Abstract
The model haloarchaeon Haloferax volcanii is polyploid with about 20 copies of its major chromosome. Recently it has been described that highly efficient intermolecular gene conversion operates in H. volcanii to equalize the chromosomal copies. In the current study, 24 genes were selected that encode proteins with orthologs involved in gene conversion or homologous recombination in archaea, bacteria, or eukaryotes. Single gene deletion strains of 22 genes and a control gene were constructed in two parent strains for a gene conversion assay; only radA and radB were shown to be essential. Protoplast fusions were used to generate strains that were heterozygous for the gene HVO_2528, encoding an enzyme for carotinoid biosynthesis. It was revealed that a lack of six of the proteins did not influence the efficiency of gene conversion, while sixteen mutants had severe gene conversion defects. Notably, lack of paralogous proteins of gene families had very different effects, e.g., mutant Δrad25b had no phenotype, while mutants Δrad25a, Δrad25c, and Δrad25d were highly compromised. Generation of a quadruple rad25 and a triple sph deletion strain also indicated that the paralogs have different functions, in contrast to sph2 and sph4, which cannot be deleted simultaneously. There was no correlation between the severity of the phenotypes and the respective transcript levels under non-stressed conditions, indicating that gene expression has to be induced at the onset of gene conversion. Phylogenetic trees of the protein families Rad3/25, MutL/S, and Sph/SMC/Rad50 were generated to unravel the history of the paralogous proteins of H. volcanii. Taken together, unselected intermolecular gene conversion in H. volcanii involves at least 16 different proteins, the molecular roles of which can be studied in detail in future projects.
Keywords:
archaea; Haloferax volcanii; polyploidy; gene conversion; homologous recombination; DNA repair; holiday junction; MutL; Hjc; NucS 1. Introduction
Gene conversion is defined as the non-reciprocal flow of information between two homologous but not identical DNA sequences. Gene conversion occurs in all three domains of life, archaea, bacteria, and eukaryotes. By far the most studies have been performed with eukaryotes, which have led to several thousand publications. In eukaryotes, gene conversion is involved, e.g., in meiosis and the development of the immune system. However, gene conversion in eukaryotes is not part of this study, and reviews about various aspects are available for those interested in this topic [1,2,3,4,5,6,7,8].
Gene conversion in bacteria is involved in several biological processes. One such process is antigenic variation, which enables pathogens to escape the immune system of the host [9]. To this end, the genome of the pathogen contains one expression site for a surface protein and several to many silent paralogous genes lacking a promoter. With low frequencies, the gene at the expression site is converted by one of the silent genes, leading to a change in the surface structure of the pathogen, which makes the immune response obsolete. The number of silent copies can vary widely even within one genus, e.g., there are about 6 silent copies in Anaplasma marginale and more than 100 silent copies in A. phagocytophilum [10]. The eukaryotic pathogen Trypanosoma brucei contains even more than 1500 silent copies of the variant surface glycoprotein [11]. In antigenic variation, gene conversion results in generating diversity within the population of pathogens. The number of possible variants is even much higher than the number of silent sites because the gene conversion tracts are very short and therefore only parts of the silent copies are involved in gene conversion, leading to novel hybrid genes at the expression site. At least in bacteria, gene conversion during antigenic variation occurs at different sites of the same chromosome and is thus an intramolecular event.
Another example for intramolecular gene conversion is the concerted evolution of gene families, which also occurs in species of various phylogenetic groups [9,12,13]. In stark contrast to antigenic variation, gene conversion in concerted evolution leads to the equalization of several copies of a gene that are present in one genome. One example are the genes for the ribosomal RNAs (rRNAs). Most bacteria contain more than one rRNA operon [12]. For example, Escherichia coli contains seven rRNA operons, and in other bacterial species the number can be as high as fifteen. Concerted evolution has also been observed in families of protein-coding genes, e.g., the tuf genes in Salmonella or the nif genes in Rhizobium [14,15]. Also, in concerted evolution, the gene conversion frequencies are very low, so that selection steps have to be involved for the identification and characterization of gene conversion events, and similar to antigenic variation, the gene conversion tract lengths in concerted evolution are rather short, so that only fractions of genes are converted in one event. On the one hand, concerted evolution can correct mutations that have occurred in a single of several gene copies and preserve the wildtype sequence, while on the other hand, it can also spread advantageous mutations from the gene copy in which they have occurred to the other copies of the gene family.
Another process that results in equalization of gene copies is intermolecular gene conversion between different copies of the chromosome in polyploid prokaryotes. Until now, this process has only been studied in halophilic archaea [16,17,18], in methanogenic archaea [19], in chloroplasts [20,21,22,23], and in mitochondria [24,25,26]. Intermolecular gene conversion in polyploid bacteria has not been studied yet. However, it can be expected to also operate in bacteria and to result in the equalization of genome copies, because (1) genome sequencing projects yield unambiguous sequences and do not point to heterozygosity, (2) heterozygous cells have been obtained with several bacterial species, but laboratory selection was needed for their generation [27,28,29,30], and (3) homozygous mutants can easily be generated in polyploid archaea and bacteria, although initially only one of many copies is mutagenized [31,32]. However, one exception has been reported, the giant bacterium Achromatium oxaliferum was shown to be naturally heterozygous [33].
In contrast to antigenic variation and concerted evolution, intermolecular gene conversion in polyploid archaea has a high efficiency, which enables its direct characterization without a selection step that enriches for a very seldom event. Recently, an experimental approach for studying unselected intermolecular gene conversion between different genome copies has been reported for H. volcanii [18]. To this end, two strains were generated that contained two different versions of a gene encoding an enzyme involved in carotinoid biosynthesis, i.e., an active version leading to red cells and an inactive version leading to white cells. The two strains were combined via protoplast fusion, resulting in heterozygous cells containing both types of genomes. The efficiency of gene conversion can be analyzed via the color of the resulting recombinants (cells that became homozygous for the inactive copy are white) or via PCR analyses. The analyses not only confirmed the high efficiency of gene conversion but also revealed that the conversion tracts are much longer than in antigenic variation or concerted evolution and can in fact span several thousand base pairs [18]. In addition, it was found that gene conversion was triggered by very small differences between the genome copies and that even sequences from very different species become converted.
In the present study, we applied this approach to identify proteins that are involved in and important for intermolecular gene conversion in H. volcanii. More than 20 genes were selected that encoded proteins that might be involved in homologous recombination or different DNA repair pathways. Single gene deletion mutants of all genes (with the exception of two essential genes) were generated, and the effect on the efficiency of gene conversion was quantified. In addition, one triple mutant and one quadruple mutant of paralogous genes were also generated and tested. The results revealed that several proteins did not affect the efficiency of gene conversion, while the majority had a very profound effect. To unravel the history of the paralogous H. volcanii members of three protein families, phylogenetic trees were generated with proteins from archaea, bacteria, and eukaryotes.
2. Materials and Methods
2.1. Strains, Media, and Growth Conditions
All strains used in this study are derivatives of the H. volcanii wildtype DS2 [34]. All strains contain a deletion of the pyrE2 (HVO_0333) encoding an enzyme for uracil biosynthesis, which is necessary for the efficient construction of further mutants [35]. One of the two parent strains (H53) used for protoplast fusion (see below) contains a further deletion in trpA (HVO_0789), making it auxotrophic for tryptophan [31]. The other parent strain (So0994) contains a deletion in thyA, making it auxotrophic for thymidine, and crtD (HVO_2528), leading to white colonies. Further mutants were generated in the framework of this project, as described below.
The H. volcanii strains were grown in complex medium or in synthetic medium with 0.5% (w/v) glucose as sole carbon and energy source [36]. If necessary, the medium was supplemented with 50 µg/mL uracil, 50 µg/mL tryptophan, or/and 20 µg/mL thymidine to allow the growth of auxotrophic strains. The cultures were grown at 42 °C with good aeration (250 rpm). Solid media contained 1.4% (w/v) agar.
The E. coli strain XL1-blue MRF’ (Agilent Technologies, Waldbronn, Germany) was used for cloning. It was grown in SOB complex medium [37].
2.2. Generation of In-Frame Deletion Mutants
The in-frame deletion mutants were generated using the so-called Pop-In-Pop-Out method as described previously [31,38]. In short, for each gene two PCR fragments were generated that contained, respectively, the upstream region and a short 5′-part of the gene, and a short 3′-part of the gene and the downstream region. All primers are listed in Supplementary Table S1. The two PCR fragments were fused and cloned into the vector pMH101 via restriction selection cloning [39]. The resulting plasmids were verified by sequencing and used to transform the two parent strains H53 and So0994. Cultivation in medium lacking uracil selected for clones that had integrated the plasmids at the respective genomic sites (Pop-In). Subsequent cultivation of the Pop-In clones in medium with 5-FOA (5-fluororotic acid) and uracil selected for clones that had lost the integrated vector based on a second recombination event (Pop-Out). Colony PCR was used to identify clones that contained the deletion version in the genome. Because H. volcanii is polyploid, clones can be heterozygous in spite of the high efficiency of gene conversion. Therefore, genomic DNA was isolated and analyzed via Southern blotting and with a PCR with 40 cycles. Table 1 summarizes the 23 genes that were deleted in both parent strains.

Table 1.
List of genes included in this study with protein names and annotated functions.
2.3. Growth Analyses
H. volcanii can be grown in microtiter plates, which enables the characterization of many strains in parallel [36]. Growth of all 46 strains was monitored in complex medium as well as in synthetic medium with 0.5% (w/v) glucose as the sole source of carbon and energy. In short, precultures were grown to the mid-exponential growth phase and used to inoculate test cultures with an OD600 of 0.05. The microtiter plates were incubated at 42 °C on a Heidolph Titramax 1000 rotary shaker with 1100 rpm. The OD600 was determined at the indicated time points using a Spectramax 340 photometer (Molecular Devices, Ismaning, Germany). Three biological replicates were performed, and average values and their standard deviations were calculated.
2.4. Analyses of Gene Conversion Efficiencies
To test the possible influence of the 22 selected proteins and the control protein DHFR (Table 1) on the efficiency of intermolecular gene conversion, pairs of cultures were grown to mid-exponential growth phase that contained the deletion of a specific gene in the two parent strains H53 and So0994. A total of 8 × 108 cells of both strains were fused by protoplast fusion as described earlier [18,40,41]. Fused cells were selected in synthetic medium lacking tryptophan and thymidine, which does not allow growth of the parent strains. This generates cells that contain about 20 copies of each of the two genomes, which contained a wildtype copy of the gene HVO_2528 (carotinoid biosynthesis proficient) and a deletion version of the gene (carotinoid biosynthesis deficient). Directly after protoplast fusion, many clones are still heterozygous, and plating results in a high fraction of sectored colonies. To allow completion of gene conversion, the mixture of fused cells was inoculated in 30 mL synthetic medium and incubated for 24 h. Then, serial tenfold dilutions were generated and spread on agar plates with synthetic medium. After five days of incubation at 42 °C, colonies had been formed. The plates were moved to room temperature and incubated for a further 5 days, which facilitates the differentiation between white colonies, red colonies, and sectored colonies. The three types of colonies were counted separately using plates with a suitable dilution (50–500 colonies per plate), and the copy number of the undiluted culture was calculated. At least four biological replicates of all protoplast fusion were performed, and average values and their standard deviations were calculated. The numbers were normalized to the copy numbers of control protoplast fusions with the two parent strains. The significance of observed values between the test strains and the parent strains was calculated using an unpaired two-tailed t-test.
2.5. Bioinformatic Analyses
The genome database Halolex [42] was used to retrieve sequences from the genome of H. volcanii and to address the newest genome annotation. The clone manager Professional Suite version 8 (Sci Ed Software, Westminster, CO, USA) was used for the experimental design. The program MEGA X with the algorithm MUSCLE was used to generate multiple sequence alignments of various orthologues and paralogues of the studied proteins. The phylogenetic trees were constructed on the basis of multiple sequence alignments. The program MEGA X [43] was used for tree construction, and in each case the maximum likelihood, maximum parsimony, and neighbor-joining approaches were used. A total of 1000 bootstrap repetitions were performed, and the results (%) were added to selected nodes. In addition, individual neighbor-joining trees were generated, which allowed us to include the actual branch lengths.
The results from previous RNA and dRNA sequencing studies [44,45] were analyzed to gain insights into the expression of selected genes. The Integrated Genome Browser [46] was used to visualize the data.
3. Results
3.1. Experimental Design for the Analysis of Unselected Gene Conversion
Recently, we have developed an experimental approach to quantify the efficiency of unselected intermolecular gene conversion in H. volcanii [18]. An extended version was used to analyze the importance of more than 20 selected proteins on gene conversion. A schematic overview of the approach is given in Figure 1. In short, two parent strains were generated that have an intact or an inactivated version of the gene HVO_2528, respectively. The gene encodes an enzyme involved in carotinoid biosynthesis and the strains are thus red and white, respectively. In addition, the strains contain a deletion either in trpA (essential for tryptophan biosynthesis) or thyA (essential for thymidine biosynthesis). In the absence of tryptophan and thymidine, neither of the two parent strains can grow. However, after protoplast fusion of the two parent strains, heterozygous fusion cells contain a wildtype copy of both trpA and thyA, are prototrophic for tryptophan and thymidine, and can be selected for in the absence of both substances. Unselected gene conversion can occur at the HVO_2528 locus either in the direction of the wildtype version or in the direction of the deletion version. If the deletion version becomes homozygous, the cells become white. Therefore, the fraction of white colonies can be taken as a quantitative value for the efficiency of gene conversion in the direction of the deletion version. This allows the very fast and easy analysis of gene conversion efficiencies in one direction in thousands of colonies. In contrast, red colonies can either be heterozygous (as directly after protoplast fusion) or homozygous for the wildtype version after gene conversion in this direction. To discriminate between these two possibilities, PCR analysis has to be applied. As colony PCR is established for H. volcanii, hundreds (but not thousands) or clones can easily be analyzed.
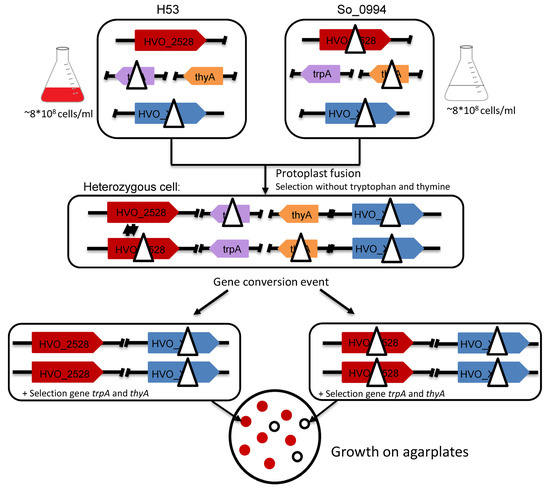
Figure 1.
Schematic overview of the experimental design of gene conversion experiments with H. volcanii. Two parent strains with nearly identical genomes are generated, the only difference is that H53 contains a deletion in the trpA gene and So_0994 carries deletions in the genes thyA and HVO_2528, as indicated. To test the possible importance of selected proteins on the efficiency of gene conversion, the respective genes (indicated in blue) were deleted in both parent strains. The two parent strains were fused by protoplast fusion, and fused heterozygous cells (middle) were selected by growth in the absence of tryptophan and thymidine. After gene conversion, two different homozygous cells (concerning HVO_2528) are possible, which are indicated in the lower part of the figure. The genes are color-coded to enable an easy overview.
For the analysis of the importance of selected proteins for intermolecular gene conversion, the cognate genes have to be deleted in both parent strains. After protoplast fusion, the fusion cells are heterozygous for HVO_2528 but homozygous for the deletion version of the gene under investigation, and the effect of the absence of the encoded protein can be quantified. Initially, it was expected that the fraction of white colonies could be taken as readout. However, during the course of the project, it turned out that the number of surviving cells was more informative (see below).
3.2. Overview of Generated and Analyzed In-Frame Deletion Mutants
A literature search was performed to identify proteins that have been shown to be involved in “gene conversion” or “homologous recombination” in archaea, bacteria, or eukaryotes. If a clear homolog was encoded in the genome of H. volcanii, the respective gene was considered as a candidate for the analysis. The gene dhfr, encoding dihydrofolate reductase, was added as a negative control because it was assumed that this metabolic enzyme should not play a role in the molecular mechanism of gene conversion. Table 1 gives an overview of the selected genes and the annotated functions of the encoded proteins.
Single gene deletion mutants were generated for the 23 genes in both parent strains H53 and So_0994, resulting in a set of 46 strains. Only the genes radA and radB could not be deleted and were thus regarded as essential. The so-called Pop-In-Pop-Out method was applied for mutant construction [31,39,47]. Supplementary Table S1 lists the primers that were used for mutant generation. H. volcanii is polyploid with about 20 copies of its major chromosome [48]; therefore, deletion strains sometimes retain one or a few copies of the wildtype allele. To exclude this possibility, all deletion mutants were checked with both PCR analysis with 40 cycles and with Southern blot analysis. The analysis of deletion mutant ΔHVO_0191 is shown in Figure 2A (PCR analysis) and Figure 2B (Southern blot analysis) to exemplify the results. Homologous deletions could be verified for all 46 strains.
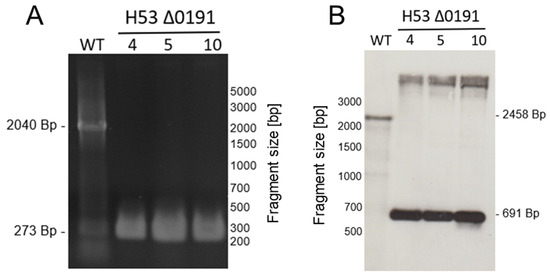
Figure 2.
Verification of homozygosity of newly generated deletion mutants. The deletion mutants were compared with the wildtype using two different approaches, i.e., multicycle PCR and Southern blotting. (A) Results of a multicycle PCR for the wildtype and three clones of mutant Δ0191. (B) Results of a Southern blot for the wildtype and three clones of mutant Δ0191. Mutant Δ0191 in the parent strain H53 is shown to exemplify the analyses. The two analyses were performed for all 46 mutants.
3.3. Growth Analysis of Deletion Mutants
Growth of all deletion strains was compared with that of the wildtype. To this end, the strains were grown in microtiter plates as described [36]. Three biological replicates were performed. Figure 3A shows the results of the growth experiments for the 23 deletion mutants in the parent strain H53 in complex medium. With one exception, all strains grew indistinguishably. Only deletion strain ΔHVO_B0118, lacking the gene for the protein Sph2, had a slightly reduced growth rate, but it reached the same growth yield as all other strains. The same result was obtained with the 23 deletion strains in the parent strain So_0994. Figure 3B shows the growth of the 23 deletion mutants and the wildtype in synthetic medium with glucose as the carbon and energy source. This was tested because the selection after protoplast fusion has to be performed in synthetic medium. Again, only strain ΔHVO_B0118 had a slight growth defect, while all other strains grew indistinguishably. The same result was obtained with the 23 deletion strains in the parent strain So_0994. These results revealed that under optimal growth conditions, none of the 22 DNA repair and recombination proteins is so important that its absence results in a severe growth defect. Of course, the importance of several of these proteins might be much higher after the application of mutagenic conditions or stress, which, however, were not part of the current project. Therefore, all mutants were suited to analyzing the effect of the lack of the proteins on the efficiency of gene conversion.
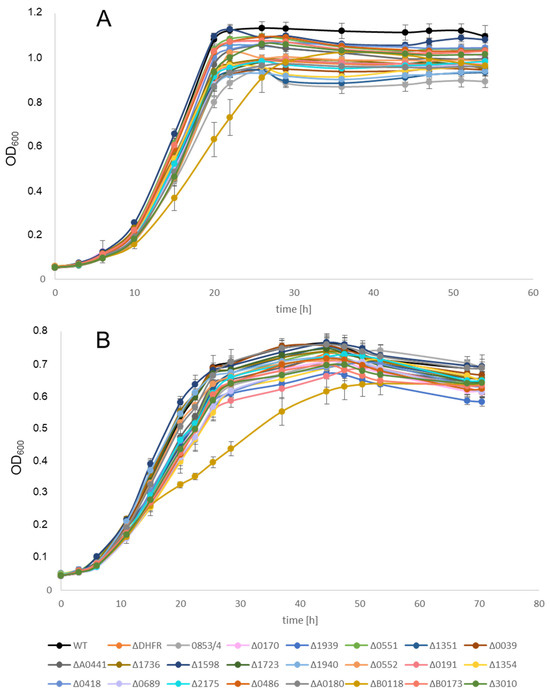
Figure 3.
Growth curves of the wildtype and 23 deletion mutants (in the parent strain H53). The growth was performed in microtiter plates. Three biological replicates were performed. Average values and their standard deviation are shown. (A) Growth in complex medium. (B) Growth in synthetic medium with glucose as carbon and energy source.
3.4. Gene Conversion Efficiencies of Wildtype and Mutants
Gene conversion experiments were performed with the two parent strains and the 23 deletion mutants of each of the two parent strains. The generation of—very sensitive—protoplasts, their fusion in the presence of PEG, and the selection of fusion cells might have a higher variance than other methods that do not compromise the integrity of the S-layer; therefore, four biological replicates were performed for each gene conversion experiment. It was expected that lack of a protein with an important function for gene conversion would lead to a lower gene conversion efficiency, which would result in a lower fraction of white colonies (with complete conversion of HVO_2528 to the deletion variant). Surprisingly, this was not observed. Figure 4 summarizes the fractions of red, white, and sectored colonies after all 24 gene conversion experiments. There was no significant difference between the 2 controls (parent strains, parent strains lacking the metabolic enzyme DHFR) and the 22 gene conversion experiments with strains lacking a protein annotated to be involved in homologous recombination or DNA repair. It is highly unlikely that none of the 22 proteins are involved in gene conversion; therefore, the fraction of white colonies was obviously not informative about the efficiency of gene conversion, in contrast to the expectation.
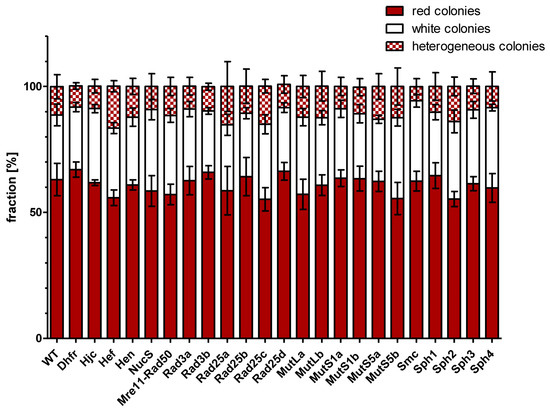
Figure 4.
The fractions of red, white, and sectored colonies after gene conversion experiments are shown for the parent strains (wt) and 23 deletion mutants. Average values of four biological replicates and their standard deviation are shown.
However, we noticed that the number of clones that survived the gene conversion experiments was highly variable and that the reduced survival rates were clone-specific and highly reproducible. The possibility can be excluded that the allele of the carotenoid biosynthesis gene HVO_2528 was related to the differences in survival because the two parent strains with the native and the inactivated allele grew equally well and because carotenoids do not have a vital function during non-stressed growth in the dark. The most plausible explanation is that our initial expectation was wrong that the absence of an important protein should result in a higher fraction of cells that do not experience gene conversion and remain heterozygous. It seems that all heterozygous fusion cells had initiated gene conversion. Gene conversion includes the interconnection between two genome molecules that act as donor and acceptor. If gene conversion cannot be terminated because an important protein is missing, these intertwined molecules might interfere with replication, genome segregation, and cell division. This might lead to cell death or at least prevent the outgrowth of clones that have initiated but failed to terminate gene conversion, resulting in reduced apparent survival rates in the gene conversion assay.
Therefore, the numbers of surviving cells were taken as an alternative readout for the efficiency of unselected intermolecular gene conversion. Supplementary Table S2 lists the average values and their standard deviations of the four biological replicates (more than 20 replicates for the wildtype), and Figure 5 visualizes the results after normalization to the control gene conversion experiment with the two parent strains. The survival rate of the control lacking the DHFR was very similar to that of the two parent strains. The same was true for six of the test strains, indicating that the proteins lacking in these strains are not involved in or at least not important for gene conversion. In stark contrast, seven of the test strains had severe deficits, with survival rates of 10% or less compared to that of the parent strains (dark red in Figure 5). In all cases, the differences compared to the control were highly significant (p < 0.001). In an additional nine cases, the survival rates were between 10% and 40% compared to the control (light red and yellow in Figure 5). Depending on the variance, the differences were significant (p < 0.01) or highly significant (p < 0.001), as indicated. Noteworthy is the fact that, in several cases, the results were very different for paralogous proteins of the same family, e.g., the lack of one of the four paralogs of Rad25 did not result in reduced survival (Rad25b) or resulted in normalized survival rates of 26%, 12%, and 10%, respectively. In addition, the lack of SMC did not influence survival, while the lack of the four SMC-like proteins (Sph1–4) resulted in drastic reduction of the survival rates down to about 20% (Sph1 and Sph3) or to about 5% (Sph2 and Sph4). Taken together, the gene conversion assay resulted in drastically different survival rates, and a lack of 16 of the 22 selected proteins had a profound effect. If the harsh treatment of the cells in the assay is taken into account, the reproducibility of the four biological replicates was quite high in the majority of cases, with a few exceptions.

Figure 5.
Normalized survival rates after gene conversion experiments are shown for the parent strains (normalized to 100%) and 23 deletion mutants. Families of paralogous proteins are boxed. The significances of the observed differences were calculated using an unpaired two-tailed t-test, with the following p-values: * < 0.05, ** < 0.01, *** < 0.001.
3.5. Analysis of Gene Expression
Recently, we performed a dRNA-Seq analysis to obtain an overview of transcription start sites and a mixed RNA-Seq analysis to obtain an overview of the transcriptome of H. volcanii [44,45]. The mixed RNA-Seq results represent four conditions, i.e., exponential growth in complex and synthetic medium at the optimal salt concentration, exponential growth at low salt, and stationary phase in complex medium. The results for all genes in this study were analyzed to reveal whether the genes are expressed under non-stressed conditions and whether any correlation exists between the severity of the effects and the expression levels. Screenshots for the first four proteins are shown in Supplementary Figure S1. In these and the following Figures, the annotated genes are shown in blue, the dRNA-Seq results in green, and the RNA-Seq results in red. The genes hjc and nucS were not expressed under the analyzed conditions, while hef and hen were highly expressed. The results for the rad genes are shown in Supplementary Figure S2. Of the four rad25 paralogs, only rad25b is highly expressed, while the deletion mutant Δrad25b had no gene conversion defect. The expression levels of the other three paralogs are very low, while all three respective deletion mutants had severe survival defects. Similarly, the transcript level of rad3b under the tested conditions was high, while the deletion mutant had no deficit, and the opposite was true for rad3a. The results for the mut genes are shown in Supplementary Figure S3. The expression level of all six genes was very low, while the survival rates of the respective deletion mutants varies from 3% to 156%. The results for the sph genes (+ smc + rad50) are shown in Supplementary Figure S4. Only the genes smc and sph2 were highly expressed, while the other four genes had a rather low expression level. Taken together, there was no correlation between the transcript levels and the severity of the gene conversion defect in the respective deletion mutants. Thus, expression of the 22 genes under the four conditions tested in mixed RNA-Seq was not indicative for the importance of the encoded proteins for gene conversion. It might be assumed that the expression of the genes important for gene conversion has to be induced when the process is initiated.
3.6. Bioinformatic Analyses of Paralogous Protein Families
Various genes included in this study encoded paralogous members of protein families. In three cases, multiple sequence alignments were generated and used to construct phylogenetic trees. Figure 6 shows a tree of the Sph protein family together with SMC and Rad50 proteins. It is a consensus tree obtained after 1000 bootstrap replications using the maximum likelihood algorithm. At selected nodes, the bootstrap values (%) are indicated that were obtained using the maximum likelihood, maximum parsimony, and neighbor-joining algorithms, respectively. An individual neighbor-joining tree is shown in Supplementary Figure S5, which includes information about the branch length but does not include information about the robustness of the tree.

Figure 6.
Consensus phylogenetic tree of the Sph/SMC/Rad50 protein family. Selected proteins from archaea, bacteria, and eukaryotes were used to construct a multiple sequence alignment. The alignment was used to generate the phylogenetic tree using the neighbor-joining option of the program MEGA X [43]. A total of 1000 bootstrap replications were performed and the bootstrap values (%) are shown at the respective nodes. Trees with 1000 bootstrap replications were also generated using the maximum parsimony and the maximum likelihood algorithms, and the bootstrap values are also included in the figure (in the order maximum likelihood/maximum parsimony/neighbor-joining) The numbered nodes are discussed in the text. The paralogs from H. volcanii are highlighted by red boxes.
The trees contain 64 proteins, mostly from halophilic Archaea, but also from other Archaea, Bacteria, and Eukaryotes. “SMC-like” (Sph) proteins occur only in halophilic archaea. All Sph proteins originated from on common ancestor (node 1 in Figure 6). All haloarchaeal SMC proteins also form a coherent group (node 2) but are rather far from the Sph proteins. The support for the monophyly of all archaeal, bacterial, and eukaryotic SMC proteins is somewhat lower (node 3), probably due to eukaryotic proteins SMC5 and SMC6, which are distant from all other SMC proteins. Similarly, all haloarchaeal Rad50 proteins form one well-supported group (node 4). The monophyly of all Rad50 proteins is not well supported, because the distinct position of the eukaryotic ortholog. Within the group of Sph proteins, two well-supported subgroups were found, on the one hand Sph1 (node 5), and on the other hand Sph2-4 (node 6). Within the latter group, the support for the monophyly of Sph3 and Sph4 is high (nodes 7 and 8), while the Sph2 do not form a monophyletic group.
A Rad25 maximum likelihood consensus tree including the bootstrap values of all three approaches is shown in Supplementary Figure S6. An individual neighbor-joining tree is shown in Supplementary Figure S9. The numbers of Rad25 paralogs vary widely, e.g., H. volcanii contains four paralogs, while Haloferax mediterranei from the same genus contains only two paralogs. Designation of the paralogs during genome annotation was typically not based on phylogenetic analysis, but two paralogs were named a and b and four paralogs were named a to d. Therefore, paralog designations are not informative about phylogenetic relationships. The tree indicates that the last common ancestor of all haloarchaea contained two paralogs (node 1 and 2 Supplementary Figure S6). In the lower part of the tree, which contains the H. volcanii paralog Rad25d, no further gene duplications occurred. However, during further evolution of halophilic archaea another gene duplication occurred, which can be observed in the upper part of the tree (node 3 and 4). In contrast to most other species, H. volcanii experienced an additional gene duplication (node 5), which generated Rad25a and Rad25b.
A maximum likelihood consensus tree of selected Mut proteins is shown in Supplementary Figure S8. An individual neighbor-joining tree is shown in Supplementary Figure S9. The upper part contains all prokaryotic MutL sequences together with their eukaryotic homologs that are named MLH (node 1), while the lower part contains all MutS sequences together with their eukaryotic homologs named MSH (node 2). In the MutL branch, a gene duplication occurred and resulted in the two paralogs, MutLa and MutLb, in genera like Haloferax and Haloquadratum, which did not occur in genera like Natrialba, Natrinema or Natronomonas. In the MutS branch, a gene duplication in archaea led to the split into the MutS5 group (node 3) and the MutS1 group including the eukaryotic MSHs (node 4). The latter group divided into the archaeal MutS1 group (node 5) and the eukaryotic MSH group (node 6). In each of the two archaeal branches, a further gene duplication event led to the generation of two further paralogs (nodes 7 + 8 and nodes 9 + 10). This evolutionary scenario can explain the occurrence of six Mut paralogs in H. volcanii.
Taken together, in all three protein families, like in other protein families present in H. volcanii, the present paralogs arose through gene duplications that occurred at very different phylogenetic levels, some of which are confined to single genera or even species. Haloarchaea and especially H. volcanii are characterized by the presence of a higher number of paralogs in many protein families than in other evolutionary lineages.
3.7. Generation and Characterization of Mutants with Multiple Deletions
H. volcanii has four paralogs of the Rad25 and Sph protein families; therefore, we decided to generate mutants with multiple deletions. This approach aimed to elucidate whether the paralogs have redundant functions and can functionally replace one another or whether all paralogs have evolved to fulfill different functions. In the former case, it can be expected that the survival rates would be much lower in multiple than in single mutants, while in the latter case, the survival rate of a multiple mutant should be similar to the lowest survival rate of any of the single mutants.
In the rad25 family, it was indeed possible to generate quadruple mutants combining the four deletions in rad25a-d in both parent strains. The quadruple mutants did not exhibit a growth defect in complex or synthetic medium. A gene conversion experiment was performed with four biological replicates, and the survival rate was 14%. This survival rate was very similar to the survival rates of Δrad25a and Δrad25c (12% and 10%), the two paralogs with the lowest survival rates of the four single mutants. Therefore, it seems that the four Rad25 paralogs have evolved to fulfill separate functions and are not redundant to one another.
A quadruple deletion mutant of the sph genes could not be generated because it turned out that sph2 and sph4 cannot be deleted simultaneously. This indicates that Sph2 and Sph4 are at least partially redundant and fulfill an essential role in H. volcanii for growth and survival under optimal conditions. However, a triple Δsph1/3/4 could be generated, and the triple mutant did not exhibit any growth defect. In gene conversion experiments, the triple mutant had an average survival rate of 10% (± 7%), similar to the survival rates of the Δsph2 and Δsph4 single mutants with survival rates 3% and 6%. This indicates that Sph1 and Sph3 do not have overlapping functions with Sph2/4, at least in the molecular mechanism of intermolecular gene conversion.
3.8. Further Characterization of the Sph-Deletion Mutants
Recently, it has been reported that Sph3 is involved in cell shape determination and that a Δsph3 mutant has a defect in swarming [49]. To unravel whether this might also be true for the other Sph proteins, the swarming abilities of the four single mutants and the triple mutant were compared to that of the wildtype. The results are shown in Supplementary Figure S10. In fact, the Δsph3 mutant (and the triple mutant) did not swarm at all for the first 48 h, in agreement with the recent report [49]. The swarming ability after 48 h differed for all four Δsph mutants (Supplementary Figure S10). Notable, the lack of Sph4 led to a severe swarming deficit, while the lack of Sph2 resulted in an increase in swarming, compared to the wildtype. These results revealed that Sph2 and Sph4 have a redundant function in gene conversion (see above) but that they fulfill opposite functions during swarming. After 48 h, all mutants started to swarm with velocities not very different from that of the wildtype.
It has also been reported that the swarming ability is correlated with the cell shape, i.e., that only rod-shaped cells are able to swarm [49]. In addition, it is known that wildtype cells are rod-shaped only in the very early exponential phase and transform to pleiomorphic cells during further growth [50]. Therefore, the cells shapes of the sph-deletion mutants were compared to that of the wildtype at three different times during the growth curve: at OD600 of 0.03 representing the very early exponential phase, at OD600 of 0.3 representing the mid-exponential phase, and at OD600 of 1.6 representing the stationary phase. The results are summarized in Supplementary Figure S11. In agreement with the recent report [49], we found that the sph3-deletion mutant does not form rods, even in the very early exponential phase. The opposite phenotype was observed for the sph2-deletion mutant, which formed predominantly rods even during the mid-exponential and stationary phases. These observations were in excellent agreement with the swarming deficit of Δsph3 and the over-swarmer phenotype of Δsph2 described above. The morphologies of the sph1 and sph4 were very similar to that of the wildtype. Similar to the results of the swarming assay, the cell shape analysis revealed that Δsph2 and Δsph4 had different phenotypes, in contrast to the gene conversion experiments, which revealed that the two genes are synthetically lethal. This can be taken as another indication that Sph2 and Sph4 have overlapping but not identical functions.
4. Discussion
The high efficiency of unselected intermolecular gene conversion enabled us to establish an experimental approach for the direct characterization of different aspects of gene conversion [18]. In stark contrast, experimental approaches for the characterization of intramolecular gene conversion in antigenic variation or concerted evolution of gene families required prior selection schemes because these processes occur only very rarely [9]. The current study aimed to unravel which of 22 proteins with DNA-related functions are involved in and important for gene conversion in H. volcanii. It had been expected that the absence of proteins with functional relevance would inhibit or slow down gene conversion and that this would lead to a higher fraction of heterozygous cells. Because completed gene conversion in the direction of the mutated copy of the reporter gene HVO_2528 results in a loss of carotenoid biosynthesis, this should be directly visible based on a reduced fraction of white colonies. Unexpectedly, the fraction of white colonies was very similar in all mutants and the wildtype. PCR analyses of red colonies revealed that more than 98% of them were homozygous for the wildtype copy of HVO_2528 and thus had experienced completed gene conversion (unpublished data). Therefore, the initially anticipated higher fraction of heterozygous cells that had not experienced gene conversion was not observed. This indicates that nearly all heterozygous cells obtained after protoplast fusion had initiated gene conversion. However, when an important protein was missing, the progression of gene conversion stalled and gene conversion intermediates remained, e.g., unresolved Holliday junctions. Such intermediates are probably incompatible with replication, genome segregation, and cell division. When such intermediates cannot be resolved at all, no apparent survivors of the gene conversion assay can be expected. In this model, the apparent survival rates are correlated to the remaining efficiency to resolve gene conversion intermediates and complete gene conversion in spite of the lack of an important protein. The higher the importance of the protein for the molecular mechanism of gene conversion in H. volcanii, the lower the normalized apparent survival rate should be.
In fact, it was found that lack of only 6 of the 22 proteins did not change the survival rate, while the lack of 16 of the 22 proteins led to a considerably reduced survival, which was less than 10% in 6 of the cases. This represents the first comprehensive study addressing the importance of proteins for gene conversion in archaea. A systematic generation of mutants has also been used for the identification of important proteins for antigenic variation in Borrelia burgdorfei (17 genes, [51]) and Neisseria meningitides (5 genes, [52]) or various steps in meiotic DNA processing in Saccharomyces cerevisiae, including gene conversion (81 genes, [53]).
Notably, in the current study, the radA gene could not be deleted. This result confirmed the results of earlier attempts of our group to delete the radA gene, which exclusively resulted in wild-type clones (in each case, more than 100 Pop_Out clones were screened; unpublished data). Therefore, in our hands, radA is an essential gene for H. volcanii. RadA is orthologous to the bacterial RecA protein and the eukaryotic Rad51 protein. Orthologs are present ubiquitously in all species of all three domains of life, and they are involved in the first step of homologous recombination, i.e., the search for homologous sequences in two DNA molecules [54,55]. Therefore, it is tempting to speculate that RadA should also be involved in early steps of gene conversion in H. volcanii, i.e., the search for homologous sites on different genome copies with sequence differences. However, despite the expectation that RadA or the bacterial ortholog RecA should be involved in gene conversion, this is not always the case. For example, antigenic variation in B. burgdorfei was found to be independent of RecA [56,57]. In contrast, RecA is essential for antigenic variation in N. meningitides [52]. Therefore, the necessity to clarify whether RadA is involved in gene conversion in H. volcanii persists, but alternative experimental approaches have to be used, e.g., the characterization of protein–protein interaction networks of important gene conversion proteins.
While the role of RadA remains unclear, the survival rates of six deletion mutants indicated that the encoded proteins are not involved in gene conversion or that their functions can be redundantly performed also be other proteins. The latter explanation might well hold true for three genes that are members of paralogous gene families, and these cases (rad3b, rad25b, mutS1a) are discussed below. However, this explanation is much less likely for the three single genes hen, mre11-rad50, and smc (discussed below). Hen is a homing endonuclease, and it has been shown that the Hen of H. volcanii is active and can insert an intein-encoding sequence into the chromosome by gene conversion [58]. While this results in the equalization of a intein-containing donor genome and a prior-to-intein-homing intein-lacking acceptor genome, this is a very specific activity, and it might not be surprising that Hen is not involved in general gene conversion.
However, it was unexpected that the Mre11-Rad50 protein complex is not important for gene conversion in H. volcanii. The homologous eukaryotic Mre11-Rad50 complex is important for various processes involving gene conversion, e.g., one pathway (of several) of double strand break repair [59], immunoglobulin gene diversification [60,61], and recombination during meiosis [62]. Contrasting the roles in eukaryotes, the Mre11-Rad50 complex of H. volcanii has been shown to inhibit the repair of double strand breaks by homologous recombination, favoring repair by microhomology-mediated end-joining [55]. In addition, the complex is involved in nucleoid compaction after DNA damage, underscoring its involvement in DNA repair [63]. Our finding that Mre11-Rad50 is not involved in gene conversion indicates that this process is independent from double-strand break-induced repair pathways.
Importantly, the current study revealed that 16 of the gene deletion mutants had severe defects in gene conversion. Three genes were single genes, i.e., hjc, hef, and nucS. Hjc is a holiday junction resolvase that is highly conserved in archaea. It specifically binds to four-way-junction DNA structures and induces endonucleolytic cleavages to initiate junction resolution. The crystal structures of Hjc orthologs from several different archaea have been solved, and models for the DNA binding have been generated [64,65,66,67]. In addition, biochemical and genetic studies with enzymes from different archaeal species have been performed [68,69,70,71,72]. This biological role fits well to the nearly complete loss of gene conversion in the hjc-deletion mutant (Figure 5).
Another protein that acts on branched DNA structures is Hef (helicase-associated endonuclease for fork-structured DNA). Hef is conserved in archaea and has been shown to be involved in the repair of stalled replication forks in various archaeal species [73,74,75,76]. The H. volcanii Hef has been characterized with genetic and biochemical approaches, and a review by Lestini et al. gives an overview of the results [77]. Eukaryotes contain a homolog to the archaeal Hef protein, and the human protein is called FANCM, because it is one protein of a protein complex that is related to Fanconi anemia [78,79]. Fanconi anemia is a recessive genetic disease that is characterized by genomic instability and cancer predisposition. Both archaeal Hef and eukaryotic FANCM interact with PCNA, indicating that their role in replication fork is conserved [80]. In H. volcanii, it has been shown that hef and hjc can both be deleted individually but that Hef becomes essential when Hjc is missing [81]. Both single mutants did not have a growth defect, in agreement with our results (Figure 3). However, the single mutants were also not compromised in a homologous recombination assay [81], in stark contrast to the nearly complete loss in gene conversion (Figure 5). This is another indication that in H. volcanii intermolecular gene conversion involves enzymes that have function in homologous recombination or other DNA repair pathways (see below) but that the molecular mechanism is separate from these pathways.
The third single gene with a severe gene conversion defect in the deletion mutant was nucS. NucS (also called EndoMS) is an endonuclease that acts on both sides of a DNA mismatch, thus generating a double strand break. It has been discovered in thermophilic archaea and is thought to be involved in a non-canonical pathway of DNA mismatch repair (MMS), and thus it is specifically important for species that lack the canonical rep MMS repair pathway involving MutL and MutS. Three recent reviews summarize the characterization of NucS from archaea and from actinobacteria [82,83,84]. Deletion of the nucS gene in Sulfolobus islandicus led to a 1000-fold increase in the spontaneous mutation rate, underscoring the important role of NucS for genome integrity [85]. Like the two proteins discussed above, Hjc and Hef, NucS also interacts with the replication clamp PCNA. The severe phenotypes of all three mutants indicate that Hjc, Hef, and NucS have important functions in gene conversion and that they are not redundant but all have non-overlapping essential roles.
The remaining 13 genes with severe gene conversion defects in the respective deletion mutants were members of several families of paralogous genes. H. volcanii and other haloarchaea are characterized by expansions of various genes, which have only a single or very few copies in other archaeal groups. For example, H. volcanii contains 12 orc (origin recognition complex genes), while Pyrococcus has a single orc gene [40]. Similarly, H. volcanii has four paralogs of TBP (TATA box-binding protein) and twelve paralogs of TFB (transcription factor B), while Pyrococcus contains only one copy of these basal transcription initiation factors (www.halolex.mpg.deaccessed at 23 February 2024). In addition, H. volcanii contains 18 transcription factors of the Lrp family, while Pyrococcus contains one (www.halolex.mpg.de (accessed on 23 February 2024)). Thus, it is not surprising that H. volcanii contains several paralogs of Rad3, Rad25, MuL, MutS, and Sph. The gene duplications occurred at different stages during evolution, some of them not before the evolution of the genus Haloferax or of the species H. volcanii (see Figure 6 and Supplementary Figures S5 and S6). Nevertheless, the drastically reduced gene conversion efficiencies of at least one member in all five families revealed that the functions of the paralogs are not redundant but that novel functions had evolved.
MutS, which binds to mismatched DNA, and the nicking endonuclease MutL constitute the canonical mismatch repair pathway. A phylogenetic analysis revealed that both proteins originated in bacteria and that archaeal homologs were obtained by lateral transfer [86]. Eukaryotes contain a higher number of paralogs, and a recent analysis including Asgard archaea indicated that they were partly obtained from an archaeal progenitor and partly from bacterial endosymbionts [87]. Deletion of the single mutL gene in Halobacterium salinarum did not increase the mutation rate, which led to the hypothesis that alternative pathways to the canonical MMR exist in haloarchaea [88]. Deletion of both mutL paralogs of H. volcanii led to a severe decrease in survival, indicating that both are involved in gene conversion (Figure 5) instead of the canonical MMR. However, the picture is complicated by the presence of four paralogs of the MutS family, two each of the subfamilies MutS1 and MutS5. MutS5 from Pyrococcus horikoshii has been shown to bind to Holliday junctions [89]. Therefore, it is tempting to speculate that MutS5 is involved in gene conversion while MutS1 is involved in canonical MMR. In agreement with this view, the mutS1a-deletion mutant was not impaired in gene conversion. However, deletion of mutS1b also led to a reduced survival in the gene conversion assay (Figure 5). Further experiments are needed to resolve this complex situation of six MutL/MutS paralogs. The bacterial MutL has been shown to be involved in antigenic variation in B. burgdorfei [56], indicating that in this species also, MutL has functions beyond MMR. The same is true for the eukaryotic homologs of MutL and MutS, which are called MLH and MSH. They have been shown to be involved in heteroduplex formation, to interact with recombination enzymes, and to promote crossing over during meiosis [90,91,92].
Eukaryotic homologs of the archaeal Rad3 and Rad25 proteins are helicases involved in nucleotide excision repair (NER) [93]. Their function in archaea is not well studied. The large differences in gene conversion efficiencies (Figure 5) indicates that at least in H. volcanii, both Rad proteins are involved in different biological processes.
The last protein family is the SMC/Sph protein family. SMC (Structural Maintenance of Chromosomes) proteins are highly conserved in archaea, bacteria, and eukaryotes [94]. Eukaryotes have six SMC proteins, SMC1-6. They form three different heterodimers (SMC1-2, SMC3-4, SMC5-6) that build complexes with additional proteins. The SMC protein complexes are involved in DNA condensation, DNA cohesion, and additional genome-related functions [95]. A recent phylogenetic analysis proposed that the six eukaryotic SMC proteins evolved from archaeal SMC proteins via several gene duplication events [96]. Archaea and bacteria typically contain a single SMC protein that forms homodimers and forms functional SMC complexes together with additional proteins. The SMC proteins are comprised of N-terminal and C-terminal globular domains that can bind DNA, two very long coiled-coil domains that are involved in dimerization, and a hinge domain in the middle. In total, SMC proteins have a length of about 1200 aa (amino acids).
More than 20 years ago, it was discovered that haloarchaea contain proteins that are comprised of the same five domains described above, which were later named SMC-like proteins of haloarchaea (Sph) [97]. It was found that heterologous overproduction of a Sph protein from H. salinarum or homologous overproduction of a H. volcanii Sph in H. volcanii results in the formation of very long rods, i.e., that the respective Sphs were involved in cell shape determination. Sequencing of the H. volcanii genome revealed that its genome contained four sph genes [34]. The Sph protein family is confined to the haloarchaea; however, a recent phylogenetic analysis revealed that several additional subfamilies of SMC-related protein exist [98].
The functions of the Sph proteins remain elusive; however, their SMC-like structure indicate that their globular N- and C-termini might bind DNA and the Sph proteins might be involved in genome-related functions. Therefore, the smc gene and all four sph genes were included in the present study. The SMC protein is apparently not involved in gene conversion, while, in stark contrast, all four Sph proteins are very important because the respective deletion mutants had severely reduced survival rates of about 10% (Δsph1 and Δsph3) or near 0% (Δsph2 and Δsph4). Sph2 and Sph4 seem to have overlapping functions, because a double mutant could not be obtained, but also paralog-specific functions based on the very severe phenotype of the single mutants. In a recent study, it was observed that a sph3-deletion mutant had lost the ability to form rod-shaped cells [49]. This observation could be verified, and in addition, it was found that the sph2-deletion mutant had the opposite phenotype and formed exclusively rod-shaped cells (Supplementary Figure S11). In agreement with earlier reports, the cell shape correlated with the swarming velocity, and Δsph3 failed to swarm during the first two days, while Δsph 2 was an overswarmer (Supplementary Figure S10). Remarkably, Δsph4 behaved exceptionally in this respect, i.e., it had a swarming deficit during the first two days, but the fraction of rod-shaped cells was higher than in the wildtype in exponential phase cultures. Together, these results indicate that the Sph proteins have multiple functions and that they act in the same direction in one biological process (gene conversion) while they have opposing functions in another (cell shape determination).
The current study used a comprehensive deletion analysis to identify proteins that are involved in intermolecular gene conversion in H. volcanii. To deepen the understanding, in future studies, alternative approaches should be used to complement the results. For example, co-affinity isolation has been successfully used to unravel the protein–protein interaction network of translation initiation [99], and this approach would be very promising for unravelling the protein–protein interaction network of gene conversion. Notably, this approach would allow researchers to detect the involvement of essential proteins like RadA, which could not be studied with the deletion analysis. It would also unravel whether or not the Sph proteins form specific heterodimers like the eukaryotic SMC proteins. As discussed above, many of the proteins’ specific functions could be predicted, e.g., binding to structured DNA forms or the activity of a helicase or nuclease. The purified proteins could not only be used for the interaction analysis but also to analyze with biochemical assays whether these predictions hold true.
5. Conclusions
The aim of the current study was the identification of proteins that are involved in the molecular mechanisms of the highly efficient intermolecular gene conversion of H. volcanii. To this end, 22 single-gene-deletion mutants were generated in two different parent strains. This enabled us to compare the efficiency of gene conversion of the mutants with that of the wildtype in heterozygous cells that were obtained by protoplast fusion. While 6 of the proteins were apparently not involved, 16 mutants were severely deficient in intermolecular gene conversion. Remarkably, mutants of paralogous genes of several gene families had very different phenotypes. The results indicated that the mechanism of gene conversion is separate from homologous recombination, even if several proteins are involved in both processes. For the first time, a biological function of the four members of the Sph protein family could be observed. Identification of the 16 gene conversion proteins paves the way for further experimental approaches to understand the molecular mechanism.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes15070861/s1, Figure S1: dRNA-Seq and RNA-Seq results for hjc, hef, hen, nucS; Figure S2: dRNA-Seq and RNA-Seq results for six rad genes, Figure S3: dRNA-Seq and RNA-Seq results for six mut genes; Figure S4: dRNA-Seq and RNA-Seq results for six sph/smc/rad50 genes; Figure S5: Individual neighbor-joining tree of the SMC/Sph protein family, Figure S6: Consensus tree of selected Rad25 proteins, including bootstrap values obtained using three algorithms, Figure S7: Individual neighbor-joining tree of the Rad25 protein family, Figure S8: Consensus tree of selected Mut proteins, including bootstrap values obtained using three algorithms; Figure S9: Individual neighbor-joining tree of the Mut protein family, Figure S10: Swarming of sph-deletion mutants and wildtype; Figure S11: cell morphologies of sph mutants and wildtype at different optical densities; Table S1: List of all oligonucleotides used for deletion mutant generation; Table S2: Results of the protoplast fusions of the parent strains, the control strains, and the test strains with deletions of genes possibly important for gene conversion.
Author Contributions
Conceptualization, D.W. and J.S.; methodology, H.Ö., D.W., L.S. and J.S.; formal analysis, H.Ö., D.W. and L.S.; investigation, H.Ö., D.W. and L.S.; data curation, D.W. and J.S.; writing—original draft preparation, J.S.; writing—review and editing, H.Ö., D.W., L.S. and J.S.; visualization, H.Ö., D.W. and J.S.; supervision, J.S.; project administration, J.S.; funding acquisition, J.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the German Research Council (Deutsche Forschungsgemeinschaft, DFG), grant number So264/29.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All H. volcanii mutant strains generated in this study (or other studies by the Soppa group) are freely available upon request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Fayos, I.; Frouin, J.; Meynard, D.; Vernet, A.; Herbert, L.; Guiderdoni, E. Manipulation of Meiotic Recombination to Hasten Crop Improvement. Biology 2022, 11, 369. [Google Scholar] [CrossRef] [PubMed]
- Daugherty, M.D.; Zanders, S.E. Gene conversion generates evolutionary novelty that fuels genetic conflicts. Curr. Opin. Genet. Dev. 2019, 58–59, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Underwood, C.J.; Choi, K. Heterogeneous transposable elements as silencers, enhancers and targets of meiotic recombination. Chromosoma 2019, 128, 279–296. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, J.A.; Innan, H. The Role of Gene Conversion between Transposable Elements in Rewiring Regulatory Networks. Genome Biol. Evol. 2019, 11, 1723–1729. [Google Scholar] [CrossRef] [PubMed]
- Korunes, K.L.; Noor, M.A.F. Gene conversion and linkage: Effects on genome evolution and speciation. Mol. Ecol. 2017, 26, 351–364. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, R.; Morrison, L.J.; Hall, J.P.J. DNA Recombination Strategies during Antigenic Variation in the African Trypanosome. Microbiol. Spectr. 2015, 3, 409–435. [Google Scholar] [CrossRef] [PubMed]
- Duret, L.; Galtier, N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu. Rev. Genom. Hum. Genet. 2009, 10, 285–311. [Google Scholar] [CrossRef] [PubMed]
- Tang, E.S.; Martin, A. Immunoglobulin gene conversion: Synthesizing antibody diversification and DNA repair. DNA Repair 2007, 6, 1557–1571. [Google Scholar] [CrossRef] [PubMed]
- Santoyo, G.; Romero, D. Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol. Rev. 2005, 29, 169–183. [Google Scholar] [CrossRef]
- Foley, J. Mini-review: Strategies for Variation and Evolution of Bacterial Antigens. Comput. Struct. Biotechnol. J. 2015, 13, 407–416. [Google Scholar] [CrossRef]
- Vink, C.; Rudenko, G.; Seifert, H.S. Microbial antigenic variation mediated by homologous DNA recombination. FEMS Microbiol. Rev. 2012, 36, 917–948. [Google Scholar] [CrossRef] [PubMed]
- Espejo, R.T.; Plaza, N. Multiple Ribosomal RNA Operons in Bacteria; Their Concerted Evolution and Potential Consequences on the Rate of Evolution of Their 16S rRNA. Front. Microbiol. 2018, 9, 1232. [Google Scholar] [CrossRef] [PubMed]
- Liao, D. Gene conversion drives within genic sequences: Concerted evolution of ribosomal RNA genes in bacteria and archaea. J. Mol. Evol. 2000, 51, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Cuna, F.O.; Castellanos, M.; Romero, D. Biased Gene Conversion in Rhizobium etli Is Caused by Preferential Double-Strand Breaks on One of the Recombining Homologs. J. Bacteriol. 2016, 198, 591–599. [Google Scholar] [CrossRef]
- Paulsson, J.; El Karoui, M.; Lindell, M.; Hughes, D. The processive kinetics of gene conversion in bacteria. Mol. Microbiol. 2017, 104, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Lange, C.; Zerulla, K.; Breuert, S.; Soppa, J. Gene conversion results in the equalization of genome copies in the polyploid haloarchaeon Haloferax volcanii. Mol. Microbiol. 2011, 80, 666–677. [Google Scholar] [CrossRef]
- Soppa, J. Evolutionary advantages of polyploidy in halophilic archaea. Biochem. Soc. Trans. 2013, 41, 339–343. [Google Scholar] [CrossRef]
- Wasser, D.; Borst, A.; Hammelmann, M.; Ludt, K.; Soppa, J. Characterization of Non-selected Intermolecular Gene Conversion in the Polyploid Haloarchaeon Haloferax volcanii. Front. Microbiol. 2021, 12, 680854. [Google Scholar] [CrossRef]
- Hildenbrand, C.; Stock, T.; Lange, C.; Rother, M.; Soppa, J. Genome copy numbers and gene conversion in methanogenic archaea. J. Bacteriol. 2011, 193, 734–743. [Google Scholar] [CrossRef]
- Ruhlman, T.A.; Zhang, J.; Blazier, J.C.; Sabir, J.S.M.; Jansen, R.K. Recombination-dependent replication and gene conversion homogenize repeat sequences and diversify plastid genome structure. Am. J. Bot. 2017, 104, 559–572. [Google Scholar] [CrossRef]
- Gong, L.; Olson, M.; Wendel, J.F. Cytonuclear evolution of rubisco in four allopolyploid lineages. Mol. Biol. Evol. 2014, 31, 2624–2636. [Google Scholar] [CrossRef]
- Hao, W.; Richardson, A.O.; Zheng, Y.; Palmer, J.D. Gorgeous mosaic of mitochondrial genes created by horizontal transfer and gene conversion. Proc. Natl. Acad. Sci. USA 2010, 107, 21576–21581. [Google Scholar] [CrossRef]
- Khakhlova, O.; Bock, R. Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 2006, 46, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Dahal, S.; Dubey, S.; Raghavan, S.C. Homologous recombination-mediated repair of DNA double-strand breaks operates in mammalian mitochondria. Cell. Mol. Life Sci. 2018, 75, 1641–1655. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Hao, W. Horizontal transfer and gene conversion as an important driving force in shaping the landscape of mitochondrial introns. G3 Genes Genomes Genet. 2014, 4, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R.; Keeling, P.J. Gene conversion shapes linear mitochondrial genome architecture. Genome Biol. Evol. 2013, 5, 905–912. [Google Scholar] [CrossRef]
- Suh, M.H.; Pulakat, L.; Gavini, N. Isolation and characterization of nifDK:kanamycin and nitrogen fixation proficient Azotobacter vinelandii strain, and its implication on the status of multiple chromosomes in Azotobacter. Genetica 2000, 110, 101–107. [Google Scholar] [CrossRef]
- Nodop, A.; Pietsch, D.; Höcker, R.; Becker, A.; Pistorius, E.K.; Forchhammer, K.; Michel, K.-P. Transcript profiling reveals new insights into the acclimation of the mesophilic fresh-water cyanobacterium Synechococcus elongatus PCC 7942 to iron starvation. Plant Physiol. 2008, 147, 747–763. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Takahama, K.; Matsuoka, M.; Nagahama, K.; Ogawa, T. Construction and analysis of a recombinant cyanobacterium expressing a chromosomally inserted gene for an ethylene-forming enzyme at the psbAI locus. J. Biosci. Bioeng. 2003, 95, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Spence, E.; Bailey, S.; Nenninger, A.; Møller, S.G.; Robinson, C. A homolog of Albino3/OxaI is essential for thylakoid biogenesis in the cyanobacterium Synechocystis sp. PCC6803. J. Biol. Chem. 2004, 279, 55792–55800. [Google Scholar] [CrossRef]
- Allers, T.; Ngo, H.-P.; Mevarech, M.; Lloyd, R.G. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 2004, 70, 943–953. [Google Scholar] [CrossRef] [PubMed]
- Nagy, C.; Thiel, K.; Mulaku, E.; Mustila, H.; Tamagnini, P.; Aro, E.-M.; Pacheco, C.C.; Kallio, P. Comparison of alternative integration sites in the chromosome and the native plasmids of the cyanobacterium Synechocystis sp. PCC 6803 in respect to expression efficiency and copy number. Microb. Cell Fact. 2021, 20, 130. [Google Scholar] [CrossRef] [PubMed]
- Ionescu, D.; Bizic-Ionescu, M.; de Maio, N.; Cypionka, H.; Grossart, H.-P. Community-like genome in single cells of the sulfur bacterium Achromatium oxaliferum. Nat. Commun. 2017, 8, 455. [Google Scholar] [CrossRef] [PubMed]
- Hartman, A.L.; Norais, C.; Badger, J.H.; Delmas, S.; Haldenby, S.; Madupu, R.; Robinson, J.; Khouri, H.; Ren, Q.; Lowe, T.M.; et al. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS ONE 2010, 5, e9605. [Google Scholar] [CrossRef] [PubMed]
- Bitan-Banin, G.; Ortenberg, R.; Mevarech, M. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 2003, 185, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Jantzer, K.; Zerulla, K.; Soppa, J. Phenotyping in the archaea: Optimization of growth parameters and analysis of mutants of Haloferax volcanii. FEMS Microbiol. Lett. 2011, 322, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef] [PubMed]
- Nagel, C.; Machulla, A.; Zahn, S.; Soppa, J. Several One-Domain Zinc Finger µ-Proteins of Haloferax Volcanii Are Important for Stress Adaptation, Biofilm Formation, and Swarming. Genes 2019, 10, 361. [Google Scholar] [CrossRef] [PubMed]
- Hammelmann, M.; Soppa, J. Optimized generation of vectors for the construction of Haloferax volcanii deletion mutants. J. Microbiol. Methods 2008, 75, 201–204. [Google Scholar] [CrossRef]
- Ludt, K.; Soppa, J. Influence of Origin Recognition Complex Proteins on the Copy Numbers of Three Chromosomes in Haloferax volcanii. J. Bacteriol. 2018, 200, e00161-18. [Google Scholar] [CrossRef]
- Rosenshine, I.; Tchelet, R.; Mevarech, M. The mechanism of DNA transfer in the mating system of an archaebacterium. Science 1989, 245, 1387–1389. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, F.; Broicher, A.; Gillich, T.; Klee, K.; Mejía, J.; Rampp, M.; Oesterhelt, D. Genome information management and integrated data analysis with HaloLex. Arch. Microbiol. 2008, 190, 281–299. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Babski, J.; Haas, K.A.; Näther-Schindler, D.; Pfeiffer, F.; Förstner, K.U.; Hammelmann, M.; Hilker, R.; Becker, A.; Sharma, C.M.; Marchfelder, A.; et al. Genome-wide identification of transcriptional start sites in the haloarchaeon Haloferax volcanii based on differential RNA-Seq (dRNA-Seq). BMC Genom. 2016, 17, 629. [Google Scholar] [CrossRef] [PubMed]
- Laass, S.; Monzon, V.A.; Kliemt, J.; Hammelmann, M.; Pfeiffer, F.; Förstner, K.U.; Soppa, J. Characterization of the transcriptome of Haloferax volcanii, grown under four different conditions, with mixed RNA-Seq. PLoS ONE 2019, 14, e0215986. [Google Scholar] [CrossRef] [PubMed]
- Nicol, J.W.; Helt, G.A.; Blanchard, S.G.; Raja, A.; Loraine, A.E. The Integrated Genome Browser: Free software for distribution and exploration of genome-scale datasets. Bioinformatics 2009, 25, 2730–2731. [Google Scholar] [CrossRef]
- Jaschinski, K.; Babski, J.; Lehr, M.; Burmester, A.; Benz, J.; Heyer, R.; Dörr, M.; Marchfelder, A.; Soppa, J. Generation and phenotyping of a collection of sRNA gene deletion mutants of the haloarchaeon Haloferax volcanii. PLoS ONE 2014, 9, e90763. [Google Scholar] [CrossRef]
- Breuert, S.; Allers, T.; Spohn, G.; Soppa, J. Regulated polyploidy in halophilic archaea. PLoS ONE 2006, 1, e92. [Google Scholar] [CrossRef]
- Schiller, H.; Hong, Y.; Kouassi, J.; Rados, T.; Kwak, J.; DiLucido, A.; Safer, D.; Marchfelder, A.; Pfeiffer, F.; Bisson, A.; et al. Identification of structural and regulatory cell-shape determinants in Haloferax volcanii. Nat. Commun. 2024, 15, 1414. [Google Scholar] [CrossRef]
- de Silva, R.T.; Abdul-Halim, M.F.; Pittrich, D.A.; Brown, H.J.; Pohlschroder, M.; Duggin, I.G. Improved growth and morphological plasticity of Haloferax volcanii. Microbiology 2021, 167, 001012. [Google Scholar] [CrossRef]
- Dresser, A.R.; Hardy, P.-O.; Chaconas, G. Investigation of the genes involved in antigenic switching at the vlsE locus in Borrelia burgdorferi: An essential role for the RuvAB branch migrase. PLoS Pathog. 2009, 5, e1000680. [Google Scholar] [CrossRef]
- Xu, J.; Seifert, H.S. Analysis of Pilin Antigenic Variation in Neisseria meningitidis by Next-Generation Sequencing. J. Bacteriol. 2018, 200, e00465-18. [Google Scholar] [CrossRef]
- Jordan, P.W.; Klein, F.; Leach, D.R.F. Novel roles for selected genes in meiotic DNA processing. PLoS Genet. 2007, 3, e222. [Google Scholar] [CrossRef]
- Seitz, E.M.; Brockman, J.P.; Sandler, S.J.; Clark, A.J.; Kowalczykowski, S.C. RadA protein is an archaeal RecA protein homolog that catalyzes DNA strand exchange. Genes Dev. 1998, 12, 1248–1253. [Google Scholar] [CrossRef] [PubMed]
- RadB acts in homologous recombination in the archaeon Haloferax volcanii, consistent with a role as recombination mediator. DNA Repair 2017, 55, 7–16. [CrossRef]
- Castellanos, M.; Verhey, T.B.; Goldstein, M.; Chaconas, G. The Putative Endonuclease Activity of MutL Is Required for the Segmental Gene Conversion Events That Drive Antigenic Variation of the Lyme Disease Spirochete. Front. Microbiol. 2022, 13, 888494. [Google Scholar] [CrossRef]
- Verhey, T.B.; Castellanos, M.; Chaconas, G. Antigenic Variation in the Lyme Spirochete: Insights into Recombinational Switching with a Suggested Role for Error-Prone Repair. Cell Rep. 2018, 23, 2595–2605. [Google Scholar] [CrossRef] [PubMed]
- Naor, A.; Lazary, R.; Barzel, A.; Papke, R.T.; Gophna, U. In vivo characterization of the homing endonuclease within the polB gene in the halophilic archaeon Haloferax volcanii. PLoS ONE 2011, 6, e15833. [Google Scholar] [CrossRef][Green Version]
- Krishna, S.; Wagener, B.M.; Liu, H.P.; Lo, Y.-C.; Sterk, R.; Petrini, J.H.J.; Nickoloff, J.A. Mre11 and Ku regulation of double-strand break repair by gene conversion and break-induced replication. DNA Repair 2007, 6, 797–808. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, M.; Sonoda, E.; Nojima, K.; Sale, J.E.; Takenaka, K.; Kikuchi, K.; Taniguchi, Y.; Nakamura, K.; Sumitomo, Y.; Bree, R.T.; et al. Genetic evidence for single-strand lesions initiating Nbs1-dependent homologous recombination in diversification of Ig v in chicken B lymphocytes. PLoS Genet. 2009, 5, e1000356. [Google Scholar] [CrossRef]
- Yabuki, M.; Fujii, M.M.; Maizels, N. The MRE11-RAD50-NBS1 complex accelerates somatic hypermutation and gene conversion of immunoglobulin variable regions. Nat. Immunol. 2005, 6, 730–736. [Google Scholar] [CrossRef] [PubMed]
- Farah, J.A.; Cromie, G.; Steiner, W.W.; Smith, G.R. A novel recombination pathway initiated by the Mre11/Rad50/Nbs1 complex eliminates palindromes during meiosis in Schizosaccharomyces pombe. Genetics 2005, 169, 1261–1274. [Google Scholar] [CrossRef] [PubMed]
- Delmas, S.; Duggin, I.G.; Allers, T. DNA damage induces nucleoid compaction via the Mre11-Rad50 complex in the archaeon Haloferax volcanii. Mol. Microbiol. 2013, 87, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Middleton, C.L.; Parker, J.L.; Richard, D.J.; White, M.F.; Bond, C.S. Substrate recognition and catalysis by the Holliday junction resolving enzyme Hje. Nucleic Acids Res. 2004, 32, 5442–5451. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nishino, T.; Komori, K.; Tsuchiya, D.; Ishino, Y.; Morikawa, K. Crystal structure of the archaeal holliday junction resolvase Hjc and implications for DNA recognition. Structure 2001, 9, 197–204. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nishino, T.; Komori, K.; Ishino, Y.; Morikawa, K. Dissection of the regional roles of the archaeal Holliday junction resolvase Hjc by structural and mutational analyses. J. Biol. Chem. 2001, 276, 35735–35740. [Google Scholar] [CrossRef] [PubMed]
- Bond, C.S.; Kvaratskhelia, M.; Richard, D.; White, M.F.; Hunter, W.N. Structure of Hjc, a Holliday junction resolvase, from Sulfolobus solfataricus. Proc. Natl. Acad. Sci. USA 2001, 98, 5509–5514. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Mayaka, J.B.; Zhong, Q.; Zhang, C.; Hou, G.; Ni, J.; Shen, Y. Phosphorylation of the Archaeal Holliday Junction Resolvase Hjc Inhibits Its Catalytic Activity and Facilitates DNA Repair in Sulfolobus islandicus REY15A. Front. Microbiol. 2019, 10, 1214. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Li, Y.; Zeng, C.; Song, T.; Yan, Z.; Ni, J.; She, Q.; Shen, Y. Genetic analysis of the Holliday junction resolvases Hje and Hjc in Sulfolobus islandicus. Extremophiles 2015, 19, 505–514. [Google Scholar] [CrossRef]
- Bolt, E.L.; Lloyd, R.G.; Sharples, G.J. Genetic analysis of an archaeal Holliday junction resolvase in Escherichia coli. J. Mol. Biol. 2001, 310, 577–589. [Google Scholar] [CrossRef]
- Komori, K.; Sakae, S.; Daiyasu, H.; Toh, H.; Morikawa, K.; Shinagawa, H.; Ishino, Y. Mutational analysis of the Pyrococcus furiosus holliday junction resolvase hjc revealed functionally important residues for dimer formation, junction DNA binding, and cleavage activities. J. Biol. Chem. 2000, 275, 40385–40391. [Google Scholar] [CrossRef]
- Komori, K.; Sakae, S.; Fujikane, R.; Morikawa, K.; Shinagawa, H.; Ishino, Y. Biochemical characterization of the hjc holliday junction resolvase of Pyrococcus furiosus. Nucleic Acids Res. 2000, 28, 4544–4551. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lestini, R.; Laptenok, S.P.; Kühn, J.; Hink, M.A.; Schanne-Klein, M.-C.; Liebl, U.; Myllykallio, H. Intracellular dynamics of archaeal FANCM homologue Hef in response to halted DNA replication. Nucleic Acids Res. 2013, 41, 10358–10370. [Google Scholar] [CrossRef] [PubMed]
- Fujikane, R.; Ishino, S.; Ishino, Y.; Forterre, P. Genetic analysis of DNA repair in the hyperthermophilic archaeon, Thermococcus kodakaraensis. Genes Genet. Syst. 2010, 85, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Nishino, T.; Komori, K.; Ishino, Y.; Morikawa, K. Structural and functional analyses of an archaeal XPF/Rad1/Mus81 nuclease: Asymmetric DNA binding and cleavage mechanisms. Structure 2005, 13, 1183–1192. [Google Scholar] [CrossRef] [PubMed]
- Komori, K.; Hidaka, M.; Horiuchi, T.; Fujikane, R.; Shinagawa, H.; Ishino, Y. Cooperation of the N-terminal Helicase and C-terminal endonuclease activities of Archaeal Hef protein in processing stalled replication forks. J. Biol. Chem. 2004, 279, 53175–53185. [Google Scholar] [CrossRef]
- Lestini, R.; Delpech, F.; Myllykallio, H. DNA replication restart and cellular dynamics of Hef helicase/nuclease protein in Haloferax volcanii. Biochimie 2015, 118, 254–263. [Google Scholar] [CrossRef]
- Meetei, A.R.; Medhurst, A.L.; Ling, C.; Xue, Y.; Singh, T.R.; Bier, P.; Steltenpool, J.; Stone, S.; Dokal, I.; Mathew, C.G.; et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 2005, 37, 958–963. [Google Scholar] [CrossRef] [PubMed]
- Kalb, R.; Neveling, K.; Nanda, I.; Schindler, D.; Hoehn, H. Fanconi anemia: Causes and consequences of genetic instability. Genome Dyn. 2006, 1, 218–242. [Google Scholar] [CrossRef]
- Rohleder, F.; Huang, J.; Xue, Y.; Kuper, J.; Round, A.; Seidman, M.; Wang, W.; Kisker, C. FANCM interacts with PCNA to promote replication traverse of DNA interstrand crosslinks. Nucleic Acids Res. 2016, 44, 3219–3232. [Google Scholar] [CrossRef]
- Lestini, R.; Duan, Z.; Allers, T. The archaeal Xpf/Mus81/FANCM homolog Hef and the Holliday junction resolvase Hjc define alternative pathways that are essential for cell viability in Haloferax volcanii. DNA Repair 2010, 9, 994–1002. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Zhang, L.; Wu, M.; Jiang, D.; Li, Z.; Yang, Z. Repair of Hypoxanthine in DNA Revealed by DNA Glycosylases and Endonucleases From Hyperthermophilic Archaea. Front. Microbiol. 2021, 12, 736915. [Google Scholar] [CrossRef] [PubMed]
- Cebrián-Sastre, E.; Martín-Blecua, I.; Gullón, S.; Blázquez, J.; Castañeda-García, A. Control of Genome Stability by EndoMS/NucS-Mediated Non-Canonical Mismatch Repair. Cells 2021, 10, 1314. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jiang, D.; Wu, M.; Yang, Z.; Oger, P.M. New Insights Into DNA Repair Revealed by NucS Endonucleases from Hyperthermophilic Archaea. Front. Microbiol. 2020, 11, 1263. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Huang, Q.; Ni, J.; Xiao, Y.; Yang, Y.; Shen, Y. Functional Analysis of the NucS/EndoMS of the Hyperthermophilic Archaeon Sulfolobus islandicus REY15A. Front. Microbiol. 2020, 11, 607431. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Nei, M.; Ma, H. The origins and early evolution of DNA mismatch repair genes--multiple horizontal gene transfers and co-evolution. Nucleic Acids Res. 2007, 35, 7591–7603. [Google Scholar] [CrossRef] [PubMed]
- Hofstatter, P.G.; Lahr, D.J.G. Complex Evolution of the Mismatch Repair System in Eukaryotes is Illuminated by Novel Archaeal Genomes. J. Mol. Evol. 2021, 89, 12–18. [Google Scholar] [CrossRef]
- Busch, C.R.; DiRuggiero, J. MutS and MutL are dispensable for maintenance of the genomic mutation rate in the halophilic archaeon Halobacterium salinarum NRC-1. PLoS ONE 2010, 5, e9045. [Google Scholar] [CrossRef] [PubMed]
- Ohshita, K.; Fukui, K.; Sato, M.; Morisawa, T.; Hakumai, Y.; Morono, Y.; Inagaki, F.; Yano, T.; Ashiuchi, M.; Wakamatsu, T. Archaeal MutS5 tightly binds to Holliday junction similarly to eukaryotic MutSγ. FEBS J. 2017, 284, 3470–3483. [Google Scholar] [CrossRef]
- Abdullah, M.F.F.; Hoffmann, E.R.; Cotton, V.E.; Borts, R.H. A role for the MutL homologue MLH2 in controlling heteroduplex formation and in regulating between two different crossover pathways in budding yeast. Cytogenet. Genome Res. 2004, 107, 180–190. [Google Scholar] [CrossRef]
- Alani, E.; Reenan, R.A.; Kolodner, R.D. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics 1994, 137, 19–39. [Google Scholar] [CrossRef] [PubMed]
- Hunter, N.; Borts, R.H. Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 1997, 11, 1573–1582. [Google Scholar] [CrossRef] [PubMed]
- Prakash, S.; Prakash, L. Nucleotide excision repair in yeast. Mutat. Res. 2000, 451, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Soppa, J. Prokaryotic structural maintenance of chromosomes (SMC) proteins: Distribution, phylogeny, and comparison with MukBs and additional prokaryotic and eukaryotic coiled-coil proteins. Gene 2001, 278, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Hoencamp, C.; Rowland, B.D. Genome control by SMC complexes. Nat. Rev. Mol. Cell Biol. 2023, 24, 633–650. [Google Scholar] [CrossRef] [PubMed]
- Yoshinaga, M.; Inagaki, Y. Ubiquity and Origins of Structural Maintenance of Chromosomes (SMC) Proteins in Eukaryotes. Genome Biol. Evol. 2021, 13, evab256. [Google Scholar] [CrossRef] [PubMed]
- Ruepp, A.; Wanner, G.; Soppa, J. A 71-kDa protein from Halobacterium salinarium belongs to a ubiquitous P-loop ATPase superfamily with head-rod-tail structure. Arch. Microbiol. 1998, 169, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yoshinaga, M.; Nakayama, T.; Inagaki, Y. A novel structural maintenance of chromosomes (SMC)-related protein family specific to Archaea. Front. Microbiol. 2022, 13, 913088. [Google Scholar] [CrossRef]
- Schramm, F.; Borst, A.; Linne, U.; Soppa, J. Elucidation of the Translation Initiation Factor Interaction Network of Haloferax volcanii Reveals Coupling of Transcription and Translation in Haloarchaea. Front. Microbiol. 2021, 12, 742806. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).