EDA Missense Variant in a Cat with X-Linked Hypohidrotic Ectodermal Dysplasia
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Clinical Examination
2.3. DNA Isolation and Whole-Genome Sequencing
2.4. Variant Filtering
2.5. In Silico Pathogenicity Prediction
3. Results
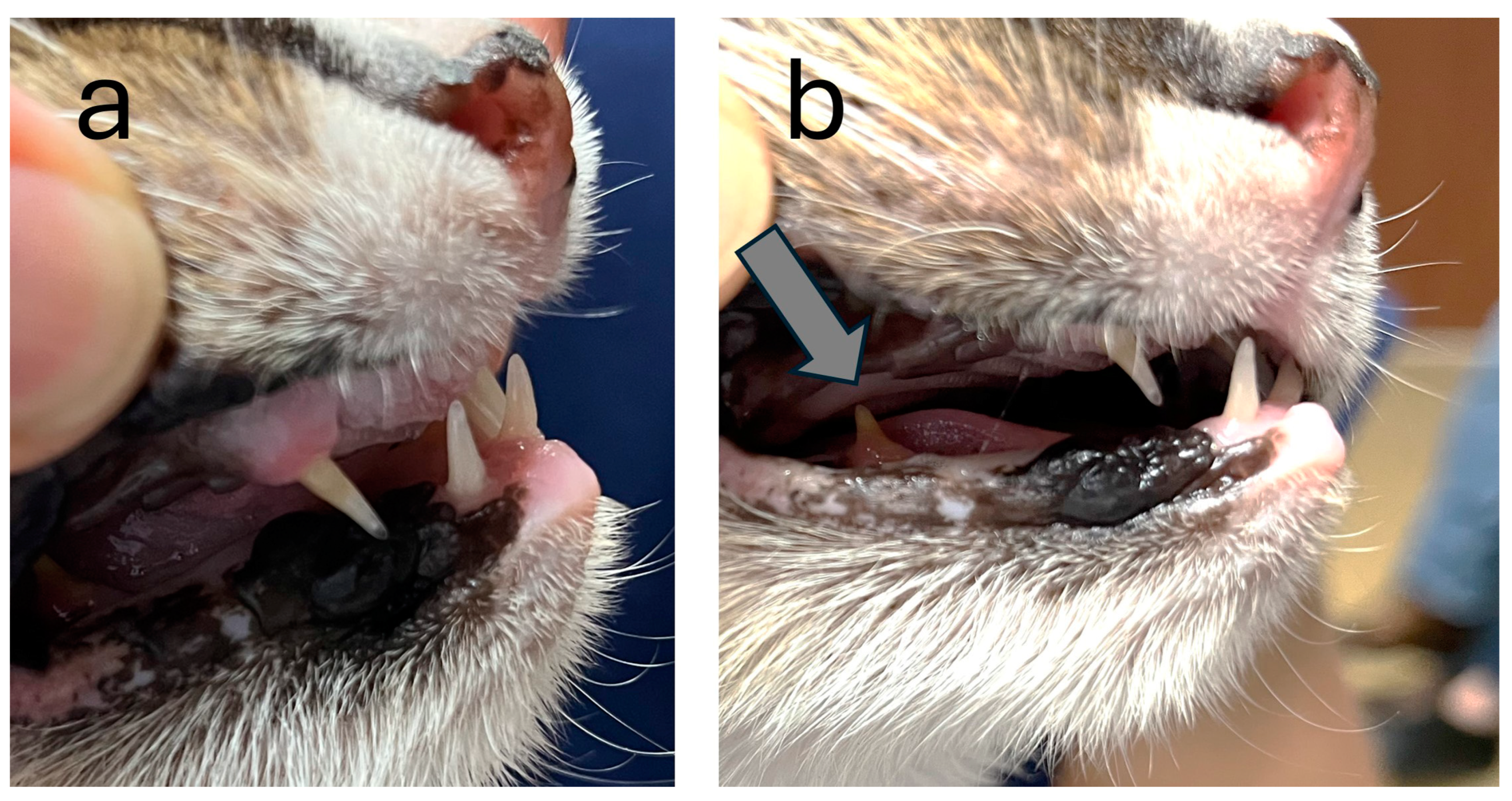
3.1. Clinical Phenotype
3.2. Genetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Darwin, C.R. The Variation of Animals and Plants under Domestication, 2nd ed.; John Murray: London, UK, 1875; Volume 2, Chapter XXV. [Google Scholar]
- Reyes-Reali, J.; Mendoza-Ramos, M.I.; Garrido-Guerrero, E.; Méndez-Catalá, C.F.; Méndez-Cruz, A.R.; Pozo-Molina, G. Hypohidrotic ectodermal dysplasia: Clinical and molecular review. Int. J. Dermatol. 2018, 57, 965–972. [Google Scholar] [CrossRef]
- Wright, J.T.; Fete, M.; Schneider, H.; Zinser, M.; Koster, M.I.; Clarke, A.J.; Hadj-Rabia, S.; Tadini, G.; Pagnan, N.; Visinoni, A.F.; et al. Ectodermal dysplasias: Classification and organization by phenotype, genotype and molecular pathway. Am. J. Med. Genet. A 2019, 179, 442–447. [Google Scholar] [CrossRef]
- Kere, J.; Srivastava, A.K.; Montonen, O.; Zonana, J.; Thomas, N.; Ferguson, B.; Munoz, F.; Morgan, D.; Clarke, A.; Baybayan, P.; et al. X-linked anhidrotic (hypohidrotic) ectodermal dysplasia is caused by mutation in a novel transmembrane protein. Nat. Genet. 1996, 13, 409–416. [Google Scholar] [CrossRef]
- Ferguson, B.M.; Brockdorff, N.; Formstone, E.; Ngyuen, T.; Kronmiller, J.E.; Zonana, J. Cloning of Tabby, the murine homolog of the human EDA gene: Evidence for a membrane-associated protein with a short collagenous domain. Hum. Mol. Genet. 1997, 6, 1589–1594. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Pispa, J.; Hartung, A.J.; Du, Y.; Ezer, S.; Jenks, T.; Shimada, T.; Pekkanen, M.; Mikkola, M.L.; Ko, M.S.; et al. The Tabby phenotype is caused by mutation in a mouse homologue of the EDA gene that reveals novel mouse and human exons and encodes a protein (ectodysplasin-A) with collagenous domains. Proc. Natl. Acad. Sci. USA 1997, 94, 13069–13074. [Google Scholar] [CrossRef]
- Monreal, A.W.; Zonana, J.; Ferguson, B. Identification of a new splice form of the EDA1 gene permits detection of nearly all X-linked hypohidrotic ectodermal dysplasia mutations. Am. J. Hum. Genet. 1998, 63, 380–389. [Google Scholar] [CrossRef]
- Yan, M.; Wang, L.C.; Hymowitz, S.G.; Schilbach, S.; Lee, J.; Goddard, A.; de Vos, A.M.; Gao, W.Q.; Dixit, V.M. Two-amino acid molecular switch in an epithelial morphogen that regulates binding to two distinct receptors. Science 2000, 290, 523–527. [Google Scholar] [CrossRef]
- Mikkola, M.L. Molecular aspects of hypohidrotic ectodermal dysplasia. Am. J. Med. Genet. A 2009, 149A, 2031–2036. [Google Scholar] [CrossRef]
- Monreal, A.W.; Ferguson, B.M.; Headon, D.J.; Street, S.L.; Overbeek, P.A.; Zonana, J. Mutations in the human homologue of mouse dl cause autosomal recessive and dominant hypohidrotic ectodermal dysplasia. Nat. Genet. 1999, 22, 366–369. [Google Scholar] [CrossRef]
- Headon, D.J.; Emmal, S.A.; Ferguson, B.M.; Tucker, A.S.; Justice, M.J.; Sharpe, P.T.; Zonana, J.; Overbeek, P.A. Gene defect in ectodermal dysplasia implicates a death domain adapter in development. Nature 2001, 414, 913–916. [Google Scholar] [CrossRef]
- LOVD v3.0 Build 29. Available online: https://databases.lovd.nl/shared/genes/EDA (accessed on 4 April 2024).
- Casal, M.L.; Scheidt, J.L.; Rhodes, J.L.; Henthorn, P.S.; Werner, P. Mutation identification in a canine model of X-linked ectodermal dysplasia. Mamm. Genome 2005, 16, 524–531. [Google Scholar] [CrossRef]
- Waluk, D.P.; Zur, G.; Kaufmann, R.; Welle, M.M.; Jagannathan, V.; Drögemüller, C.; Müller, E.J.; Leeb, T.; Galichet, A. A splice defect in the EDA gene in dogs with an X-linked hypohidrotic ectodermal dysplasia (XLHED) phenotype. G3 Genes Genomes Genet. 2016, 6, 2949–2954. [Google Scholar] [CrossRef]
- Hadji Rasouliha, S.; Bauer, A.; Dettwiler, M.; Welle, M.M.; Leeb, T. A frameshift variant in the EDA gene in Dachshunds with X-linked hypohidrotic ectodermal dysplasia. Anim. Genet. 2018, 49, 651–654. [Google Scholar] [CrossRef]
- Vasiliadis, D.; Hewicker-Trautwein, M.; Klotz, D.; Fehr, M.; Ruseva, S.; Arndt, J.; Metzger, J.; Distl, O. A de novo EDA-variant in a litter of shorthaired standard Dachshunds with X-linked hypohidrotic ectodermal dysplasia. G3 Genes Genomes Genet. 2019, 9, 95–104. [Google Scholar] [CrossRef]
- Drögemüller, C.; Distl, O.; Leeb, T. Partial deletion of the bovine ED1 gene causes anhidrotic ectodermal dysplasia in cattle. Genome Res. 2001, 11, 1699–1705. [Google Scholar] [CrossRef][Green Version]
- Drögemüller, C.; Peters, M.; Pohlenz, J.; Distl, O.; Leeb, T. A single point mutation within the ED1 gene disrupts correct splicing at two different splice sites and leads to anhidrotic ectodermal dysplasia in cattle. J. Mol. Med. 2002, 80, 319–323. [Google Scholar] [CrossRef]
- Barlund, C.S.; Clark, E.G.; Leeb, T.; Drögemüller, C.; Palmer, C.W. Congenital hypotrichosis and partial anodontia in a crossbred beef calf. Can. Vet. J. 2007, 48, 612–614. [Google Scholar]
- Ogino, A.; Kohama, N.; Ishikawa, S.; Tomita, K.; Nonaka, S.; Shimizu, K.; Tanabe, Y.; Okawa, H.; Morita, M. A novel mutation of the bovine EDA gene associated with anhidrotic ectodermal dysplasia in Holstein cattle. Hereditas 2011, 148, 46–49. [Google Scholar] [CrossRef]
- Karlskov-Mortensen, P.; Cirera, S.; Nielsen, O.L.; Arnbjerg, J.; Reibel, J.; Fredholm, M.; Agerholm, J.S. Exonization of a LINE1 fragment implicated in X-linked hypohidrotic ectodermal dysplasia in cattle. Anim. Genet. 2011, 42, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Gargani, M.; Valentini, A.; Pariset, L. A novel point mutation within the EDA gene causes an exon dropping in mature RNA in Holstein Friesian cattle breed affected by X-linked anhidrotic ectodermal dysplasia. BMC Vet. Res. 2011, 7, 35. [Google Scholar] [CrossRef] [PubMed]
- Ogino, A.; Shimizu, K.; Tanabe, Y.; Morita, M. De novo mutation of the bovine EDA gene associated with anhidrotic ectodermal dysplasia in Japanese Black cattle. Anim. Genet. 2012, 43, 646. [Google Scholar] [CrossRef] [PubMed]
- Escouflaire, C.; Rebours, E.; Charles, M.; Orellana, S.; Cano, M.; Rivière, J.; Grohs, C.; Hayes, H.; Capitan, A. A de novo 3.8-Mb inversion affecting the EDA and XIST genes in a heterozygous female calf with generalized hypohidrotic ectodermal dysplasia. BMC Genom. 2019, 20, 715. [Google Scholar] [CrossRef]
- O’Toole, D.; Häfliger, I.M.; Leuthard, F.; Schumaker, B.; Steadman, L.; Murphy, B.; Drögemüller, C.; Leeb, T. X-linked hypohidrotic ectodermal dysplasia in crossbred beef cattle due to a large deletion in EDA. Animals 2021, 11, 657. [Google Scholar] [CrossRef]
- Capuzzello, G.; Jacinto, J.G.P.; Häfliger, I.M.; Chapman, G.E.; Martin, S.S.; Viora, L.; Jonsson, N.N.; Drögemüller, C. A large deletion encompassing exon 2 of the ectodysplasin A (EDA) gene in a British blue crossbred calf with hypohidrotic ectodermal dysplasia. Acta Vet. Scand. 2022, 64, 23. [Google Scholar] [CrossRef]
- Reinartz, S.; Weiß, C.; Heppelmann, M.; Hewicker-Trautwein, M.; Hellige, M.; Willen, L.; Feige, K.; Schneider, P.; Distl, O. A missense mutation in the collagen triple helix of EDA is associated with X-linked recessive hypohidrotic ectodermal dysplasia in Fleckvieh cattle. Genes 2023, 15, 8. [Google Scholar] [CrossRef]
- Krull, F.; Bleyer, M.; Schäfer, J.; Brenig, B. A missense mutation in the highly conserved TNF-like domain of ectodysplasin A is the candidate causative variant for X-linked hypohidrotic ectodermal dysplasia in Limousin cattle: Clinical, histological, and molecular analyses. PLoS ONE 2024, 19, e0291411. [Google Scholar] [CrossRef]
- Casal, M.L.; Lewis, J.R.; Mauldin, E.A.; Tardivel, A.; Ingold, K.; Favre, M.; Paradies, F.; Demotz, S.; Gaide, O.; Schneider, P. Significant correction of disease after postnatal administration of recombinant ectodysplasin A in canine X-linked ectodermal dysplasia. Am. J. Hum. Genet. 2007, 81, 1050–1056. [Google Scholar] [CrossRef]
- Schneider, H.; Faschingbauer, F.; Schuepbach-Mallepell, S.; Körber, I.; Wohlfart, S.; Dick, A.; Wahlbuhl, M.; Kowalczyk-Quintas, C.; Vigolo, M.; Kirby, N.; et al. Prenatal correction of X-linked hypohidrotic ectodermal dysplasia. N. Engl. J. Med. 2018, 378, 1604–1610. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, V.; Drögemüller, C.; Leeb, T.; Aguirre, G.; André, C.; Bannasch, D.; Becker, D.; Davis, B.; Ekenstedt, K.; Faller, K.; et al. A Comprehensive Biomedical Variant Catalogue Based on Whole Genome Sequences of 582 Dogs and Eight Wolves. Anim. Genet. 2019, 50, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Bendl, J.; Stourac, J.; Salanda, O.; Pavelka, A.; Wieben, E.D.; Zendulka, J.; Brezovsky, J.; Damborsky, J. PredictSNP: Robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput. Biol. 2014, 10, e1003440. [Google Scholar] [CrossRef] [PubMed]
- Pejaver, V.; Urresti, J.; Lugo-Martinez, J.; Pagel, K.A.; Lin, G.N.; Nam, H.; Mort, M.; Cooper, D.N.; Sebat, J.; Iakoucheva, L.M.; et al. Inferring the molecular and phenotypic impact of amino acid variants with MutPred2. Nat. Commun. 2020, 11, 5918. [Google Scholar] [CrossRef] [PubMed]
- Mol* 3D Viewer. Available online: https://www.rcsb.org/3d-view (accessed on 18 April 2024).
- Hymowitz, S.G.; Compaan, D.M.; Yan, M.; Wallweber, H.J.; Dixit, V.M.; Starovasnik, M.A.; de Vos, A.M. The crystal structures of EDA-A1 and EDA-A2: Splice variants with distinct receptor specificity. Structure 2003, 11, 1513–1520. [Google Scholar] [CrossRef] [PubMed]
- Moura, E.; Cirio, S.M. Clinical and genetic aspects of X-linked ectodermal dysplasia in the dog—A review including three new spontaneous cases. Vet. Dermatol. 2004, 15, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.R.; Reiter, A.M.; Mauldin, E.A.; Casal, M.L. Dental abnormalities associated with X-linked hypohidrotic ectodermal dysplasia in dogs. Orthod. Craniofac. Res. 2010, 13, 40–47. [Google Scholar] [CrossRef] [PubMed]
- 99 Lives. Available online: https://cvm.missouri.edu/research/feline-genetics-and-comparative-medicine-laboratory/99-lives/ (accessed on 18 April 2024).
- McElroy, A.; Gray-Edwards, H.; Coghill, L.M.; Lyons, L.A. Precision medicine using whole genome sequencing in a cat identifies a novel COL5A1 variant for classical Ehlers-Danlos syndrome. J. Vet. Intern. Med. 2023, 37, 1716–1724. [Google Scholar] [CrossRef] [PubMed]
- Bayés, M.; Hartung, A.J.; Ezer, S.; Pispa, J.; Thesleff, I.; Srivastava, A.K.; Kere, J. The anhidrotic ectodermal dysplasia gene (EDA) undergoes alternative splicing and encodes ectodysplasin-A with deletion mutations in collagenous repeats. Hum. Mol. Genet. 1998, 7, 1661–1669. [Google Scholar] [CrossRef]
- Lexner, M.O.; Bardow, A.; Juncker, I.; Jensen, L.G.; Almer, L.; Kreiborg, S.; Hertz, J.M. X-linked hypohidrotic ectodermal dysplasia. Genetic and dental findings in 67 Danish patients from 19 families. Clin. Genet. 2008, 74, 252–259. [Google Scholar] [CrossRef]
- Daniel, E.; McCurdy, E.A.; Shashi, V.; McGuirt, W.F., Jr. Ectodermal dysplasia: Otolaryngologic manifestations and management. Laryngoscope 2002, 112, 962–967. [Google Scholar] [CrossRef]
- Seeliger, F.; Drögemüller, C.; Tegtmeier, P.; Baumgärtner, W.; Distl, O.; Leeb, T. Ectodysplasin-1 deficiency in a German Holstein bull associated with loss of respiratory mucous glands and chronic rhinotracheitis. J. Comp. Pathol. 2005, 132, 346–349. [Google Scholar] [CrossRef]
- Cooper, D.N.; Krawczak, M. The mutational spectrum of single base-pair substitutions causing human genetic disease: Patterns and predictions. Hum. Genet. 1990, 85, 55–74. [Google Scholar] [CrossRef]


| Filtering Step | Heterozygous Variants | Homozygous Variants 1 |
|---|---|---|
| All variants | 8,698,639 | 4,662,163 |
| Private variants | 101,930 | 3933 |
| Private protein-changing variants | 536 | 20 |
| Private protein-changing variants in three candidate genes | 0 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rietmann, S.J.; Cochet-Faivre, N.; Dropsy, H.; Jagannathan, V.; Chevallier, L.; Leeb, T. EDA Missense Variant in a Cat with X-Linked Hypohidrotic Ectodermal Dysplasia. Genes 2024, 15, 854. https://doi.org/10.3390/genes15070854
Rietmann SJ, Cochet-Faivre N, Dropsy H, Jagannathan V, Chevallier L, Leeb T. EDA Missense Variant in a Cat with X-Linked Hypohidrotic Ectodermal Dysplasia. Genes. 2024; 15(7):854. https://doi.org/10.3390/genes15070854
Chicago/Turabian StyleRietmann, Stefan J., Noëlle Cochet-Faivre, Helene Dropsy, Vidhya Jagannathan, Lucie Chevallier, and Tosso Leeb. 2024. "EDA Missense Variant in a Cat with X-Linked Hypohidrotic Ectodermal Dysplasia" Genes 15, no. 7: 854. https://doi.org/10.3390/genes15070854
APA StyleRietmann, S. J., Cochet-Faivre, N., Dropsy, H., Jagannathan, V., Chevallier, L., & Leeb, T. (2024). EDA Missense Variant in a Cat with X-Linked Hypohidrotic Ectodermal Dysplasia. Genes, 15(7), 854. https://doi.org/10.3390/genes15070854








