The Whole Mitochondrial Genome Sequence of Dendrobium loddigesii Rolfe, an Endangered Orchid Species in China, Reveals a Complex Multi-Chromosome Structure
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and DNA Sequencing
2.2. Assembly and Annotation
2.3. Repeat Sequences Analysis
2.4. Synonymous Codon Usage Analysis
2.5. RNA Editing Analyses and Chloroplast to Mitochondrion DNA Transfer
2.6. Synteny and Phylogenetic Analysis
3. Results
3.1. Genomic Features of the D. loddigesii Mitogenome
3.2. Repeat Sequence Analysis
3.3. Codon Usage Analysis
3.4. RNA Editing Site Analysis
3.5. Chloroplast to Mitochondrion DNA Transfer
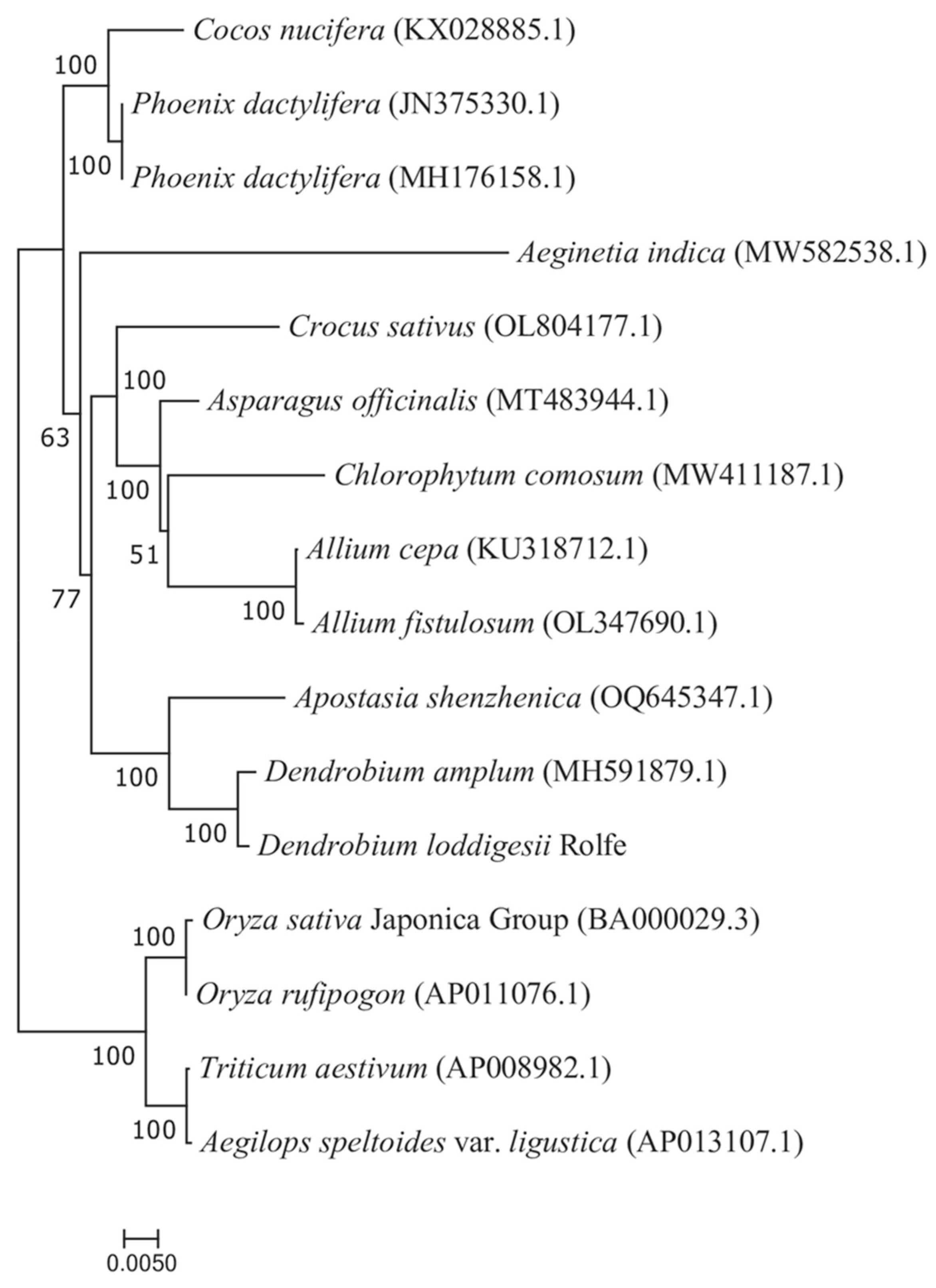
3.6. Synteny and Phylogenetic Analysis
4. Discussion
4.1. Characterization of the D. lodigesii Mitogenome
4.2. The Repeat Sequences in the D. lodigesii Mitogenome
4.3. RNA Editing in the D. lodigesii Mitogenome
4.4. MTPTs in the D. lodigesii Mitogenome
4.5. Synteny and Phylogenetic Analyses in the D. lodigesii Mitogenome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, D.; Cheng, Z.-Q.; Yang, L.; Hou, B.; Yang, J.; Li, X.-N.; Zi, C.-T.; Dong, F.-W.; Liu, Z.-H.; Zhou, J. Seco-dendrobine-type alkaloids and bioactive phenolics from Dendrobium findlayanum. J. Nat. Prod. 2018, 81, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.-M.; Zheng, C.-J.; Gan, L.-S.; Chen, G.-Y.; Zhang, X.-P.; Song, X.-P.; Li, G.-N.; Sun, C.-G. Bioactive phenanthrene and bibenzyl derivatives from the stems of Dendrobium nobile. J. Nat. Prod. 2016, 79, 1791–1797. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.-J.; Yang, L.; Bai, X.; Li, J.-Y.; Yuan, M.-Y.; Wang, Y.-Q.; Xie, Y.; Hu, J.-M.; Zhou, J. Phenolic constituents with antioxidative, tyrosinase inhibitory and anti-aging activities from Dendrobium loddigesii Rolfe. Nat. Prod. Bioprospecting 2019, 9, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Pharmacopoeia, C. Pharmacopoeia of the People’s Republic of China; People’s Medical Publishing House: Beijing, China, 2015. [Google Scholar]
- Zhang, K.-H.; Wang, M.-Q.; Wei, L.-L.; Feng, C.-J.; Zhang, Y.-S.; Teng, J.-B. Investigation of the Effects and Mechanisms of Dendrobium loddigesii Rolfe Extract on the Treatment of Gout. Evid. Based Complement. Altern. Med. 2020, 2020, 4367347. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Hirata, Y.; Xu, G.; Niwa, M.; Wu, H. Studies on the chemical constituents of Dendrobium loddigesii rolfe. Yao Xue Xue Bao Acta Pharm. Sin. 1991, 26, 307–310. [Google Scholar]
- Shen, X.; Liu, C.; Pan, K. Reproductive Biological Characteristics of Dendrobium Species; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Singh, S.; Naik, J.; Pandey, A. Genetics of Plant Organelles: Plastid and Mitochondrial Genomes. In Plant Genomics for Sustainable Agriculture; Springer: Berlin/Heidelberg, Germany, 2022; pp. 313–330. [Google Scholar]
- Ye, N.; Wang, X.; Li, J.; Bi, C.; Xu, Y.; Wu, D.; Ye, Q. Assembly and comparative analysis of complete mitochondrial genome sequence of an economic plant Salix suchowensis. PeerJ 2017, 5, e3148. [Google Scholar] [CrossRef] [PubMed]
- Chevigny, N.; Schatz-Daas, D.; Lotfi, F.; Gualberto, J.M. DNA repair and the stability of the plant mitochondrial genome. Int. J. Mol. Sci. 2020, 21, 328. [Google Scholar] [CrossRef] [PubMed]
- Nosek, J.; Tomáška, Ľ. Mitochondrial genome diversity: Evolution of the molecular architecture and replication strategy. Curr. Genet. 2003, 44, 73–84. [Google Scholar] [CrossRef]
- Backert, S.; Börner, T. Phage T4-like intermediates of DNA replication and recombination in the mitochondria of the higher plant Chenopodium album (L.). Curr. Genet. 2000, 37, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Zardoya, R. Recent advances in understanding mitochondrial genome diversity. F1000Research 2020, 9. [Google Scholar] [CrossRef]
- Unseld, M.; Marienfeld, J.R.; Brandt, P.; Brennicke, A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 1997, 15, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, Y.; Watase, Y.; Nagase, M.; Makita, N.; Yagura, S.; Hirai, A.; Sugiura, M. The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: Comparative analysis of mitochondrial genomes in higher plants. Mol. Gene. Genom. 2005, 272, 603–615. [Google Scholar] [CrossRef] [PubMed]
- Clifton, S.W.; Minx, P.; Fauron, C.M.-R.; Gibson, M.; Allen, J.O.; Sun, H.; Thompson, M.; Barbazuk, W.B.; Kanuganti, S.; Tayloe, C. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 2004, 136, 3486–3503. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.T.; Hou, Z.Y.; Li, C.; Yang, J.P.; Niu, Z.T.; Xue, Q.Y.; Liu, W.; Ding, X.Y. Rapid structural evolution of Dendrobium mitogenomes and mito-nuclear phylogeny discordances in Dendrobium (Orchidaceae). J. Syst. Evol. 2023, 61, 790–805. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Qiao, Y.; Zhang, X.; Li, Z.; Song, Y.; Sun, Z. Assembly and comparative analysis of the complete mitochondrial genome of Bupleurum chinense DC. BMC Genom. 2022, 23, 664. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Peden, J.F. Analysis of Codon Usage. Ph.D. Thesis, University of Nottingham, Nottingham, UK, 2000; pp. 73–74. [Google Scholar]
- Chateigner-Boutin, A.-L.; Small, I. Plant RNA editing. RNA Biol. 2010, 7, 213–219. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Tong, C.; Shi, J. Analysis of codon usage between different poplar species. J. Genet. Genom. 2007, 34, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Salojärvi, J.; Smolander, O.-P.; Nieminen, K.; Rajaraman, S.; Safronov, O.; Safdari, P.; Lamminmäki, A.; Immanen, J.; Lan, T.; Tanskanen, J. Genome sequencing and population genomic analyses provide insights into the adaptive landscape of silver birch. Nat. Genet. 2017, 49, 904–912. [Google Scholar] [CrossRef] [PubMed]
- Omelchenko, D.O.; Makarenko, M.S.; Kasianov, A.S.; Schelkunov, M.I.; Logacheva, M.D.; Penin, A.A. Assembly and analysis of the complete mitochondrial genome of Capsella bursa-pastoris. Plants 2020, 9, 469. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Zhang, Q.; Li, F.; Huang, J.; Zhang, M. Assembly and comparative analysis of the complete mitochondrial genome of Ilex metabaptista (Aquifoliaceae), a Chinese endemic species with a narrow distribution. BMC Plant Biol. 2023, 23, 393. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-C.; Su, Y.-Y.; Wu, C.-H.; Liu, Y.-C.; Huang, C.-H.; Chang, C.-C. Analysis of mitochondrial genomics and transcriptomics reveal abundant RNA edits and differential editing status in moth orchid, Phalaenopsis aphrodite subsp. formosana. Sci. Hortic. 2020, 267, 109304. [Google Scholar] [CrossRef]
- Wang, M.; Yu, W.; Yang, J.; Hou, Z.; Li, C.; Niu, Z.; Zhang, B.; Xue, Q.; Liu, W.; Ding, X. Mitochondrial genome comparison and phylogenetic analysis of Dendrobium (Orchidaceae) based on whole mitogenomes. BMC Plant Biol. 2023, 23, 586. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.H.; Wu, H.; Fujihara, T.; Fang, S.G. Which genetic marker for which conservation genetics issue? Electrophoresis 2004, 25, 2165–2176. [Google Scholar] [CrossRef] [PubMed]
- Guichoux, E.; Lagache, L.; Wagner, S.; Chaumeil, P.; Léger, P.; Lepais, O.; Lepoittevin, C.; Malausa, T.; Revardel, E.; Salin, F. Current trends in microsatellite genotyping. Mol. Ecol. Resour. 2011, 11, 591–611. [Google Scholar] [CrossRef]
- Li, L.; Fang, Z.; Zhou, J.; Chen, H.; Hu, Z.; Gao, L.; Chen, L.; Ren, S.; Ma, H.; Lu, L. An accurate and efficient method for large-scale SSR genotyping and applications. Nucleic Acids Res. 2017, 45, e88. [Google Scholar] [CrossRef]
- Børsting, C.; Morling, N. Next generation sequencing and its applications in forensic genetics. Forensic Sci. Int. Genet. 2015, 18, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Kadirvel, P.; Senthilvel, S.; Geethanjali, S.; Sujatha, M.; Varaprasad, K. Genetic markers, trait mapping and marker-assisted selection in plant breeding. In Plant Biology and Biotechnology, Volume II: Plant Genomics and Biotechnology; Springer: New Delhi, India, 2015; pp. 65–88. [Google Scholar] [CrossRef]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Luo, C. Molecular and functional diversity of RNA editing in plant mitochondria. Mol. Biotechnol. 2018, 60, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Edera, A.A.; Gandini, C.L.; Sanchez-Puerta, M.V. Towards a comprehensive picture of C-to-U RNA editing sites in angiosperm mitochondria. Plant Mol. Biol. 2018, 97, 215–231. [Google Scholar] [CrossRef] [PubMed]
- Gott, J.M.; Emeson, R.B. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 2000, 34, 499–531. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Zhou, P.; Tong, C.; Bi, C.; Xu, L.a. Assembly and analysis of the Populus deltoides mitochondrial genome: The first report of a multicircular mitochondrial conformation for the genus Populus. J. For. Res. 2023, 34, 717–733. [Google Scholar] [CrossRef]
- Bi, C.; Lu, N.; Xu, Y.; He, C.; Lu, Z. Characterization and analysis of the mitochondrial genome of common bean (Phaseolus vulgaris) by comparative genomic approaches. Int. J. Mol. Sci. 2020, 21, 3778. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, S.; Wang, W.; Chen, X.; Zhao, Y.; Mo, Z.; Zhu, C. Chloroplast genome sequencing of Carya Illinoinensis cv. Xinxuan-4, a new pecan pollinated cultivar. Fruit Res. 2024, 4, e012. [Google Scholar] [CrossRef]
- Tsunewaki, K. Interorganellar DNA transfer in wheat: Dynamics and phylogenetic origin. Proc. JPN. Acad. Ser. B 2011, 87, 529–549. [Google Scholar] [CrossRef][Green Version]
- Wei, L.; Liu, T.-J.; Hao, G.; Ge, X.-J.; Yan, H.-F. Comparative analyses of three complete Primula mitogenomes with insights into mitogenome size variation in Ericales. BMC Genom. 2022, 23, 770. [Google Scholar] [CrossRef]
- Shan, Y.; Li, J.; Zhang, X.; Yu, J. The complete mitochondrial genome of Amorphophallus albus and development of molecular markers for five Amorphophallus species based on mitochondrial DNA. Front. Plant Sci. 2023, 14, 1180417. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, S.E.; Smith, J.F.; Davidson, C.; Buerki, S. Phylogenetics and comparative plastome genomics of two of the largest genera of angiosperms, Piper and Peperomia (Piperaceae). Mol. Phylogenetics Evol. 2021, 163, 107229. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-C.; Chen, H.; Yang, D.; Liu, C. Diversity of mitochondrial plastid DNAs (MTPTs) in seed plants. Mitochondrial DNA Part A 2018, 29, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Alverson, A.J.; Wei, X.; Rice, D.W.; Stern, D.B.; Barry, K.; Palmer, J.D. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol. 2010, 27, 1436–1448. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Feng, Y.-L.; Wang, J.; Zhu, S.-S.; Jin, X.-J.; Wu, Z.-Q.; Zhang, Y.-H. Comprehensive Analysis of the Complete Mitochondrial Genome of Rehmannia chingii: An Autotrophic Species in the Orobanchaceae Family. Genes 2024, 15, 98. [Google Scholar] [CrossRef] [PubMed]
- Duminil, J.; Besnard, G. Utility of the mitochondrial genome in plant taxonomic studies. Mol. Plant Taxon. Methods Protoc. 2021, 2222, 107–118. [Google Scholar]
- Wang, J.; Kan, S.; Liao, X.; Zhou, J.; Tembrock, L.R.; Daniell, H.; Jin, S.; Wu, Z. Plant organellar genomes: Much done, much more to do. Trends Plant Sci. 2024. [Google Scholar] [CrossRef]
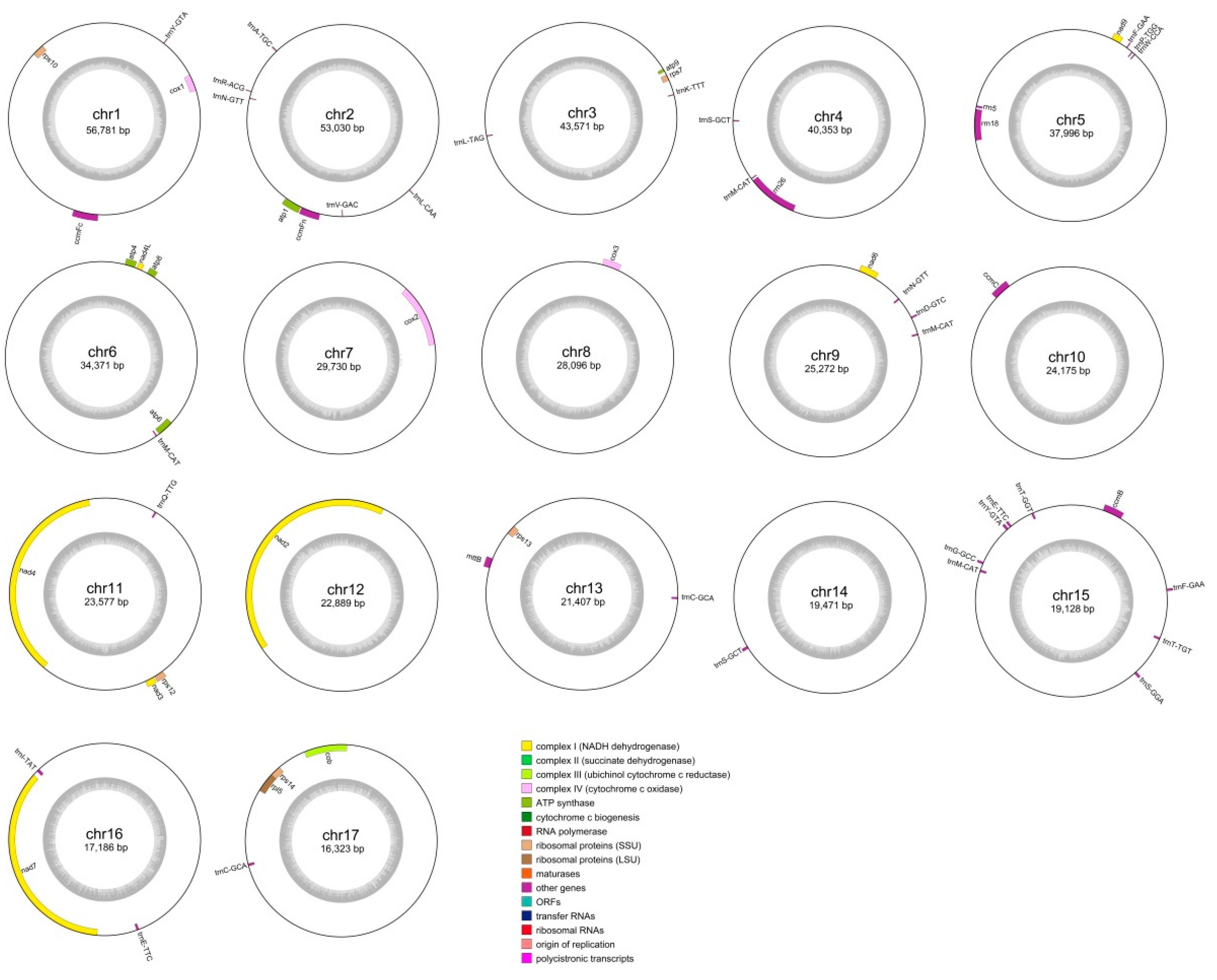


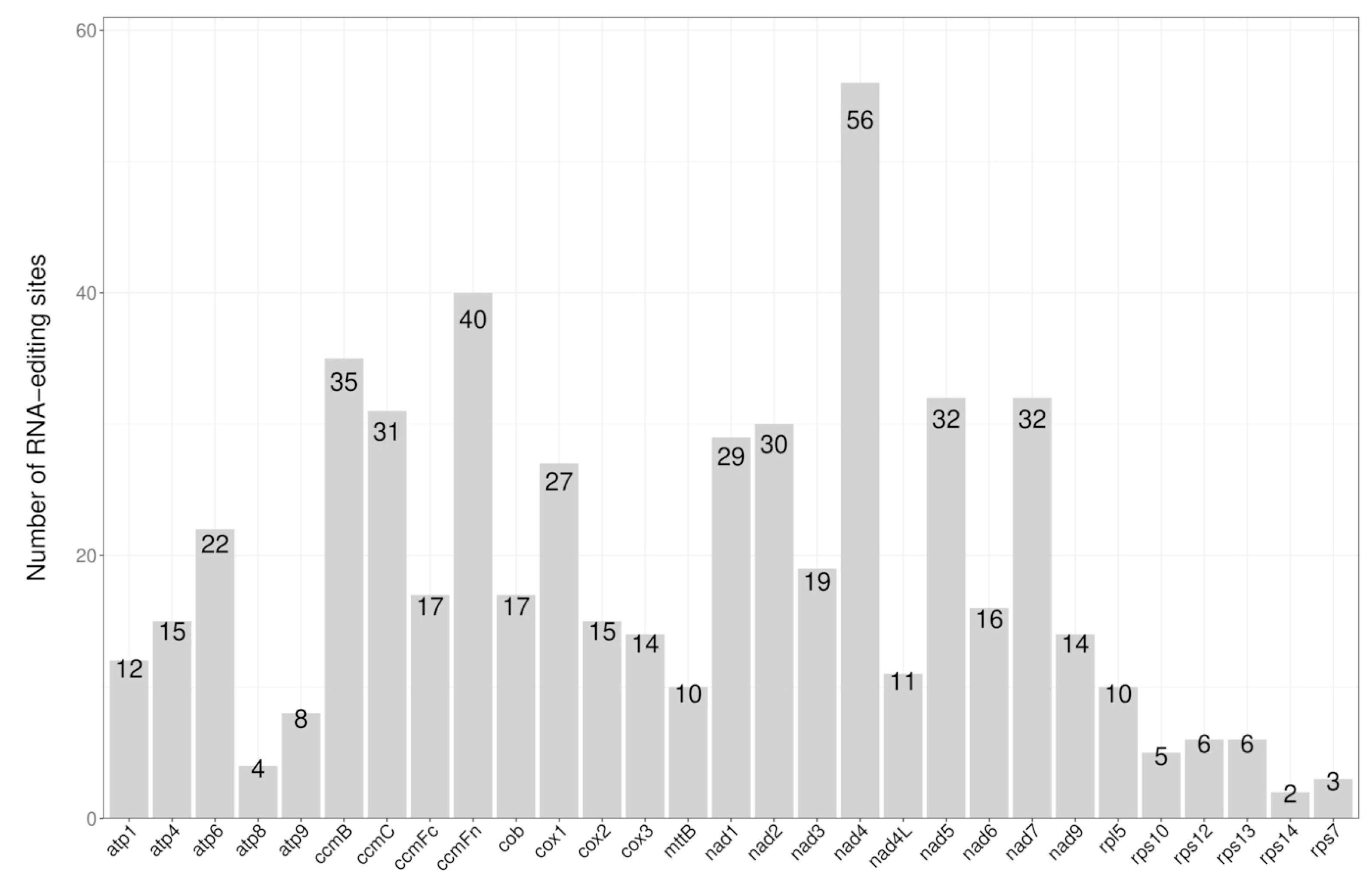

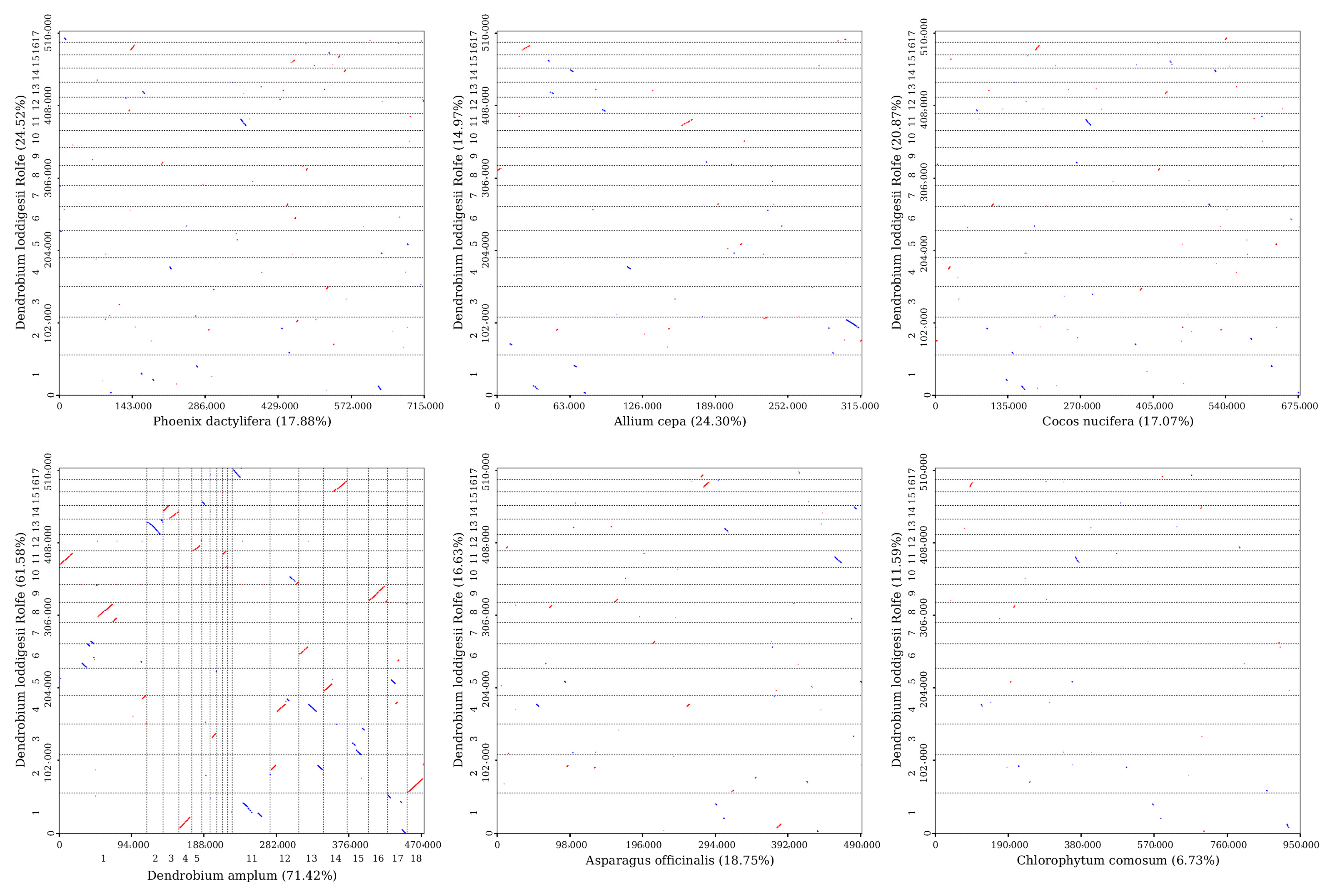
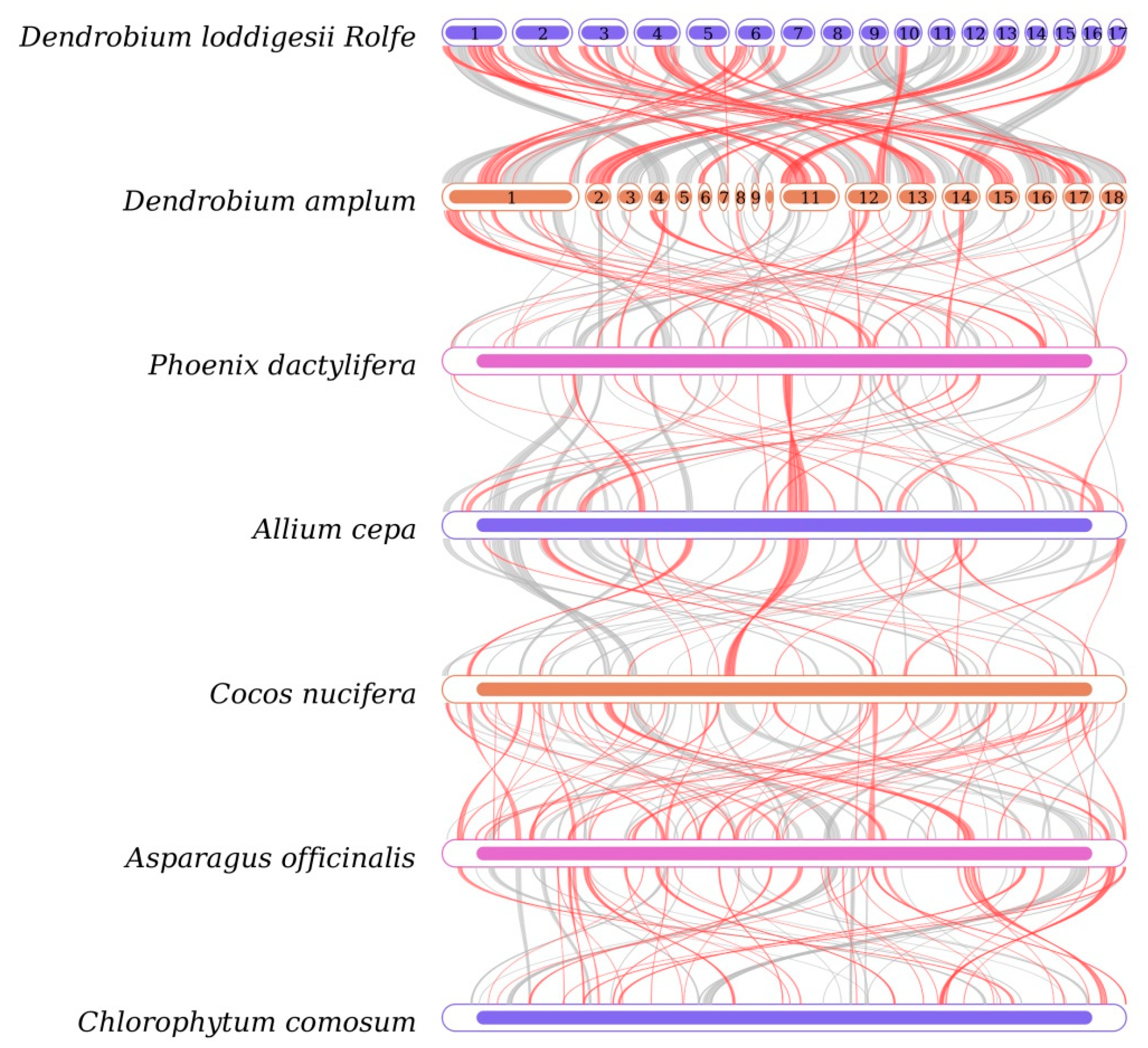
| ID | Accession Number | Length (bp) | GC (%) |
|---|---|---|---|
| Circular 1 | PP829175 | 56,781 | 44.42 |
| Circular 2 | PP829176 | 53,030 | 43.36 |
| Circular 3 | PP829177 | 43,571 | 41.19 |
| Circular 4 | PP829178 | 40,353 | 44.24 |
| Circular 5 | PP829179 | 37,996 | 43.02 |
| Circular 6 | PP829180 | 34,371 | 42.63 |
| Circular 7 | PP829181 | 29,730 | 43.32 |
| Circular 8 | PP829182 | 28,096 | 43.44 |
| Circular 9 | PP829183 | 25,272 | 44.40 |
| Circular 10 | PP829184 | 24,175 | 43.87 |
| Circular 11 | PP829185 | 23,577 | 45.93 |
| Circular 12 | PP829186 | 22,889 | 40.28 |
| Circular 13 | PP829187 | 21,407 | 45.55 |
| Circular 14 | PP829188 | 19,471 | 45.01 |
| Circular 15 | PP829189 | 19,128 | 38.27 |
| Circular 16 | PP829190 | 17,186 | 44.82 |
| Circular 17 | PP829191 | 16,323 | 45.08 |
| Total | - | 513,356 | 43.41 |
| Group of Genes | Name of Genes |
|---|---|
| ATP synthase | atp1, atp4, atp6, atp8, atp9 |
| Cytochrome c biogenesis | ccmB, ccmC, ccmFc *, ccmFn |
| Ubiquinol cytochrome c reductase | cob |
| Cytochrome c oxidase | cox1, cox2 **, cox3 |
| Maturases | matR |
| Transport membrane protein | mttB |
| NADH dehydrogenase | nad1 ****, nad2 ****, nad3, nad4 ***, nad4L, nad5 ****, nad6, nad7 ****, nad9 |
| Large subunit of ribosome | rpl5 |
| Small subunit of ribosome | rps10 *, rps12, rps13, rps14, rps7 |
| Succinate dehydrogenase | sdh4 |
| Ribosomal RNAs | rrn18, rrn26, rrn5 |
| Transfer RNAs | trnA-TGC *, trnC-GCA(2), trnD-GTC, trnE-TTC(2), trnF-GAA(2), trnG-GCC, trnI-TAT *, trnK-TTT, trnL-CAA, trnL-TAG, trnM-CAT(4), trnN-GTT(2), trnP-TGG, trnQ-TTG, trnR-ACG, trnS-GCT(2), trnS-GGA, trnT-GGT, trnT-TGT, trnV-GAC, trnW-CCA, trnY-GTA(2) |
| Type | RNA Editing | Number | Percentage |
|---|---|---|---|
| Hydrophilic–hydrophilic | CAC (H) => TAC (Y) | 8 | |
| CAT (H) => TAT (Y) | 18 | ||
| CGC (R) => TGC (C) | 12 | ||
| CGT (R) => TGT (C) | 32 | ||
| Total | 70 | 13.01% | |
| Hydrophilic–hydrophobic | ACA (T) => ATA (I) | 5 | |
| ACG (T) => ATG (M) | 8 | ||
| ACT (T) => ATT (I) | 4 | ||
| CGG (R) => TGG (W) | 34 | ||
| TCA (S) => TTA (L) | 77 | ||
| TCC (S) => TTC (F) | 35 | ||
| TCG (S) => TTG (L) | 44 | ||
| TCT (S) => TTT (F) | 55 | ||
| Total | 262 | 48.70% | |
| Hydrophilic–stop | CGA (R) => TGA (X) | 2 | |
| Total | 2 | 0.37% | |
| Hydrophobic–hydrophilic | CCA (P) => TCA (S) | 8 | |
| CCC (P) => TCC (S) | 14 | ||
| CCG (P) => TCG (S) | 6 | ||
| CCT (P) => TCT (S) | 20 | ||
| Total | 48 | 8.92% | |
| Hydrophobic–hydrophobic | CCA (P) => CTA (L) | 46 | |
| CCC (P) => CTC (L) | 8 | ||
| CCC (P) => TTC (F) | 6 | ||
| CCG (P) => CTG (L) | 27 | ||
| CCT (P) => CTT (L) | 28 | ||
| CCT (P) => TTT (F) | 14 | ||
| CTC (L) => TTC (F) | 6 | ||
| CTT (L) => TTT (F) | 12 | ||
| GCA (A) => GTA (V) | 1 | ||
| GCC (A) => GTC (V) | 1 | ||
| GCG (A) => GTG (V) | 4 | ||
| GCT (A) => GTT (V) | 3 | ||
| Total | 156 | 29.00% | |
| All | 538 | 100% |
| Fragments | Alignment Length (bp) | Identity (%) | CP Start | CP End | Mt Start | Mt End | Genes |
|---|---|---|---|---|---|---|---|
| 1 | 8595 | 99.162 | 89,287 | 97,869 | 51,672 | 43,104 | trnL-CAA |
| 2 | 8595 | 99.162 | 138,714 | 147,296 | 43,104 | 51,672 | trnL-CAA |
| 3 | 6080 | 99.266 | 125,537 | 131,590 | 25,997 | 19,973 | trnR-ACG, trnN-GUU |
| 4 | 5009 | 99.406 | 104,993 | 109,983 | 19,973 | 24,944 | trnR-ACG, trnN-GUU |
| 5 | 3855 | 97.588 | 98,032 | 101,870 | 43,117 | 39,293 | trnV-GAC |
| 6 | 3855 | 97.588 | 134,713 | 138,551 | 39,293 | 43,117 | trnV-GAC |
| 7 | 892 | 97.534 | 131,912 | 132,795 | 19,983 | 19,101 | trnA-UGC |
| 8 | 892 | 97.534 | 103,788 | 104,671 | 19,101 | 19,983 | trnA-UGC |
| 9 | 564 | 91.844 | 111,119 | 111,682 | 23,525 | 22,986 | trnL-UAG |
| 10 | 4124 | 91.844 | 42,520 | 46,619 | 14,032 | 18,149 | trnS-GGA, trnT-UGU |
| 11 | 1332 | 98.836 | 36,289 | 37,620 | 8109 | 9435 | trnG-GCC, trnM-CAU |
| 12 | 526 | 95.627 | 31,811 | 32,336 | 6418 | 5908 | trnT-GGU |
| 13 | 438 | 97.26 | 31,381 | 31,810 | 7143 | 6706 | trnE-UUC, trnY-GUA |
| 14 | 393 | 79.898 | 47,966 | 48,331 | 1 | 368 | trnF-GAA (partical: 72.60%) |
| 15 | 728 | 85.714 | 149,723 | 150,413 | 29,180 | 28,493 | trnM-CAU |
| 16 | 728 | 85.714 | 86,170 | 86,860 | 28,493 | 29,180 | trnM-CAU |
| 17 | 466 | 84.335 | 65,206 | 65,666 | 4931 | 5365 | trnW-CCA, trnP-UGG |
| 18 | 887 | 74.183 | 101,742 | 102,605 | 19,636 | 18,778 | rrn18 (partical: 43.25%) |
| 19 | 887 | 74.183 | 133,978 | 134,841 | 18,778 | 19,636 | rrn18 (partical: 43.25%) |
| 20 | 419 | 79.475 | 48,009 | 48,396 | 5401 | 5794 | trnF-GAA |
| 21 | 329 | 90.274 | 6532 | 6847 | 3650 | 3977 | trnQ-UUG |
| 22 | 649 | 82.897 | 7774 | 8386 | 11,710 | 11,096 | trnS-GCU |
| 23 | 29 | 88.235 | 45,071 | 45,099 | 11,438 | 11,466 | trnS-GCU (partical: 32.95%) |
| 24 | 83 | 96.386 | 126,909 | 126,991 | 2873 | 2791 | trnN-GUU |
| 25 | 83 | 96.386 | 109,592 | 109,674 | 2791 | 2873 | trnN-GUU |
| 26 | 97 | 85.567 | 105,841 | 105,937 | 27,256 | 27,160 | rrn26 (partical: 2.83%) |
| 27 | 97 | 85.567 | 130,646 | 130,742 | 27,160 | 27,256 | rrn26 (partical: 2.83%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tong, W.; Yu, D.; Zhu, X.; Le, Z.; Chen, H.; Hu, F.; Wu, S. The Whole Mitochondrial Genome Sequence of Dendrobium loddigesii Rolfe, an Endangered Orchid Species in China, Reveals a Complex Multi-Chromosome Structure. Genes 2024, 15, 834. https://doi.org/10.3390/genes15070834
Tong W, Yu D, Zhu X, Le Z, Chen H, Hu F, Wu S. The Whole Mitochondrial Genome Sequence of Dendrobium loddigesii Rolfe, an Endangered Orchid Species in China, Reveals a Complex Multi-Chromosome Structure. Genes. 2024; 15(7):834. https://doi.org/10.3390/genes15070834
Chicago/Turabian StyleTong, Wenjun, Dandan Yu, Xiaojing Zhu, Zhifang Le, Hui Chen, Feilong Hu, and Shengmin Wu. 2024. "The Whole Mitochondrial Genome Sequence of Dendrobium loddigesii Rolfe, an Endangered Orchid Species in China, Reveals a Complex Multi-Chromosome Structure" Genes 15, no. 7: 834. https://doi.org/10.3390/genes15070834
APA StyleTong, W., Yu, D., Zhu, X., Le, Z., Chen, H., Hu, F., & Wu, S. (2024). The Whole Mitochondrial Genome Sequence of Dendrobium loddigesii Rolfe, an Endangered Orchid Species in China, Reveals a Complex Multi-Chromosome Structure. Genes, 15(7), 834. https://doi.org/10.3390/genes15070834





