Circular ANRIL isoforms switch from repressors to activators of p15/CDKN2B expression during RAF1 oncogene-induced senescence. RNA Biol. 2021, 18, 404–420. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Gius, D.; Onyango, P.; Muldoon-Jacobs, K.; Karp, J.; Feinberg, A.P.; Cui, H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 2008, 451, 202–206. [Google Scholar] [CrossRef] [PubMed]
- Pasmant, E.; Laurendeau, I.; Héron, D.; Vidaud, M.; Vidaud, D.; Bièche, I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: Identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007, 67, 3963–3969. [Google Scholar] [CrossRef] [PubMed]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Hsieh, C.H.; Alonso, L.C. ANRIL: A lncRNA at the CDKN2A/B Locus with Roles in Cancer and Metabolic Disease. Front. Endocrinol. 2018, 9, 405. [Google Scholar] [CrossRef]
- Wan, G.; Mathur, R.; Hu, X.; Liu, Y.; Zhang, X.; Peng, G.; Lu, X. Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cell Signal. 2013, 25, 1086–1095. [Google Scholar] [CrossRef] [PubMed]
- Menon, D.R.; Das, S.; Krepler, C.; Vultur, A.; Rinner, B.; Schauer, S.; Kashofer, K.; Wagner, K.; Zhang, G.; Rad, E.B.; et al. A stress-induced early innate response causes multidrug tolerance in melanoma. Oncogene 2015, 34, 4448–4459. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Shaffer, S.M.; Dunagin, M.C.; Torborg, S.R.; Torre, E.A.; Emert, B.; Krepler, C.; Beqiri, M.; Sproesser, K.; Brafford, P.A.; Xiao, M.; et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 2017, 546, 431–435. [Google Scholar] [CrossRef]
- Pauli, A.; Rinn, J.L.; Schier, A.F. Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 2011, 12, 136–149. [Google Scholar] [CrossRef]
- Mehravar, M.; Ghaemimanesh, F.; Poursani, E.M. An Overview on the Complexity of OCT4: At the Level of DNA, RNA and Protein. Stem Cell Rev. Rep. 2021, 17, 1121–1136. [Google Scholar] [CrossRef] [PubMed]
- Shahryari, A.; Jazi, M.S.; Samaei, N.M.; Mowla, S.J. Long non-coding RNA SOX2OT: Expression signature, splicing patterns, and emerging roles in pluripotency and tumorigenesis. Front. Genet. 2015, 6, 196. [Google Scholar] [CrossRef] [PubMed]
- Fantes, J.; Ragge, N.K.; Lynch, S.-A.; McGill, N.I.; Collin, J.R.O.; Howard-Peebles, P.N.; Hayward, C.; Vivian, A.J.; Williamson, K.; van Heyningen, V.; et al. Mutations in SOX2 cause anophthalmia. Nat. Genet. 2003, 33, 462–463. [Google Scholar] [CrossRef] [PubMed]
- Shahryari, A.; Rafiee, M.R.; Fouani, Y.; Oliae, N.A.; Samaei, N.M.; Shafiee, M.; Semnani, S.; Vasei, M.; Mowla, S.J. Two Novel Splice Variants of SOX2OT, SOX2OT-S1, and SOX2OT-S2 are Coupregulated with SOX2 and OCT4 in Esophageal Squamous Cell Carcinoma. Stem Cells 2014, 32, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Zhao, W.; Zhou, J.; Shen, L.; Zhan, P.; Xu, C.; Chang, C.; Bi, H.; Zou, J.; Yao, X.; et al. A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. Int. J. Biochem. Cell Biol. 2014, 53, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; He, L.; Du, Y.; Zhu, P.; Huang, G.; Luo, J.; Yan, X.; Ye, B.; Li, C.; Xia, P.; et al. The Long Noncoding RNA lncTCF7 Promotes Self-Renewal of Human Liver Cancer Stem Cells through Activation of Wnt Signaling. Cell Stem Cell 2015, 16, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Jiang, Y.-Y.; Xie, J.-J.; Mayakonda, A.; Hazawa, M.; Chen, L.; Xiao, J.-F.; Li, C.-Q.; Huang, M.-L.; Ding, L.-W.; et al. Co-activation of super-enhancer-driven CCAT1 by TP63 and SOX2 promotes squamous cancer progression. Nat. Commun. 2018, 9, 3619. [Google Scholar] [CrossRef] [PubMed]
- Flores, E.R.; Tsai, K.Y.; Crowley, D.; Sengupta, S.; Yang, A.; McKeon, F.; Jacks, T. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 2002, 416, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Jen, J.; Tang, Y.A.; Lu, Y.H.; Lin, C.C.; Lai, W.W.; Wang, Y.C. Oct4 transcriptionally regulates the expression of long non-coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol. Cancer 2017, 16, 104. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Yang, Z.; Long, F.; Luo, L.; Yang, B.; Zhu, R.; Sang, X.; Cao, G.; Wang, K. Long noncoding RNA MALAT1 mediates stem cell-like properties in human colorectal cancer cells by regulating miR-20b-5p/Oct4 axis. J. Cell Physiol. 2019, 234, 20816–20828. [Google Scholar] [CrossRef]
- Zhu, Y.; Hu, H.; Yuan, Z.; Zhang, Q.; Xiong, H.; Hu, Z.; Wu, H.; Huang, R.; Wang, G.; Tang, Q. LncRNA NEAT1 remodels chromatin to promote the 5-Fu resistance by maintaining colorectal cancer stemness. Cell Death Dis. 2020, 11, 962. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Chen, A.; Wu, X.; Zhou, M.; Ul Haq, I.; Mariyam, Z.; Feng, Q. NEAT1 acts as an inducer of cancer stem cell-like phenotypes in NSCLC by inhibiting EGCG-upregulated CTR1. J. Cell Physiol. 2018, 233, 4852–4863. [Google Scholar] [CrossRef] [PubMed]
- Takakuwa, H.; Yamazaki, T.; Souquere, S.; Adachi, S.; Yoshino, H.; Fujiwara, N.; Yamamoto, T.; Natsume, T.; Nakagawa, S.; Pierron, G.; et al. Shell protein composition specified by the lncRNA NEAT1 domains dictates the formation of paraspeckles as distinct membraneless organelles. Nat. Cell Biol. 2023, 25, 1664–1675. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Wu, F.; Li, Y.; Qin, C.; Zhao, Y.; Xie, M.; Dai, H.; Yao, H.; Cai, H.; Wang, Q.; et al. MALAT1 modulates alternative splicing by cooperating with the splicing factors PTBP1 and PSF. Sci. Adv. 2022, 8, eabq7289. [Google Scholar] [CrossRef]
- Guru, S.C.; Agarwal, S.K.; Manickam, P.; Olufemi, S.-E.; Crabtree, J.S.; Weisemann, J.M.; Kester, M.B.; Kim, Y.S.; Wang, Y.; Emmert-Buck, M.R.; et al. A transcript map for the 2.8-Mb region containing the multiple endocrine neoplasia type 1 locus. Genome Res. 1997, 7, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Valenstein, M.L.; Yario, T.A.; Tycowski, K.T.; Steitz, J.A. Formation of triple-helical structures by the 3’-end sequences of MALAT1 and MENβ noncoding RNAs. Proc. Natl. Acad. Sci. USA 2012, 109, 19202–19207. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Tano, K.; Mizuno, R.; Okada, T.; Rakwal, R.; Shibato, J.; Masuo, Y.; Ijiri, K.; Akimitsu, N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010, 584, 4575–4580. [Google Scholar] [CrossRef]
- Gutschner, T.; Hämmerle, M.; Eißmann, M.; Hsu, J.; Kim, Y.; Hung, G.; Revenko, A.; Arun, G.; Stentrup, M.; Groß, M.; et al. The Noncoding RNA MALAT1 Is a Critical Regulator of the Metastasis Phenotype of Lung Cancer Cells. Cancer Res. 2013, 73, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.T.; Ideue, T.; Sano, M.; Mituyama, T.; Hirose, T. MENε/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA 2009, 106, 2525–2530. [Google Scholar] [CrossRef]
- Yamazaki, T.; Nakagawa, S.; Hirose, T. Architectural RNAs for Membraneless Nuclear Body Formation. Cold Spring Harb. Symp. Quant. Biol. 2019, 84, 227–237. [Google Scholar] [CrossRef]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef]
- Fan, X.-J.; Wang, Y.-L.; Zhao, W.-W.; Bai, S.-M.; Ma, Y.; Yin, X.-K.; Feng, L.-L.; Feng, W.-X.; Wang, Y.-N.; Liu, Q.; et al. NONO phase separation enhances DNA damage repair by accelerating nuclear EGFR-induced DNA-PK activation. Am. J. Cancer Res. 2021, 11, 2838–2852. [Google Scholar]
- Levone, B.R.; Lenzken, S.C.; Antonaci, M.; Maiser, A.; Rapp, A.; Conte, F.; Reber, S.; Mechtersheimer, J.; Ronchi, A.E.; Mühlemann, O.; et al. FUS-dependent liquid-liquid phase separation is important for DNA repair initiation. J. Cell Biol. 2021, 220, e202008030. [Google Scholar] [CrossRef]
- Desai, T.J.; Brownfield, D.G.; Krasnow, M.A. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 2014, 507, 190–194. [Google Scholar] [CrossRef]
- Zepp, J.A.; Morrisey, E.E. Cellular crosstalk in the development and regeneration of the respiratory system. Nat. Rev. Mol. Cell Biol. 2019, 20, 551–566. [Google Scholar] [CrossRef]
- Zhao, W.; Luo, J.; Jiao, S. Comprehensive characterization of cancer subtype associated long non-coding RNAs and their clinical implications. Sci. Rep. 2014, 4, 6591. [Google Scholar] [CrossRef]
- Teng, F.; Zhang, J.; Chen, Y.; Shen, X.; Su, C.; Guo, Y.; Wang, P.; Shi, C.; Lei, M.; Cao, Y.; et al. LncRNA NKX2-1-AS1 promotes tumor progression and angiogenesis via upregulation of SERPINE1 expression and activation of the VEGFR-2 signaling pathway in gastric cancer. Mol. Oncol. 2021, 15, 1234–1255. [Google Scholar] [CrossRef]
- Singh, N.; Ramnarine, V.R.; Song, J.H.; Pandey, R.; Padi, S.K.R.; Nouri, M.; Olive, V.; Kobelev, M.; Okumura, K.; McCarthy, D.; et al. The long noncoding RNA H19 regulates tumor plasticity in neuroendocrine prostate cancer. Nat. Commun. 2021, 12, 7349. [Google Scholar] [CrossRef]
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic reprogramming and cancer progression. Science 2020, 368, eaaw5473. [Google Scholar] [CrossRef]
- Hangauer, M.J.; Viswanathan, V.S.; Ryan, M.J.; Bole, D.; Eaton, J.K.; Matov, A.; Galeas, J.; Dhruv, H.D.; Berens, M.E.; Schreiber, S.L.; et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 2017, 551, 247–250. [Google Scholar] [CrossRef]
- Cargill, K.R.; Hasken, W.L.; Gay, C.M.; Byers, L.A. Alternative Energy: Breaking Down the Diverse Metabolic Features of Lung Cancers. Front. Oncol. 2021, 11, 757323. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Li, Y.; Chen, H.; Xie, X.; Yang, B.; Wang, X.; Zhang, J.; Qiao, T.; Guan, J.; Qiu, Y.; Huang, Y.-X.; et al. PINK1-Mediated Mitophagy Promotes Oxidative Phosphorylation and Redox Homeostasis to Induce Drug-Tolerant Persister Cancer Cells. Cancer Res. 2023, 83, 398–413. [Google Scholar] [CrossRef]
- Roesch, A.; Vultur, A.; Bogeski, I.; Wang, H.; Zimmermann, K.M.; Speicher, D.; Körbel, C.; Laschke, M.W.; Gimotty, P.A.; Philipp, S.E.; et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell 2013, 23, 811–825. [Google Scholar] [CrossRef]
- Connerty, P.; Lock, R.B.; de Bock, C.E. Long Non-coding RNAs: Major Regulators of Cell Stress in Cancer. Front. Oncol. 2020, 10, 285. [Google Scholar] [CrossRef]
- Fan, N.; Fu, H.; Feng, X.; Chen, Y.; Wang, J.; Wu, Y.; Bian, Y.; Li, Y. Long non-coding RNAs play an important regulatory role in tumorigenesis and tumor progression through aerobic glycolysis. Front. Mol. Biosci. 2022, 9, 941653. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, H.; Mei, Y.; Wu, M. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol. Cell 2014, 53, 88–100. [Google Scholar] [CrossRef]
- Park, M.K.; Zhang, L.; Min, K.-W.; Cho, J.-H.; Yeh, C.-C.; Moon, H.; Hormaechea-Agulla, D.; Mun, H.; Ko, S.; Lee, J.W.; et al. NEAT1 is essential for metabolic changes that promote breast cancer growth and metastasis. Cell Metab. 2021, 33, 2380–2397.e9. [Google Scholar] [CrossRef]
- Zhu, Y.; Jin, L.; Shi, R.; Li, J.; Wang, Y.; Liang, C.-Z.; Narayana, V.K.; De Souza, D.P.; Thorne, R.F.; Zhang, L.R.; et al. The long noncoding RNA glycoLINC assembles a lower glycolytic metabolon to promote glycolysis. Mol. Cell 2022, 82, 542–554.e6. [Google Scholar] [CrossRef]
- Xiang, S.; Gu, H.; Jin, L.; Thorne, R.F.; Zhang, X.D.; Wu, M. LncRNA IDH1-AS1 links the functions of c-Myc and HIF1α via IDH1 to regulate the Warburg effect. Proc. Natl. Acad. Sci. USA 2018, 115, E1465–E1474. [Google Scholar] [CrossRef]
- Mao, C.; Liu, X.; Zhang, Y.; Lei, G.; Yan, Y.; Lee, H.; Koppula, P.; Wu, S.; Zhuang, L.; Fang, B.; et al. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature 2021, 593, 586–590. [Google Scholar] [CrossRef]
- Leon, L.M.; Gautier, M.; Allan, R.; Ilié, M.; Nottet, N.; Pons, N.; Paquet, A.; Lebrigand, K.; Truchi, M.; Fassy, J.; et al. The nuclear hypoxia-regulated NLUCAT1 long non-coding RNA contributes to an aggressive phenotype in lung adenocarcinoma through regulation of oxidative stress. Oncogene 2019, 38, 7146–7165. [Google Scholar] [CrossRef]
- Russo, M.; Sogari, A.; Bardelli, A. Adaptive Evolution: How Bacteria and Cancer Cells Survive Stressful Conditions and Drug Treatment. Cancer Discov. 2021, 11, 1886–1895. [Google Scholar] [CrossRef]
- Russo, M.; Pompei, S.; Sogari, A.; Corigliano, M.; Crisafulli, G.; Puliafito, A.; Lamba, S.; Erriquez, J.; Bertotti, A.; Gherardi, M.; et al. A modified fluctuation-test framework characterizes the population dynamics and mutation rate of colorectal cancer persister cells. Nat. Genet. 2022, 54, 976–984. [Google Scholar] [CrossRef]
- Juvekar, A.; Burga, L.N.; Hu, H.; Lunsford, E.P.; Ibrahim, Y.H.; Balmaña, J.; Rajendran, A.; Papa, A.; Spencer, K.; Lyssiotis, C.A.; et al. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov. 2012, 2, 1048–1063. [Google Scholar] [CrossRef] [PubMed]
- Temprine, K.; Campbell, N.R.; Huang, R.; Langdon, E.M.; Simon-Vermot, T.; Mehta, K.; Clapp, A.; Chipman, M.; White, R.M. Regulation of the error-prone DNA polymerase Polκ by oncogenic signaling and its contribution to drug resistance. Sci. Signal. 2020, 13, eaau1453. [Google Scholar] [CrossRef]
- Gay, C.M.; Balaji, K.; Byers, L.A. Giving AXL the axe: Targeting AXL in human malignancy. Br. J. Cancer 2017, 116, 415–423. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Khoshbakht, T.; Taheri, M.; Mokhtari, M. A review on the role of GAS6 and GAS6-AS1 in the carcinogenesis. Pathol. Res. Pract. 2021, 226, 153596. [Google Scholar] [CrossRef]
- Zhang, P.; Dong, Q.; Zhu, H.; Li, S.; Shi, L.; Chen, X. Long non-coding antisense RNA GAS6-AS1 supports gastric cancer progression via increasing GAS6 expression. Gene 2019, 696, 1–9. [Google Scholar] [CrossRef]
- Luo, J.; Wang, H.; Wang, L.; Wang, G.; Yao, Y.; Xie, K.; Li, X.; Xu, L.; Shen, Y.; Ren, B. lncRNA GAS6-AS1 inhibits progression and glucose metabolism reprogramming in LUAD via repressing E2F1-mediated transcription of GLUT1. Mol. Ther. Nucleic Acids 2021, 25, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.R.; Yan, L.; Liu, Y.T.; Cao, L.; Guo, Y.H.; Zhang, Y.; Yao, H.; Cai, L.; Shang, H.B.; Rui, W.W.; et al. Inhibition of mTORC1 by lncRNA H19 via disrupting 4E-BP1/Raptor interaction in pituitary tumours. Nat. Commun. 2018, 9, 4624. [Google Scholar] [CrossRef]
- Li, Z.; Li, X.; Wu, S.; Xue, M.; Chen, W. Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci. 2014, 105, 951–955. [Google Scholar] [CrossRef] [PubMed]
- Malakar, P.; Shilo, A.; Mogilevsky, A.; Stein, I.; Pikarsky, E.; Nevo, Y.; Benyamini, H.; Elgavish, S.; Zong, X.; Prasanth, K.V.; et al. Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation. Cancer Res. 2017, 77, 1155–1167. [Google Scholar] [CrossRef] [PubMed]
- Crooke, S.T.; Baker, B.F.; Crooke, R.M.; Liang, X.-H. Antisense technology: An overview and prospectus. Nat. Rev. Drug Discov. 2021, 20, 427–453. [Google Scholar] [CrossRef] [PubMed]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef] [PubMed]
- Barry, G.; Briggs, J.A.; Hwang, D.W.; Nayler, S.P.; Fortuna, P.R.J.; Jonkhout, N.; Dachet, F.; Maag, J.L.V.; Mestdagh, P.; Singh, E.M.; et al. The long non-coding RNA NEAT1 is responsive to neuronal activity and is associated with hyperexcitability states. Sci. Rep. 2017, 7, 40127. [Google Scholar] [CrossRef]
- Perez-Pinera, P.; Kocak, D.D.; Vockley, C.M.; Adler, A.F.; Kabadi, A.M.; Polstein, L.R.; Thakore, P.I.; Glass, K.A.; Ousterout, D.G.; Leong, K.W.; et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 2013, 10, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef]
- Aparicio-Prat, E.; Arnan, C.; Sala, I.; Bosch, N.; Guigó, R.; Johnson, R. DECKO: Single-oligo, dual-CRISPR deletion of genomic elements including long non-coding RNAs. BMC Genom. 2015, 16, 846. [Google Scholar] [CrossRef] [PubMed]
- Paunovska, K.; Loughrey, D.; Dahlman, J.E. Drug delivery systems for RNA therapeutics. Nat. Rev. Genet. 2022, 23, 265–280. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.J.; Revenko, A.S.; Hanson, L.L.; Ellston, R.; Staniszewska, A.; Whalley, N.; Pandey, S.K.; Revill, M.; Rooney, C.; Buckett, L.K.; et al. Targeting KRAS-dependent tumors with AZD4785, a high-affinity therapeutic antisense oligonucleotide inhibitor of KRAS. Sci. Transl. Med. 2017, 9, eaal5253. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Pant, S. Personalizing Medicine with Germline and Somatic Sequencing in Advanced Pancreatic Cancer: Current Treatments and Novel Opportunities. Am. Soc. Clin. Oncol. Educ. Book 2021, 41, e153–e165. [Google Scholar] [CrossRef] [PubMed]
- Wiesenfarth, M.; Dorst, J.; Brenner, D.; Elmas, Z.; Parlak, Ö.; Uzelac, Z.; Kandler, K.; Mayer, K.; Weiland, U.; Herrmann, C.; et al. Effects of tofersen treatment in patients with SOD1-ALS in a “real-world” setting—A 12-month multicenter cohort study from the German early access program. eClinicalMedicine 2024, 69, 102495. [Google Scholar] [CrossRef] [PubMed]
- Durcan, N.; Murphy, C.; Cryan, S.-A. Inhalable siRNA: Potential as a Therapeutic Agent in the Lungs. Mol. Pharm. 2008, 5, 559–566. [Google Scholar] [CrossRef]
- Shaffer, C. Mist begins to clear for lung delivery of RNA. Nat. Biotechnol. 2020, 38, 1110–1112. [Google Scholar] [CrossRef]
- Qiu, Y.; Lam, J.K.W.; Leung, S.W.S.; Liang, W. Delivery of RNAi Therapeutics to the Airways—From Bench to Bedside. Molecules 2016, 21, 1249. [Google Scholar] [CrossRef] [PubMed]
- Wilbie, D.; Walther, J.; Mastrobattista, E. Delivery Aspects of CRISPR/Cas for in Vivo Genome Editing. Acc. Chem. Res. 2019, 52, 1555–1564. [Google Scholar] [CrossRef]
- Abulwerdi, F.A.; Xu, W.; Ageeli, A.A.; Yonkunas, M.J.; Arun, G.; Nam, H.; Schneekloth, J.J.S.; Dayie, T.K.; Spector, D.; Baird, N.; et al. Selective Small-Molecule Targeting of a Triple Helix Encoded by the Long Noncoding RNA, MALAT1. ACS Chem. Biol. 2019, 14, 223–235. [Google Scholar] [CrossRef]
- Childs-Disney, J.L.; Yang, X.; Gibaut, Q.M.R.; Tong, Y.; Batey, R.T.; Disney, M.D. Targeting RNA structures with small molecules. Nat. Rev. Drug Discov. 2022, 21, 736–762. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.N.; Sattler, M. Challenges and perspectives for structural biology of lncRNAs-the example of the Xist lncRNA A-repeats. J. Mol. Cell Biol. 2019, 11, 845–859. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fei, Y.; Sun, L.; Zhang, Q.C. Advances and opportunities in RNA structure experimental determination and computational modeling. Nat. Methods 2022, 19, 1193–1207. [Google Scholar] [CrossRef] [PubMed]
- Flynn, R.A.; Zhang, Q.C.; Spitale, R.C.; Lee, B.; Mumbach, M.R.; Chang, H.Y. Transcriptome-wide interrogation of RNA secondary structure in living cells with icSHAPE. Nat. Protoc. 2016, 11, 273–290. [Google Scholar] [CrossRef] [PubMed]
- Townshend, R.J.L.; Eismann, S.; Watkins, A.M.; Rangan, R.; Karelina, M.; Das, R.; Dror, R.O. Geometric deep learning of RNA structure. Science 2021, 373, 1047–1051. [Google Scholar] [CrossRef] [PubMed]
- Feyder, M.; Goff, L.A. Investigating long noncoding RNAs using animal models. J. Clin. Investig. 2016, 126, 2783–2791. [Google Scholar] [CrossRef][Green Version]
- Zhang, B.; Mao, Y.S.; Diermeier, S.D.; Novikova, I.V.; Nawrocki, E.P.; Jones, T.A.; Lazar, Z.; Tung, C.-S.; Luo, W.; Eddy, S.R.; et al. Identification and Characterization of a Class of MALAT1-like Genomic Loci. Cell Rep. 2017, 19, 1723–1738. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davis, W.J.H.; Drummond, C.J.; Diermeier, S.; Reid, G. The Potential Links between lncRNAs and Drug Tolerance in Lung Adenocarcinoma. Genes 2024, 15, 906. https://doi.org/10.3390/genes15070906
Davis WJH, Drummond CJ, Diermeier S, Reid G. The Potential Links between lncRNAs and Drug Tolerance in Lung Adenocarcinoma. Genes. 2024; 15(7):906. https://doi.org/10.3390/genes15070906
Chicago/Turabian StyleDavis, William J. H., Catherine J. Drummond, Sarah Diermeier, and Glen Reid. 2024. "The Potential Links between lncRNAs and Drug Tolerance in Lung Adenocarcinoma" Genes 15, no. 7: 906. https://doi.org/10.3390/genes15070906
APA StyleDavis, W. J. H., Drummond, C. J., Diermeier, S., & Reid, G. (2024). The Potential Links between lncRNAs and Drug Tolerance in Lung Adenocarcinoma. Genes, 15(7), 906. https://doi.org/10.3390/genes15070906






